Abstract
Albumins are the most well-known globular proteins, and the most typical representatives are the serum albumins. However, less attention was paid to the albumin family, except for the human and bovine serum albumin. To characterize the features of albumin family, we have mined all the putative albumin proteins from the available genome sequences. The results showed that albumin is widely distributed in vertebrates, but not present in the bacteria and archaea. The phylogenetic analysis of vertebrate albumin family implied an evolutionary relationship between members of serum albumin, α-fetoprotein, vitamin D–binding protein, and afamin. Meanwhile, a new member from the albumin family was found, namely, extracellular matrix protein 1. The structural analysis revealed that the motifs for forming the internal disulfide bonds are highly conserved in the albumin family, despite the low overall sequence identity across the family. The domain arrangement of albumin proteins indicated that most of vertebrate albumins contain 3 characteristic domains, arising from 2 evolutionary patterns. And a significant trend has been observed that the albumin proteins in higher vertebrate species tend to possess more characteristic domains. This study has provided the fundamental information required for achieving a better understanding of the albumin distribution, phylogenetic relationship, characteristic motif, structure, and new insights into the evolutionary pattern.
Keywords: Albumin, vertebrate, characteristic motif, evolution
Introduction
Albumins are a family of globular proteins that are water soluble and moderately soluble in the concentrated salt solutions; what is more, they experience heat denaturation.1 Albumins are commonly found in animal sources, such as blood plasma,2 milk,3 and egg white,4 as well as plant sources, such as soybeans5 and grains.6
Albumins have many applications.1 For instance, albumins can act as a vehicle for the transport of metals, fatty acids, drugs, etc.7 They could also be used diagnostically for various indications of breast cancer, some solid tumors, and rheumatoid arthritis.8,9 Commercially, albumins are extracted from egg white (ovalbumin), bovine serum, and human serum albumin (HSA).10 Other alternative methods have also been developed, for instance, obtaining recombinant HSA by genetic engineering.11,12
From the albumin family in animals, several members have been identified, including serum albumin, α-fetoprotein, vitamin D–binding protein and afamin.13–15 Serum albumin is the most abundant blood plasma protein; to be more exact, HSA normally constitutes about 50% of human plasma protein.16 In human body, serum albumin aids in the regulation of osmotic pressure and pH in blood. Furthermore, HSA can mediate lipid metabolism, sequester toxins, and resist oxidative stress.17 Meanwhile, serum albumins can bind water, cations (such as Ca2+, Na+, and K+), fatty acids, hormones, pharmaceuticals, etc.18–20 α-Fetoprotein (α-fetoglobulin) is a fetal plasma protein that can bind various cations, fatty acids, and bilirubin.21 Vitamin D–binding protein is the major plasma carrier for vitamin D and its metabolites, as well as for fatty acids.22 Afamin is a unique human plasma vitamin E–binding glycoprotein, primarily expressed in liver.23,24
It is well known that serum albumins are the most common albumins and have been one of the most studied proteins, especially for HSA.25 The amino and carboxyl terminal sequences of serum albumins from human, cow, and several other species had been determined in 1950s even when sequencing was still in its infancy.26–28 The 3-dimensional structure of HSA has been determined by X-ray crystallography to a resolution of 2.5 Å.29 Human serum albumin comprises 3 homologous domains that are assembled to form a heart-shaped molecule.30 Each domain possesses certain common structural motifs, containing 5 to 6 internal disulfide bonds.
The development in the whole-genome sequences offers an opportunity to further understand the diversity of albumin family. To our knowledge, no systematical comparative study of albumin family at the genome level has been reported thus far. In this study, we have investigated the current available genomes on the National Center for Biotechnology Information (NCBI) database to identify all possible albumin family members with hidden Markov models. Furthermore, the phylogeny, characteristic motifs, and domain arrangement of these proteins have been analyzed. This study has significantly improved our understanding of the albumin family and provided foundations for related studies in the future.
