Abstract
Leptospira interrogans sensu stricto is responsible for the most frequent and severe cases of human leptospirosis. The epidemiology and clinical features of leptospirosis are usually associated with the serovars and serogroups of Leptospira. Because of the difficulties associated with serological identification of Leptospira strains, we evaluated a novel PCR-based method for typing L. interrogans serovars. Based upon the genome sequence of L. interrogans serovar Lai type strain 5660, 44 loci were analyzed by PCR for their variability in size due to the presence of variable-number tandem repeats (VNTR). Seven VNTR loci were found to be powerful markers for serovar identification, epidemiology, and phylogenetic studies of L. interrogans. This rapid and easy method should greatly contribute to a better knowledge of the epidemiology of Leptospira.
The genus Leptospira consists of a heterogeneous group of pathogenic and saprophytic species belonging to the order Spirochaetales. Pathogenic Leptospira species, currently classified in seven species based on DNA relatedness (2, 25), are the agents of leptospirosis. Transmission to humans occurs through direct or indirect contacts with urine of infected animals. Leptospira interrogans sensu stricto (25) is the main species associated with human leptospirosis. In France, L. interrogans sensu stricto is responsible for about 60% of human cases and for the most severe ones. The intraspecies taxonomy of leptospires is well established and based on antigenic determinants. Since the description of serovars in 1915, about 80 serovars have been identified in L. interrogans sensu stricto (2); among them, 60 serovars are validly described (12). Since each serovar is usually associated with a particular host, identification of serovars is essential to epidemiological studies and strategies for prevention (5). The reference method for serological identification is the microagglutination test, which is a complex and fastidious test since it requires cross adsorption of many rabbit hyperimmune sera (24).
Antigenically related serovars are grouped into serogroups. However, a given serogroup is often found in several Leptospira species. For instance, the nine validly described serovars from serogroup Bataviae are distributed among L. interrogans sensu stricto species (two serovars), L. santarosai (four serovars), L. kirschneri (one serovar), L. noguchii (one serovar), and L. borgpetersenii (one serovar). Several studies have thus shown that the system of serogroups is not related to molecular classifications. In contrast, serovars can be characterized by different molecular methods, such as restriction fragment length polymorphism-based methods (15, 22), arbitrarily primed PCR (19), and pulsed-field gel electrophoresis (PFGE) (8, 9). However, these techniques are not widely applied, because PFGE and restriction fragment length polymorphism are laborious and require significant volumes of culture and arbitrarily primed PCR results in poor reproducibility and interpretation of results. In addition, lateral genetic transfer among leptospires (18) and large chromosomal rearrangements between serovars (26) prevent the construction of species phylogenetic trees by gene sequencing (7) or discrete whole-genomic data (19).
Analysis of variable-number tandem repeats (VNTR), also called multiple-locus VNTR analysis, has proven to be a highly powerful and discriminant method to study the population structure of bacteria (17) and to characterize isolates even from monomorphic bacterial populations (6, 11, 13). The genome of L. interrogans serovar Lai has recently been sequenced (20), and this allowed us to define pairs of primers flanking some VNTR-like loci. Our goal is to determine whether VNTR analysis will be able to differentiate most of the serovar reference strains from L. interrogans sensu stricto, providing a practical and simple PCR-based method for the identification of L. interrogans serovars.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. interrogans serovars (Table 1), L. borgpetersenii serovar Castellonis strain Castellon 3, L. kirschneri serovar Grippotyphosa strain Moskva V, L. kirschneri serovar Cynopteri strain 3522C, L. biflexa serovar Patoc strain Patoc1, and L. meyeri serovar Semaranga strain Veldrat were obtained from the strain collection at the National Reference Laboratory for Leptospira at the Institut Pasteur, Paris, France. Leptospiral strains used in this study were also isolated from patients (one strain of L. interrogans serovar Canicola, six strains of L. interrogans serovar Icterohaemorrhagiae, three strains of L. interrogans serovar Pomona, and one strain of L. interrogans serovar Hardjo), dogs (seven strains of L. interrogans serovar Canicola), bovines (two strains of L. interrogans serovar Hardjo), horses (one strain of L. interrogans serovar Hardjo), and sheep (two strains of L. interrogans serovar Hardjo) in the last 20 years. Leptospira strains were grown at 30°C in EMJH (4, 10) liquid medium.
