Abstract
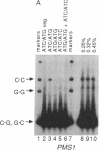
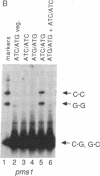
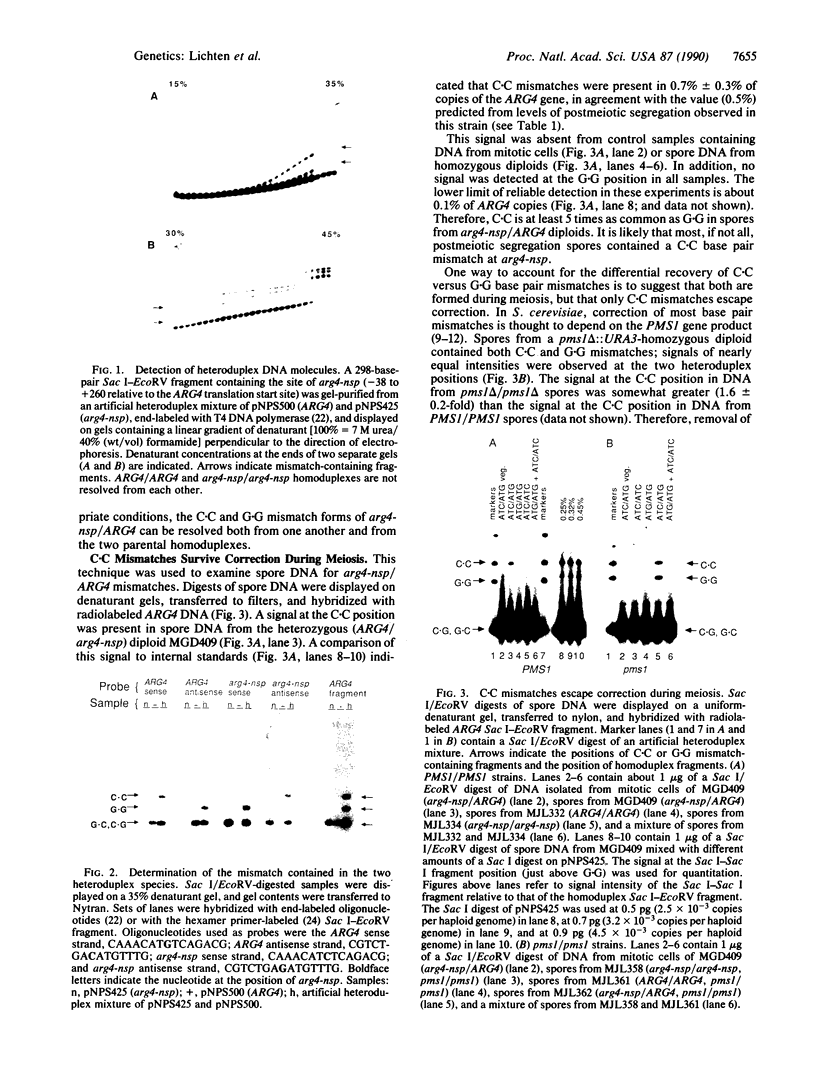
We have used denaturant-gel electrophoresis to provide a physical demonstration of heteroduplex DNA in the products of yeast meiosis. We examined heteroduplex formation at arg4-nsp, a G.C----C.G transversion that displays a moderately high level of postmeiotic segregation. Of the two possible arg4-nsp/ARG4 mismatches (G.G and C.C), only C.C was detected in spores from mismatch repair-competent (Pms1+) diploids. In contrast, C.C and G.G were present at nearly equal levels in spores from Pms1- diploids. These results confirm previous suggestions that postmeiotic segregation spores contain heteroduplex DNA at the site of the marker in question, that C.C is repaired less frequently than is G.G, and that the PMS1 gene product plays a role in mismatch correction. Combined with the observation that Pms1+ ARG4/arg4-nsp diploids produce 3 times more 3+:5m (wildtype:mutant) tetrads (+, +, +/m, m) than 5+:3m tetrads (+, +/m, m, m), these results indicate that, during meiosis, formation of heteroduplex DNA at ARG4 involves preferential transfer of the sense (nontranscribed) strand of the DNA duplex.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beacham I. R., Schweitzer B. W., Warrick H. M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984 Sep;29(3):271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Andersen J., Kolodner R. D. Specificity of mismatch repair following transformation of Saccharomyces cerevisiae with heteroduplex plasmid DNA. Proc Natl Acad Sci U S A. 1989 May;86(10):3713–3717. doi: 10.1073/pnas.86.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987 Jul 23;328(6128):362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays. 1987 May;6(5):232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M. S. Some features of genetic recombination in procaryotes. Annu Rev Genet. 1978;12:47–68. doi: 10.1146/annurev.ge.12.120178.000403. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Williamson M. S., Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989 Oct;9(10):4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson A., Silver D., Seed B. Minimal size plasmids containing an M13 origin for production of single-strand transducing particles. J Mol Appl Genet. 1984;2(6):507–517. [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Genetic evidence for preferential strand transfer during meiotic recombination in yeast. Genetics. 1990 Aug;125(4):753–761. doi: 10.1093/genetics/125.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A., Petes T. D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989 Jul 27;340(6231):318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Treco D., Schultes N. P., Szostak J. W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989 Mar 2;338(6210):35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985 May 23;315(6017):350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985 Aug;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]