Materials and Methods
Sequence data
In this study, the available genome data from the NCBI resources (http://www.ncbi.nlm.nih.gov/genome/browse/, last access August 20, 2016) were used for mining the putative albumins. In the analysis, the overall protein sequences extracted from the corresponding genomes were applied to investigate the genome-wide distribution of the albumin family. Moreover, the information of species with putative albumins is presented in Table S1 (see the supporting information).
Identification of the putative albumins
In the Pfam database, the conserved domain of albumin family has been characterized by a profile hidden Markov model (http://pfam.xfam.org/family/PF00273).31,32 As mentioned above, the overall protein sequences were used to determine whether they have the albumin domains, and then those with albumin characteristic domains were screened as the putative albumins. The identification procedure of putative albumins was the following: the albumin profile hidden Markov model (PF00273, Pfam protein families database) was used to identify the homologues by searching the selected overall protein sequences with local HMMER 3.1b2. 31,32 The hits passed MSV, Bias, Vit, and Fwd filters (see HMMER User’s Guide, http://eddylab.org/) were recognized as the putative albumins.
Construction of the phylogenetic tree
The phylogenetic tree of the annotated albumins was constructed for analyzing their evolutionary relationships. The pipeline was referred from the previous reports.33 In brief, the annotated albumin proteins were aligned to the albumin profile hidden Markov model with HMMER package. Furthermore, the generated alignment was subjected to FastTree 2.1.8 for inferring the phylogenetic tree with maximum-likelihood method.34 In the settings, FastTree used JTT model for the amino acid substitution, as well as the computed local support values with the Shimodaira-Hasegawa test to estimate the reliability of each split in the tree (for more details, see FastTree User’s Guide, http://meta.microbesonline.org/fasttree/). Finally, the generated tree data were submitted to Interactive Tree Of Life for viewing the phylogenetic trees.35,36
Visualization of the structural features
Sequence logo was generated from albumin homologous groups for revealing their primary structural features. The flow was referred from the previous reports.33 First, the homologous albumin alignments were built by being adjusted to the albumin profile hidden Markov model with the HMMER package. The mismatched residues were removed. After that, the alignments were submitted to WebLogo (http://weblogo.threeplusone.com/create.cgi) for generating their consensus logos for the primary structure by a stack of residues for each position.37,38
Results and Discussion
Putative albumins are widely distributed in the vertebrate genomes
In this study, we have searched the putative albumins throughout the available overall protein sequences, including animals, plants, fungi, bacteria, and archaea, using their profile hidden Markov model PF00273 from the Pfam database. However, our findings indicated that albumins are solely presented in vertebrates (except for an exception mentioned below). The detail distribution of albumins is presented in Table S1 (see supporting information). As mentioned previously, plants also contain albumins; however, those are not phylogenetically related to the animal albumins.39 This fact is consistent with the concept of albumin, which are water soluble, moderately soluble in concentrated salt solutions, not evolutionarily related, and experience heat denaturation.1 Therefore, just to avoid possible confusion, for the following sections of this article, albumins solely refer to those from animals if not otherwise specified.
We summarized albumins in animal species (Table S1) and found that no albumins were identified in flatworm and roundworm. Insects also showed no albumins, except that a homologue was found in Culex quinquefasciatus. Culex quinquefasciatus is a kind of house mosquito, and a blood meal on mammals and birds is necessary for their egg development.40 As the phylogenetic analysis revealed later, this albumin protein is evolutionarily close to its fish counterparts. Probably, this gene was horizontally transferred from fish. Among different fishes, about half of them contain albumin gene with the average count of 3 (Table 1). For the groups of amphibians, reptiles, birds, and mammals, all the tested species contain albumin genes. It needs to be pointed out that the mammals have significantly higher count of albumin genes than other groups, about double count of those in the fishes, reptiles, and birds, suggesting their crucial requirement in mammals because they are the main constituents in blood.16
Table 1.