TABLE 1.
Strains used in this study
| Serovar | Strain | Serogroupc | Abbreviation | Country | Source |
|---|---|---|---|---|---|
| Australisa | Ballicob | Australis | A1 | Australia | Human |
| Bangkok | Bangkok-D92 | Australis | A2 | Thailand | Dog |
| Bratislavaa | Jez-Bratislavab | Australis | A3 | Czechoslovakia | Hedgehog |
| Fugisa | Fudgeb | Australis | A4 | Malaysia | Human |
| Jalnaa | Jalnab | Australis | A6 | Czechoslovakia | Mouse |
| Muenchena | Munchen C 90b | Australis | A8 | Germany | Human |
| Autumnalisa | Akiyami Ab | Autumnalis | Au1 | Japan | Human |
| Carlosa | C-3b | Autumnalis | Au4 | Philippines | Toad |
| Moorisa | Mooresb | Autumnalis | Au5 | Malaysia | Human |
| Bataviaea | Van Tienen | Bataviae | B1 | Indonesia | Human |
| Benjaminia | Benjaminb | Canicola | C1 | Indonesia | Human |
| Bindjeia | Bindjeib | Canicola | C2 | Indonesia | Human |
| Broomia | Pataneb | Canicola | C3 | Australia | Human |
| Canicolaa | Hond Utrecht IVb | Canicola | C4 | Netherlands | Dog |
| Jonsisa | Jonesb | Canicola | C6 | Malaysia | Human |
| Kuwaita | 136/2/2b | Canicola | C7 | Kuwait | Rat |
| Portlandverea | MY 1039b | Canicola | C8 | Jamaica | Human |
| Schueffneria | Vleermuis 90Cb | Canicola | C10 | Indonesia | Bat |
| Sumneria | Sumnerb | Canicola | C11 | Malaysia | Human |
| Djasimana | Djasimanb | Djasiman | D1 | Indonesia | Human |
| Gurungia | Gurungb | Djasiman | D2 | Malaysia | Human |
| Grippotyphosaa | Andaman | Grippo | G1 | NDd | ND |
| Liangguang | 1880 | Grippo | G2 | China | Rat |
| Muelleria | RM 2b | Grippo | G3 | Malaysia | Rat |
| Valbuzzia | Valbuzzib | Grippo | G4 | Australia | Human |
| Hebdomadisa | Hebdomadisb | Hebdomadis | H1 | Japan | Human |
| Kremastosa | Kremastosb | Hebdomadis | H2 | Australia | Human |
| Birkini | Birkinb | Ictero | I1 | Malaysia | Human |
| Copenhagenia | M20b | Ictero | I3 | Denmark | Human |
| Copenhageni | Fiocruz L1-130 | Ictero | I3b | Brazil | Human |
| Copenhageni | Wijnberg | Ictero | I3c | Holland | Human |
| Gema | Simonb | Ictero | I4 | Sri Lanka | Human |
| Icterohaemorrhagiaea | RGA | Ictero | I6 | Belgium | Human |
| Laia | Laib | Ictero | I7 | China | Human |
| Naama | Naamb | Ictero | I11 | Indonesia | Human |
| Smithia | Smithb | Ictero | I13 | Malaysia | Human |
| Lankaa | R 740b | Louisiana | L0 | Sri Lanka | Human |
| Kennewicki | LT 1026 | Pomona | P2 | United States | Bovine |
| Pomonaa | Pomonab | Pomona | P4 | Australia | Human |
| Abramisa | Abrahamb | Pyrogenes | PY1 | Malaysia | Human |
| Biggisa | Biggsb | Pyrogenes | PY2 | Malaysia | Human |
| Camloa | LT 64-67b | Pyrogenes | PY3 | Vietnam | Human |
| Manilaea | LT 398b | Pyrogenes | PY5 | Philippines | Rat |
| Pyrogenesa | Salinemb | Pyrogenes | PY6 | Indonesia | Human |
| Robinsonia | Robinsonb | Pyrogenes | PY7 | Australia | Human |
| Evansia | 267-1348b | Ranarum | R0 | Malaysia | Water |
| Geyaweeraa | Geyaweerab | Sejroe | SE1 | Sri Lanka | Human |
| Haemolyticaa | Marshb | Sejroe | SE2 | Malaysia | Human |
| Hardjoa | Hardjoprajitnob | Sejroe | SE3 | Indonesia | Human |
| Jin | A81 | Sejroe | SE4 | China | Human |
| Ricardia | Richardsonb | Sejroe | SE7 | Malaysia | Human |
| Romanicaa | LM 294b | Sejroe | SE8 | Romania | Mus musculus |
| Wolffia | 3705b | Sejroe | SE10 | Indonesia | Human |
DNA manipulations.
Genomic DNA of Leptospira was isolated as previously described (16). Amplification was achieved with Taq polymerase (Amersham), using one cycle of denaturation (94°C for 5 min) followed by 35 cycles of amplification consisting of denaturation (94°C for 30 s), annealing (55°C for 30 s), and primer extension (72°C for 1 min 30 s) and a final extension of 10 min at 72°C. The amplified products were analyzed by 1.5% agarose gel electrophoresis. The sizes of the amplified products were estimated by comparison with a 100-bp ladder (Invitrogen). Some of the amplified products were sequenced at the Genomic Platform (Institut Pasteur).
Sequence analysis.
The large chromosome CI sequence (4,332 kb) of L. interrogans serovar Lai (20) was analyzed by using the Repeat Finder software (1) and the Tandem Repeats Database (http://iech5.igmors.u-psud.fr/GPMS/) (3). The copy number of repeats of each VNTR locus was deduced from sequencing data and sizes of the amplified products. The data were then imported into the Bionumerics software package (Applied Maths, Kortrijk, Belgium), and a phylogenetic tree was constructed by using the neighbor-joining method. The multiple phylogenetic methods showed similar tree topology. The ClustalX program (23) was used to generate nucleotide sequence alignments.
Nucleotide sequence accession numbers.
The sequences of the VNTR loci described in this report can be found in GenBank under accession numbers AY766398, AY766399, AY766400, AY766401, AY766402, and AY766403.
RESULTS
Computer-assisted analysis of VNTR-like regions in the L. interrogans genome.
At the beginning of our work, only one Leptospira genome sequence was available (at the time of this writing, another genome, that of L. interrogans serogroup serovar Copenhageni, has been sequenced [14]), preventing strain comparison and prediction of polymorphic loci. Analysis of the genome sequence of L. interrogans serovar Lai (20) enabled the rapid identification of loci containing repetitions of short sequences. Among more than 1,000 VNTR-like regions, an initial selection of 44 loci was chosen after comparing the lengths of repeats (repeats of 30 to 75 bp), sequence identities (nucleotide sequence identity between repeats of >85%), and copy numbers (between two and eight copies of the unit repeat).
PCR analysis of VNTR-like regions in L. interrogans senso stricto.
Forty-four primer pairs (sequences of the primers are available on request) were tested for their usefulness with a set of six well-characterized L. interrogans strains (strains A1, I3, I7, C4, PY6, SE3) (Table 1). The primers used for the PCR correspond to the VNTR flanking regions identified in the L. interrogans serovar Lai genome. Analysis of the amplified products by agarose gel electrophoresis revealed size variations in most of the loci. However, either no amplification or amplification of several faint bands was obtained for several loci, which were therefore excluded from this study (data not shown). This could be due to low conservation of the VNTR flanking regions among serovars. The seven most discriminative VNTR loci (i.e., VNTR4, VNTR7, VNTR9, VNTR10, VNTR11, VNTR19, and VNTR23) that exhibited a single PCR product whose size could be easily determined in 1.5% agarose gel electrophoresis for the six reference strains were further evaluated with a large collection of strains.