Summary on albumin distribution in animal species.
| SPECIES NUMBERa | PERCENTAGEb | AVERAGE GENOMIC NUMBERc | |
|---|---|---|---|
| Flatworm | 7 | 0 | − |
| Roundworm | 27 | 0 | − |
| Insects | 73 | 1.37 | + |
| Fishes | 32 | 53.1 | 3.2 |
| Amphibians | 2 | 100 | + |
| Reptiles | 9 | 100 | 3.0 |
| Birds | 56 | 100 | 3.2 |
| Mammals | 84 | 100 | 6.1 |
“−” represents the absence of albumin genetic sequence; “+” means that the limited species are not sufficient to calculate the accurate average genetic number.
Species number: the total number of the species we studied.
Percentage: the percentage of the species possessing albumin genetic sequences in the corresponding category.
Average genomic number: the average number of the albumin sequences in one species genome.
In summary, based on the distribution of albumins, we have found that albumins are unique in vertebrates and provided their evolutionary cues that evolved from sea to land.
Extracellular matrix protein 1 is a new member of albumin family
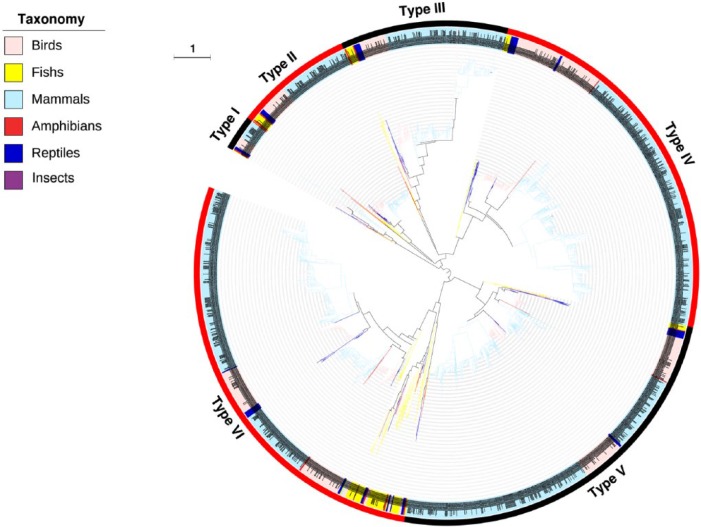
To address the evolutionary relationship of albumins, their phylogenetic tree was constructed and 6 types (I–VI) were summarized (Figure 1). Generally, albumins from the same taxonomic group tend to locate closely in the tree. However, it should be noted that the albumin family has been highly evolved in vertebrates. As recognized previously, the albumin family consists of several evolutionarily related members, including serum albumin, α-fetoprotein, vitamin D–binding protein, and afamin.13–15 In the phylogenetic tree, 6 distinct albumin types are clearly separated throughout a wide range of vertebrate taxonomic groups (Figure 1). Therefore, we annotated the albumin proteins in these 6 phylogenetic types using the NCBI database to assign them to the corresponding members (Table S2).
Figure 1.
Phylogenetic tree of albumins. The inner circle is the phylogenetic tree derived from the consensus sequences of albumins. The upper left legend shows their taxonomic groups with different colors in the branches. The middle circle is the corresponding albumins of different taxonomic groups. The dotted line links the branch with each taxon. The outer numbers indicate the 6 types based on this study; alternating black and red show their ranges in phylogenetic tree. The branch length is indicated as scale bar.
The annotation demonstrated that types I to III are vitamin D–binding proteins. Because the vitamin D–binding proteins in types I to III cover a wide range of vertebrates including mammals, birds, reptiles, fishes, and amphibians, it is reasonable to suggest that the current vitamin D–binding proteins could be divided into 3 distinct groups arising from the early divergent ancestors. Types IV to VI are 3 large groups from the albumin family covering mammals, birds, reptiles, fishes, and amphibians. However, unlike the types I to III with a clear and consistent annotation to vitamin D–binding proteins, types IV to VI consist of albumin members of α-fetoprotein, serum albumin, and afamin. On one hand, this suggests a close phylogenetic relationship between albumin members of α-fetoprotein, serum albumin, and afamin. On the other hand, it might mean that these 3 members could also be divided into 3 distinct groups.
Unexpectedly, a new member of the albumin family was found in type VI, namely, the extracellular matrix protein 1 (ECM1). The structural analysis indicated that ECM1 contains motifs with a cysteine pattern characteristic of the cysteine pattern of the ligand-binding “double-loop” domains of the albumin protein family.41 Therefore, ECM1 demonstrates a close phylogenetic relationship with the serum albumin in the type VI and could be included in the albumin family. Extracellular matrix protein 1 is involved in endochondral bone formation and angiogenesis and might promote the wound healing and repair.42 It also interacts with a variety of extracellular and structural proteins, contributing to the maintenance of skin integrity and homeostasis.43,44 However, ECM1 was associated with some pathological processes, such as lipoid proteinosis and tumor biology, and thus was considered as a potential biomarker for the diagnostic of the related diseases.45,46
Only characteristic cysteine residues are conserved across albumin family
Characteristic motifs and residues of above identified albumin types (I–VI) were delineated (Figure 2). In general, it is clearly observed that the overall sequence identities of albumin characteristic domains are very low across the family. Notably, many cysteine residues are largely conserved across the albumin family. These cysteine residues, especially for the characteristic cysteine motif CCXnC, are responsible for forming of the double-disulfide loop of the albumin family, contributing to its ligand configuration and binding.47–49
Figure 2.
Structural features of 6 albumin types (I–VI). The classification of albumins into 6 types is referred Figure 1.
As mentioned above, types I to III are vitamin D–binding proteins; however, their structural features differ. Type I shows a significant loss in its albumin characteristic domain, only presenting 1 cysteine motif, C62C63X23C88, whereas types II and III have 2 additional cysteine motifs, C140C141X11C153 and C312C313X23C331. Interestingly, except for the common characteristic cysteine motif, less sequence identities are shared within types I to III, which also suggest their early divergence. The similar situation was observed in types IV to VI as well.
Albumins in higher vertebrate groups tend to have more characteristic domains
As indicated in Figure 2, the albumin family shares a conserved characteristic domain about 176 amino acids. In the analysis, an interesting phenomenon was observed that most albumins have more than one characteristic domain. To investigate the domain architecture, the distribution and number of characteristic domains count per albumin protein in vertebrates were summarized (Table 2) and the details are presented in Table S3. A significant trend was observed that albumin proteins in more advanced vertebrate species tend to possess more characteristic domains. As demonstrated, the albumin proteins with 3 characteristic domains are the major forms, despite the fact that their percentage varied greatly from 38.9% in fishes to 90.0% in mammals. Especially, in mammals, 3 unique albumin proteins were found. One of them is from Camelus ferus with 9 characteristic domains, and the other 2 with 10 characteristic domains are from Myotis davidii and Tupaia chinensis.
Table 2.
Distribution of characteristic domain count within albumins in different taxonomic groups (%).
| DOMAIN COUNT | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | AVERAGE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| All species | 5.37 | 8.18 | 83.2 | 2.30 | 0.38 | 0.13 | – | – | 0.13 | 0.26 | 2.87 |
| Mammals | 3.14 | 3.73 | 90.0 | 1.76 | 0.59 | 0.20 | – | – | 0.20 | 0.39 | 2.97 |
| Birds | 0.56 | 11.7 | 83.3 | 4.44 | – | – | – | – | – | – | 2.92 |
| Reptiles | 7.41 | 29.6 | 59.3 | 3.70 | – | – | – | – | – | – | 2.59 |
| Amphibians | 30.0 | 20.0 | 50.0 | – | – | – | – | – | – | – | 2.20 |
| Fishes | 35.2 | 25.9 | 38.9 | – | – | – | – | – | – | – | 2.04 |
“–” represents that no albumin genetic sequence contains this number of function regions. The percentage is presented per taxonomic groups.