Sequence features of selected VNTR loci.
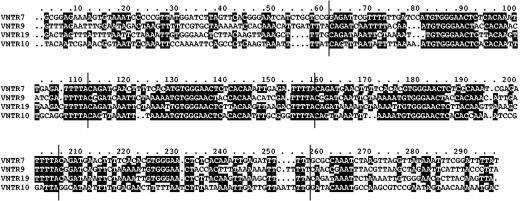
The positions of these VNTR loci were scattered in different locations in chromosome CI of L. interrogans (Table 2). Further sequence analysis indicated that none of the selected VNTR loci were located in open reading frames. However, some of the VNTR loci could contain small open reading frames. In addition, despite obvious sequence similarities between unit repeats, a closer look at the selected loci revealed sequence similarities among VNTR7, VNTR9, VNTR10, and VNTR19 (Fig. 1). These four loci share a 47-bp consensus sequence which is repeated in tandem (Fig. 1). Sequences of the repeats from the VNTR4 and VNTR23 loci also display significant similarities with repeats in other locations of the L. interrogans genome (data not shown). These results suggest that VNTR loci could be grouped in distinct families of tandem-repeat-containing loci.
TABLE 2.
VNTR loci from the L. interrogans serovar Lai genome used in this study
| VNTR locus | Primers (5′→3′) | Position in CI (bp) | Unit length (bp) | Copy no. | Total length of PCR product (bp) | No. of alleles/51 serovars | Copy no. range in L. interrogans serovars |
|---|---|---|---|---|---|---|---|
| VNTR4 | 4a (CAAAATCAGTCACTACCCTG) | 1122221-1122580 | 34 | 5 | 362 | 10 | 0-23 |
| 4b (CTTTGTTGGAGCGCAATCTC) | |||||||
| VNTR7 | 7a (TCATCTGCTCCGGAGATTCG) | 3312338-3312035 | 46 | 3 | 304 | 15 | 0-14 |
| 7b (TCCCTCCACAGGTTGTCTTG) | |||||||
| VNTR9 | 9a (TCGCTCTACAGGTCGGTGTT) | 2652531-2652151 | 46 | 4 | 381 | 13 | 1-13 |
| 9b (GGTGAAGAGCAAACCTTTGG) | |||||||
| VNTR10 | 10a (TCCAAAATTCAGCCCTCAAG) | 1666395-1666157 | 45 | 2 | 239 | 15 | 1-18 |
| 10b (GACGCTTGGCATTTGTATCC) | |||||||
| VNTR11 | 11a (ACAGAAGCCGTCTCATTTTG) | 167476-167184 | 45 | 4 | 293 | 7 | 1-11 |
| 11b (CACAGGTCGGAATTTGTCA) | |||||||
| VNTR19 | 19a (CAGAAACAAGAGGGAAGATTC) | 2877449-2877029 | 47 | 6 | 421 | 15 | 1-18 |
| 19b (ACTCTCATTTAAGAGTGGCTG) | |||||||
| VNTR23 | 23a (TTTCCAAATATACTTACTCGG) | 2179070-2178732 | 46 | 5 | 339 | 13 | 0-14 |
| 23b (GCAAGAGAATTATTGGGATGG) | |||||||
| VNTR31 | 31a (TTCATGAAGGTCCCGAAAAC) | 2729699-2730370 | 77 | 4 | 671 | NDa | ND |
| 31b (ACGTGAGTTCGACCATGATTC) |
ND, not determined (VNTR31 was used to differentiate between L. interrogans serovars Canicola and Portlandvere from serogroup Canicola).