Hence, what is the relationship among the characteristic domains within albumins? This study focuses on the albumins with 3 characteristic domains, and the phylogenetic analysis indicated 2 evolutionary patterns (Figure 3A—pattern A and Figure 3B—pattern B). Pattern A contains 3 characteristic domains from different origins, and pattern B represents 2 domains from a duplication. Furthermore, we investigated the distribution of these 2 domain evolutionary patterns in 6 albumin types (I–VI) and different vertebrate groups (Figure 3C). In general, pattern A and pattern B are mixed in 6 albumin types. As illustrated, pattern A is a major pattern in type III, reaching a percentage of about 60%; meanwhile, pattern B is significantly more presented than pattern A in other types.
Figure 3.

Evolutionary pattern of the albumin characteristic domains. The evolutionary relationship were investigated among albumins with 3 characteristic domains and summarized into 2 types: (A) pattern A, 3 characteristic domains derived from different origins and (B) pattern B, 2 characteristic domains originated from the same sequences. (C) Percentage of the evolution pattern A in albumin types I, II, III, IV, V, and VI, whereas the remaining is for the evolution pattern B. The taxonomic groups were indicated by different colors.
Conclusions
In this study, the genome-wide analysis of albumins from 290 animal species provided the fundamental understanding of albumin distribution, phylogenetic relationship, characteristic motif, structure, and evolutionary patterns. The results indicated that albumins are widely distributed in vertebrate species, especially their blooming presence in mammals with average of 6 albumin counts per genome. However, the vertebrate albumins are phylogenetically disconnected with the plant counterparts despite their similar physicochemical properties. The phylogenetic analysis revealed 6 distinct types (I–VI) of the albumin family, including the current members (vitamin D–binding proteins, α-fetoprotein, serum albumin, and afamin). Meaningfully, based on the phylogenetic relationship, ECM1 could be assigned to be a new member of the albumin family, which improves the understanding of the diversity of the albumin family. The structure analysis revealed that cysteine residues are extraordinarily presented in the albumin domain and seem to be the only conserved residues across the albumin family. These findings could further elevate our knowledge on the relationship between the structure and function of the albumin family. The investigation of characteristic domain count per albumin demonstrated a wide distribution from 1 to 10, and the most common count is 3. Moreover, a significant trend was observed that albumins in the more advanced vertebrate groups tend to have more characteristic domains. Finally, based on the phylogenetic analysis of multiple characteristic domains within each albumin protein, 2 evolutionary patterns were summarized.
Acknowledgments
The authors would like to thank Dr Wanping Chen (Huazhong Agricultural University) for his kind advice on phylogenetic analysis.
Footnotes
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2667 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is financially supported by the National Natural Science Foundation of China (nos 31401644, 31601490), Construction project of Youth Science and technology innovation leader in Corps (2016BC001), and grant from the Project of Hubei Collaborative Innovation Center for Security Precaution and Emergency Response Technology (Wuhan University of Technology) (project number: JD2016010201).
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contribution
SL and FG conceived and designed the experiments. SL, YC and FG analyzed the data; wrote the first draft of the manuscript; agreed with manuscript results and conclusions. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Puoci F. Advanced Polymers in Medicine. Berlin, Germany: Springer; 2015. [Google Scholar]
- 2.Kalra H, Adda CG, Liem M, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 3.Indyk HE, Gill BD, Woollard DC. An optical biosensor-based immunoassay for the determination of bovine serum albumin in milk and milk products. Int Dairy J. 2015;47:72–78. [Google Scholar]
- 4.Abeyrathne EDNS, Lee HY, Ahn DU. Egg white proteins and their potential use in food processing or as nutraceutical and pharmaceutical agents—a review. Poult Sci. 2013;92:3292–3299. doi: 10.3382/ps.2013-03391. [DOI] [PubMed] [Google Scholar]
- 5.Moreno FJ, Clemente A. 2S albumin storage proteins: what makes them food allergens? Open Biochem J. 2008;2:16–28. doi: 10.2174/1874091X00802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Žilić S, Barać M, Pešić M, Dodig D, Ignjatović-Micić D. Characterization of proteins from grain of different bread and durum wheat genotypes. Int J Mol Sci. 2011;12:5878–5894. doi: 10.3390/ijms12095878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Human serum albumin: from bench to bedside. Mol Aspects Med. 2012;33:209–290. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Elsadek B, Kratz F. Impact of albumin on drug delivery—new applications on the horizon. J Control Release. 2012;157:4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 9.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Elzoghby AO, Samy WM, Elgindy NA. Albumin-based nanoparticles as potential controlled release drug delivery systems. J Control Release. 2012;157:168–182. doi: 10.1016/j.jconrel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K. Summary of recombinant human serum albumin development. Biologicals. 2006;34:55–59. doi: 10.1016/j.biologicals.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Tuan Giam Chuang V, Kragh-Hansen U, Otagiri M. Pharmaceutical strategies utilizing recombinant human serum albumin. Pharm Res. 2002;19:569–577. doi: 10.1023/a:1015396825274. [DOI] [PubMed] [Google Scholar]
- 13.Haefliger DN, Moskaitis JE, Schoenberg DR, Wahli W. Amphibian albumins as members of the albumin, alpha-fetoprotein, vitamin D-binding protein multigene family. J Mol Evol. 1989;29:344–354. doi: 10.1007/BF02103621. [DOI] [PubMed] [Google Scholar]
- 14.Lichenstein HS, Lyons DE, Wurfel MM, et al. Afamin is a new member of the albumin, alpha-fetoprotein, and vitamin D-binding protein gene family. J Biol Chem. 1994;269:18149–18154. [PubMed] [Google Scholar]
- 15.Schoentgen F, Metz-Boutique M-H, Jollès J, Constans J, Jollès P. Complete amino acid sequence of human vitamin D-binding protein (group-specific component): evidence of a three-fold internal homology as in serum albumin and alpha-fetoprotein. Biochim Biophys Acta. 1986;871:189–198. doi: 10.1016/0167-4838(86)90173-1. [DOI] [PubMed] [Google Scholar]
- 16.Farrugia A. Albumin usage in clinical medicine: tradition or therapeutic? Transfus Med Rev. 2010;24:53–63. doi: 10.1016/j.tmrv.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Peters T., Jr . All About Albumin: Biochemistry, Genetics, and Medical Applications. San Diego, CA: Academic Press; 1995. [Google Scholar]
- 18.Colmenarejo G. In silico prediction of drug-binding strengths to human serum albumin. Med Res Rev. 2003;23:275–301. doi: 10.1002/med.10039. [DOI] [PubMed] [Google Scholar]
- 19.Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 20.Hage DS, Sengupta A. Characterisation of the binding of digitoxin and acetyl-digitoxin to human serum albumin by high-performance affinity chromatography. J Chromatogr B Biomed Sci Appl. 1999;724:91–100. doi: 10.1016/s0378-4347(98)00589-1. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed RAB. Evaluation of Plasma Antioxidant Levels Albumin, Uric Acid and Total Bilirubin in Benzene Station Workers in Khartoum State. Khartoum, Sudan: Sudan University of Science and Technology; 2015. [Google Scholar]