FIG. 1.
Nucleotide sequence alignment of the VNTR7, VNTR9, VNTR10, and VNTR19 loci from L. interrogans serovar Lai. The 47-bp repeated units are delineated by vertical bars.
Characterization of L. interrogans serovars by VNTR polymorphism analysis.
PCR was performed with the seven selected VNTR loci and a total of 51 serovars, clustered in 13 distinct serogroups (Table 1). The sizes of the amplified products displayed a wide range of polymorphism, suggesting variation in tandem-repeat copy numbers in the seven VNTR loci (Fig. 2). This was confirmed by sequencing of 60 amplified products from the seven loci (Fig. 3). Figure 4 shows sequences of VNTR19, which contains variable number of perfectly identical repeats. For each VNTR locus, sequence analysis of amplified products indicated a high conservation of repeat units and flanking regions among L. interrogans serovars (data not shown). Although the presence of multiples of a full-length repeat was the general rule, an absence of the unit repeat, insertions, and/or deletions (4 out of the 60 sequenced amplicons) could result in a size of the amplified product which is not compatible with the variation in copy number of a full-length repeat.
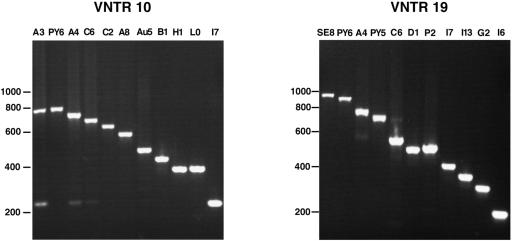
FIG. 2.
PCR analysis of the polymorphism of two representative VNTR loci. Amplification was performed on the VNTR10 and VNTR19 loci of L. interrogans strains. Lanes indicate Leptospira serovars (Table 1).
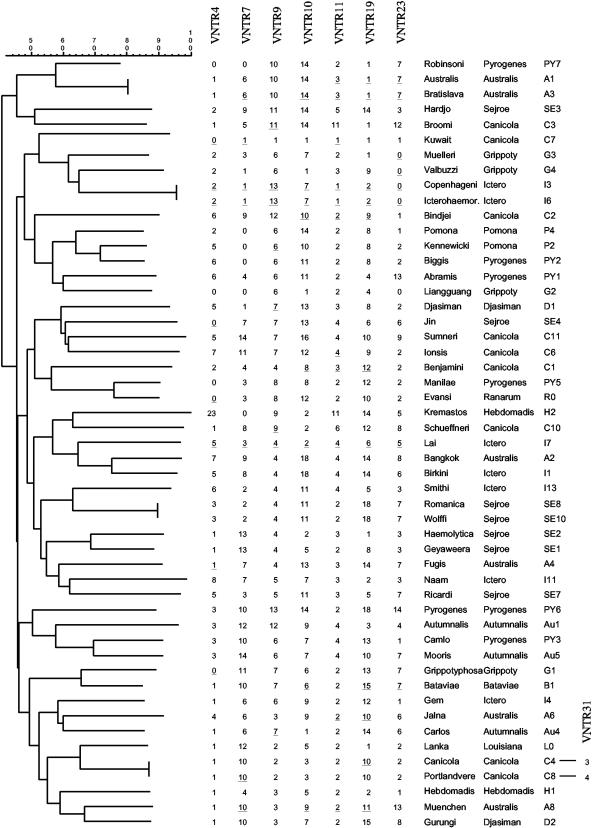
FIG. 3.
Dendrogram of the VNTR-typed serovars of L. interrogans. The copy number of each VNTR locus is indicated. The serovars and serogroups of reference strains are also indicated. PCR products that were sequenced are underlined. VNTR31was used to differentiate between L. interrogans serovars Canicola and Portlandvere from serogroup Canicola.
FIG. 4.
Sequence alignment of VNTR19 loci. Brackets on the left indicate the repeat units. Cani, L. interrogans serovar Canicola; Lai, L. interrogans serovar Lai; Grip, L. interrogans serovar Grippotyphosa; Autu, L. interrogans serovar Autumnalis; Cope, L. interrogans serovar Copenhageni; Aust, L. interrogans serovar Australis.
For each locus, the number of tandem repeats was calculated by measuring the sizes of the amplified products. The strains were typed by the numbers of variable tandem repeats in each of the seven VNTR loci. It should be noted that the value of 0 was used for amplified fragments shorter than a one-copy VNTR locus (this is the case in VNTR4, VNTR7, and VNTR23). These data could then be easily stored in databases and imported in Bionumerics for analysis.
The seven markers were able to differentiate 43 of 51 L. interrogans serovars (Fig. 3). An identical level of discrimination was obtained with only three markers, i.e., VNTR7, VNTR10, and VNTR19, that displayed the widest range of polymorphism with 15 distinct alleles among the 51 serovars (Table 2). Only four strain pairs were not differentiated whatever VNTR locus was used. The two strains within each pair belong to the same serogroup (L. interrogans serovars Copenhageni and Icterohaemorrhagiae from serogroup Icterohaemorrhagiae, L. interrogans serovars Australis and Bratislava from serogroup Australis, L. interrogans serovars Romanica and Wolffi from serogroup Sejroe, and L. interrogans serovars Canicola and Portlandvere from serogroup Canicola). L. interrogans serovars Copenhageni and Icterohaemorrhagiae were not differentiated with the 44 VNTR loci, as they were initially used as reference strains for the screening of the markers. We have undertaken PCR with VNTR loci that were previously excluded from the study to differentiate serovars that gave identical results with the seven selected markers. VNTR31 (four copies of a 77-bp repeat in L. interrogans serovar Lai) was able to differentiate between L. interrogans serovars Canicola (three copies) and Portlandvere (four copies) from serogroup Canicola. The three other pairs of strains were not differentiated whatever other VNTR was used.
Interestingly, three strains belonging to a same serovar (serovar Copenhageni strain M20 from Denmark, serovar Copenhageni strain Wijnberg from Holland, and serovar Copenhageni strain Fiocruz L1-130 from Brazil) exhibited identical results with the seven VNTR loci (data not shown). Since L. interrogans is phylogenetically related to other pathogenic species, we performed the VNTR assay with a few strains from L. kirschneri and L. borgpetersenii. Analysis of the PCR products of two of the seven markers, i.e., VNTR11 and VNTR19, exhibited a single band, variable in size, with the four strains of L. kirschneri and L. borgpetersenii. The size variation corresponded to multiples of the unit repeat identified in L. interrogans serovar Lai. These results suggest that L. interrogans, L. kirschneri, and L. borgpetersenii shared similar VNTR loci. In contrast, no amplification was obtained with DNAs from the saprophytic species L. biflexa and L. meyeri with the seven markers.
Application of our VNTR-based method to clinical strains.
To assess our PCR-based method for genotyping, we analyzed 23 clinical strains (including 11 strains isolated from humans) with the seven most discriminative VNTR loci (Table 1). The serovars of these isolates (L. interrogans serovars Icterohaemorrhagiae, Pomona, Hardjo, and Canicola) were previously identified by NotI restriction and PFGE (data not shown). All isolates displayed a pattern identical to that of the corresponding reference strain, suggesting the stability of PCR patterns in strains belonging to the same serovar. Again, the use of only three markers, i.e., VNTR7, VNTR10, and VNTR19, was enough to identify these strains at the serovar level.