- 22.Malik S, Fu L, Juras DJ, et al. Common variants of the vitamin D binding protein gene and adverse health outcomes. Crit Rev Clin Lab Sci. 2013;50:1–22. doi: 10.3109/10408363.2012.750262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieplinger H, Dieplinger B. Afamin—a pleiotropic glycoprotein involved in various disease states. Clin Chim Acta. 2015;446:105–110. doi: 10.1016/j.cca.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Jerkovic L, Voegele AF, Chwatal S, et al. Afamin is a novel human vitamin E-binding glycoprotein characterization and in vitro expression. J Proteome Res. 2005;4:889–899. doi: 10.1021/pr0500105. [DOI] [PubMed] [Google Scholar]
- 25.Anguizola J, Matsuda R, Barnaby OS, et al. Review: glycation of human serum albumin. Clin Chim Acta. 2013;425:64–76. doi: 10.1016/j.cca.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikenaka T. Studies of the N- and C-terminal amino acid sequence of human serum albumin. J Am Chem Soc. 1960;82:3180–3183. [Google Scholar]
- 27.Kusama K. On the species difference in carboxy-terminal amino acids of serum albumins from various animals. J Biochem. 1957;44:375–381. [Google Scholar]
- 28.White WF, Shields J, Robbins KC. C-terminal sequence of crystalline bovine and human serum albumins: relationship of C-terminus to antigenic determinants of bovine serum albumin. J Am Chem Soc. 1955;77:1267–1269. [Google Scholar]
- 29.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 30.He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 31.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res. 2013;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai L, Xie T, Hu Q, Deng C, Zheng R, Chen W. Genome-wide comparison of ferritin family from Archaea, Bacteria, Eukarya, and Viruses: its distribution, characteristic motif, and phylogenetic relationship. Naturwissenschaften. 2015;102:1–10. doi: 10.1007/s00114-015-1314-3. [DOI] [PubMed] [Google Scholar]
- 34.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 36.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karaki L, Da Silva P, Rizk F, et al. Genome-wide analysis identifies gain and loss/change of function within the small multigenic insecticidal albumin 1 family of Medicago truncatula. BMC Plant Biol. 2016;16:63. doi: 10.1186/s12870-016-0745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A, editors. “Bird biting” mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect Genet Evol. 2011;11:1577–1585. doi: 10.1016/j.meegid.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan I, Liu L, Hamada T, Sethuraman G, McGrath JA. The molecular basis of lipoid proteinosis: mutations in extracellular matrix protein 1. Exp Dermatol. 2007;16:881–890. doi: 10.1111/j.1600-0625.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 42.Gu M, Guan J, Zhao L, Ni K, Li X, Han Z. Correlation of ECM1 expression level with the pathogenesis and metastasis of laryngeal carcinoma. Int J Clin Exp Pathol. 2013;6:1132–1137. [PMC free article] [PubMed] [Google Scholar]
- 43.Sercu S, Lambeir AM, Steenackers E, et al. ECM1 interacts with fibulin-3 and the beta 3 chain of laminin 332 through its serum albumin subdomain-like 2 domain. Matrix Biol. 2009;28:160–169. doi: 10.1016/j.matbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Youssefian L, Vahidnezhad H, Daneshpazhooh M, et al. Lipoid proteinosis: phenotypic heterogeneity in Iranian families with c. 507delT mutation in ECM1. Exp Dermatol. 2015;24:220–222. doi: 10.1111/exd.12620. [DOI] [PubMed] [Google Scholar]
- 45.Meng X-Y, Liu J, Lv F, Liu M-Q, Wan JM. Study on the correlation between extracellular matrix protein-1 and the growth, metastasis and angiogenesis of laryngeal carcinoma. Asian Pac J Cancer Prev. 2014;16:2313–2316. doi: 10.7314/apjcp.2015.16.6.2313. [DOI] [PubMed] [Google Scholar]
- 46.Mondejar R, Garcia-Moreno JM, Rubio R, et al. Clinical and molecular study of the extracellular matrix protein 1 gene in a Spanish family with lipoid proteinosis. J Clin Neurol. 2014;10:64–68. doi: 10.3988/jcn.2014.10.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fasano M, Curry S, Terreno E, et al. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 48.Kragh-Hansen U. Structure and ligand binding properties of human serum albumin. Dan Med Bull. 1990;37:57–84. [PubMed] [Google Scholar]
- 49.Rosenoer VM, Oratz M, Rothschild MA. Albumin: Structure, Function and Uses. Amsterdam, The Netherlands: Elsevier; 2014. [Google Scholar]