DISCUSSION
Recently, a giant leap forward has been achieved with the completion of the genome sequence of L. interrogans serovar Lai (20). L. interrogans contains a 4.33-Mb large circular chromosome, a 359-kb small circular chromosome, and no extrachromosomal element (14, 20). Numerous repeated sequences have been found in its genome. For example, about 50 insertion sequences have been identified. The L. interrogans genome also contains abundant small repetitive DNA sequences. Among these DNA repeats, the structure of short sequence repeats is typical of tandem repeats. Tandem repeats consist of head-to-tail repetitions of short sequence motifs of about 10 to 100 bp. Polymorphic tandem repeats, also called VNTR, have been extensively used for fingerprinting in higher eukaryotes, including humans. Recently, the use of VNTR has also been described for phylogenetic and epidemiological studies of pathogenic bacteria.
A database of tandem repeats in more than 140 completely sequenced genomes is available (3). By using this database (http://iech5.igmors.u-psud.fr/GPMS/), genome analysis indicates that most of the tandem repeats (901 of 1,100; 82%) in L. interrogans strain Lai are between 15 and 50 bp, which is convenient for observation of polymorphism by analyzing PCR products of polymorphic loci on agarose gels. Compared to those of other bacteria, the L. interrogans genome exhibits a high number of tandem repeats with a total length greater than 100 bp (27 and 29 per Mb for strains Fiocruz and Lai, respectively). Comparative genomics shows that 131 VNTR loci (length of unit repeat, between 5 and 500 bp) were shared between the two available L. interrogans genomes (3, 14, 20). For strains within a species or genus, this number varies from 4 (strains from Chlamydia pneumoniae) up to 163 (strains from Yersinia pestis). Tandem repeats predicted to be polymorphic by genome comparison between the two serovars from serogroup Icterohaemorrhagiae indeed exhibit size variations by PCR typing of Leptospira strains.
The function of repetitive elements in bacteria is not fully understood. Similar to the Mycobacterium tuberculosis mycobacterial interspersed repetitive units (21), tandem-repeat-containing loci in L. interrogans are located in intergenic regions, are dispersed throughout the genome, and constitute subfamilies based on sequence similarities. The dissemination of homologous VNTR loci in the genome may suggest frequent intragenomic rearrangement or that these elements are (or have been) mobile elements. Studies on the structural and functional properties of these families of repetitive DNA should improve our knowledge of the role of these abundant repeat sequences in Leptospira spp.
All molecular tools described so far for the diagnosis of Leptospira suffer from significant drawbacks, such as a low discrimination level, lack of reproducibility, and requirement for large quantities of purified DNA. We therefore took advantage of the presence of VNTR-like regions to design a PCR-based test. Our data showed that VNTR typing was able to differentiate 45 out of 51 serovars of L. interrogans. Our VNTR assay shows that serovars from either the same serogroup or the same geographical area are not grouped together (Table 1; Fig. 3). This was true when the seven markers were used in combination but also when only three markers (VNTR7, -10, and -19) were used. This result confirms the heterogeneity among serovars of a given serogroup previously found by DNA relatedness (2) and PFGE (8, 9). Most serovar reference strains from L. interrogans have been isolated in Asia (2), mainly in southeast Asia (Table 1). It is noteworthy to find that strains originating from distant continents can be grouped together in the dendrogram. We also show that VNTR analysis was able to differentiate serovars among L. interrogans clinical strains, therefore demonstrating the usefulness of this PCR-based method for the identification of L. interrogans serovars.
A current method for typing Leptospira strains is macrorestriction by PFGE. This method is labor-intensive and not accessible to most laboratories in tropical and subtropical countries, where the incidence of the disease is the highest. Similar to PFGE, VNTR typing was not able to discriminate between L. interrogans serovars Icterohaemorrhagiae and Copenhageni. However, L. interrogans serovars Muenchen, Jalna, and Bratislava from serogroup Australis, which gave identical NotI macrorestriction patterns by PFGE (9), were differentiated by VNTR analysis. The use of only one VNTR locus, VNTR19 or VNTR23, was enough to discriminate serovars Muenchen, Jalna, and Bratislava (Fig. 3). In contrast, VNTR analysis of L. interrogans serovars Bratislava and Aus-tralis from serogroup Australis gave identical results (Fig. 3), but the two serovars were differentiated by PFGE (9). Distinct macrorestriction profiles of closely related strains could be due to large genomic rearrangements. For example, comparative genomics between two L. interrogans serovars from serogroup Icterohaemorrhagiae revealed a one-Mb chromosomal inversion (14).
In conclusion, this method based on VNTR polymorphism provides rapid typing as well as a highly discriminant assay to identify L. interrogans serovars. In addition, VNTR typing could be widely accessible for research and public health laboratories, particularly in developing countries. This method should also be suitable for sharing results and for the generation of databases. Further studies should include the development of a VNTR typing test with biological materials. The genome sequences of other Leptospira pathogenic strains are at different stages of completion. These sequences would greatly facilitate the development of multiple-locus VNTR assays for pathogenic Leptospira spp.
Acknowledgments
This work was supported by the Programme Transversal de Recherche (PTR no. 139), Institut Pasteur.
We thank A. Ko for the generous gift of DNA from strain Fiocruz L1-130. We are grateful to G. Vergnaud and F. Denoeud for the strain comparison database. We thank C. Gutierrez Perez for help in using Bionumerics and I. Saint Girons for her support.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. [DOI] [PubMed] [Google Scholar]
- 3.Denoeud, F., and G. Vergnaud. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource. BMC Bioinformatics 12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinghausen, H. C., and W. G. McCullough. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45-51. [PubMed] [Google Scholar]
- 5.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis. MedScience, Melbourne, Australia.
- 6.Farlow, J., D. Postic, K. L. Smith, Z. Jay, G. Baranton, and P. Keim. 2002. Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 40:4612-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haake, D. A., M. A. Suchard, M. M. Kelley, M. Dundoo, D. P. Alt, and R. L. Zuerner. 2004. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 186:2818-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrmann, J. L., C. Baril, E. Bellenger, P. Perolat, G. Baranton, and I. Saint Girons. 1991. Genome conservation in isolates of Leptospira interrogans. J. Bacteriol. 173:7582-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann, J. L., E. Bellenger, P. Perolat, G. Baranton, and I. Saint Girons. 1992. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J. Clin. Microbiol. 30:1696-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 94:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kmety, E., and H. Dikken. 1993. Classification of the species Leptospira interrogans and history of its serovars. University Press, Groningen, The Netherlands.
- 13.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perolat, P., I. Lecuyer, D. Postic, and G. Baranton. 1993. Diversity of ribosomal DNA fingerprints of Leptospira serovars provides a database for subtyping and species assignation. Res. Microbiol. 144:5-15. [DOI] [PubMed] [Google Scholar]
- 16.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189-199. [DOI] [PubMed] [Google Scholar]
- 17.Pourcel, C., Y. Vidgop, F. Ramisse, G. Vergnaud, and C. Tram. 2003. Characterization of a tandem repeat polymorphism in Legionella pneumophila and its use for genotyping. J. Clin. Microbiol. 41:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralph, D., and M. McClelland. 1994. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J. Bacteriol. 176:5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ralph, D., M. McClelland, J. Welsh, G. Baranton, and P. Perolat. 1993. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J. Bacteriol. 175:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Weng, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 21.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 22.Thiermann, A. B., A. L. Handsaker, S. L. Moseley, and B. Kingscote. 1985. New method for classification of leptospiral isolates belonging to serogroup pomona by restriction endonuclease analysis: serovar kennewicki. J. Clin. Microbiol. 21:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. 2003. Human leptospirosis: guidance for diagnosis, surveillance and control. World Health Organization, Geneva, Switzerland.
- 25.Yasuda, P. H., A. G. Steigerwalt, K. R. Sulzer, A. F. Kaufmann, F. C. Rogers, and D. J. Brenner. 1987. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int. J. Syst. Bacteriol. 37:407-415. [DOI] [PubMed] [Google Scholar]
- 26.Zuerner, R. L., J. L. Herrmann, and I. Saint Girons. 1993. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J. Bacteriol. 175:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]