Abstract
Meningococci causing New Zealand's epidemic, which began in 1991, are defined as group B, serosubtype P1.4 (subtype P1.7-2,4), belonging to the ST-41/ST-44 complex, lineage III. Of the 2,358 group B isolates obtained from disease cases from 1991 through 2003, 85.7% (2,021 of 2,358) were determined to be serosubtype P1.4. Of the remaining isolates, 156 (6.6%) were not serosubtypeable (NST). Molecular analysis of the porA gene from these B:NST meningococcal isolates was used to determine the reason. Most NST isolates (156, 88.5%) expressed a PorA that was distinct from P1.7-2,4 PorA. Fifteen isolates expressed variants of P1.7-2,4 PorA, and a further three expressed P1.7-2,4 PorA without any sequence variation. These three isolates expressed P1.7-2,4 PorA at very low levels, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, and showed variation in the porA promoter region. Among the 15 meningococcal isolates expressing variants of P1.7-2,4 PorA, 11 different sequence variations were found. Compared with the P1.7-2,4 PorA sequence, the sequences of these variants contained deletions, insertions, or single-nucleotide substitutions in the VR2 region of the protein. Multilocus restriction typing was used to assess the clonal derivations of B:NST case isolates. Meningococcal isolates expressing distinct PorA proteins belonged mostly to clonal types that were unrelated to the epidemic strain, whereas all meningococcal isolates expressing variants of P1.7-2,4 PorA belonged to the ST-41/ST-44 complex, lineage III. These results, together with those obtained serologically, demonstrate that the P1.7-2,4 PorA protein of meningococci responsible for New Zealand's epidemic has remained relatively stable over 13 years and support the use of a strain-specific outer membrane vesicle vaccine to control the epidemic.
New Zealand has experienced an ongoing epidemic of group B meningococcal disease since mid-1991 (11). The highest rate of disease to date occurred in 2001, when 650 cases were reported, giving a rate of 17.4 cases per 100,000 people (10). Elevated disease rates in excess of 14 cases per 100,000 people have continued into 2004. The increased levels of disease are attributable to group B meningococci expressing the P1.7-2,4 PorA protein (11) belonging to the ST-41/ST-44 complex, lineage III (3). Epidemics of meningococcal disease occur infrequently, and with the exception of group B epidemics can be controlled by the use of polysaccharide-based vaccines (6). As the group B polysaccharide is poorly immunogenic and an autoantigen, outer membrane vesicle (OMV) vaccines containing subcapsular antigens have been developed to control group B epidemics (6). Bactericidal antibody responses against OMV vaccines in both animal and human vaccine trials are directed mainly against PorA, and thus immunity is strain specific (17, 20, 26, 36).
In the absence of a commercially available vaccine against the New Zealand epidemic strain, a tailor-made vaccine has had to be developed. The development and manufacture of a New Zealand epidemic strain-specific vaccine (MeNZB) in sufficient quantities for a nationwide meningococcal group B immunization program for all under 20 years of age were made possible through a partnership between the New Zealand Ministry of Health and Chiron Vaccines, working in collaboration with the Norwegian Institute of Public Health (5). The decision to use such a vaccine was assisted by demonstration of the highly clonal nature of meningococci that caused the ongoing epidemic (3) and the apparent stability of the PorA protein (10, 11).
The PorA protein consists of eight surface-exposed loops, with loops 1 and 4 each containing one variable region (VR), designated VR1 and VR2, respectively (9, 29). Serosubtyping monoclonal antibodies recognize linear epitopes encoded by porA VR1 or VR2 (13, 14). The P1.7-2 and P1.4 epitopes are located in VR1 and VR2, respectively. Isolates expressing the P1.7-2 epitope have a 3-amino-acid deletion on the carboxy side of the epitope that prevents detection of the native protein by the P1.7 serosubtyping monoclonal antibody (35). The sequence encoding the P1.7-2 epitope can be identified by DNA sequence analysis. Previously, the P1.7-2,4 PorA protein has been described as P1.7h,4 (30) and P1.7b,4 (35), where P1.7-2, P1.7h, and P1.7b denote the same genetic variation.
From 1991 to the end of 2003, there were 2,358 serogroup B case isolates. All were subjected to serologic typing. However, for 156 (6.6%) isolates, the PorA subtype could not be determined with the monoclonal antibodies used. This study was undertaken to investigate the porA VR1 and VR2 sequences in these 156 group B nonserosubtypeable (B:NST) isolates. We wished to determine the reason for their failure to be serosubtyped and, in particular, if mutations in the regions encoding the P1.7-2 and P1.4 epitopes could be the source. The results of this study will provide baseline information against which any porA variation in clinical isolates, both during and following delivery of the strain-specific OMV vaccine, can be compared.
MATERIALS AND METHODS
Meningococcal isolates.
Meningococci used in this study originated from clinical cases of meningococcal disease and were referred to the Meningococcal Reference Laboratory, Institute of Environmental Science and Research (ESR), under New Zealand's national surveillance program. All meningococci were assigned an isolate number which consisted of the last two numbers of the year in which they were isolated, followed by a unique identifying number. The letters “NZ” were placed before the isolate number to indicate their country of origin.
Referred meningococcal isolates were maintained at −70°C in glycerol broth suspensions (Trypticase soy broth, 15% [vol/vol] glycerol). Cultures were grown on 5% sheep blood agar plates (Fort Richard Laboratories, Auckland, New Zealand) at 36°C, in an atmosphere of 5% CO2, for 18 h. All case isolates were routinely serogrouped, serotyped, and serosubtyped (1). Monoclonal antibodies that recognize serotypes 1, 2a, 2b, 4, 14, and 15 and serosubtypes P1.1, P1.2, P1.4, P1.5, P1.6, P1.7, P1.9, P1.10, P1.12, P1.13, P1.14, P1.15, and P1.16 (RIVM, Bilthoven, The Netherlands) were used. All NST meningococci were typed by using standard DNA-DNA hybridization methodology (19), with P1.2, P1.4, P1.7, P1.13, P1.15, P1.16, and P1.19 as the oligonucleotide probes (Table 1). Those isolates found to be negative by DNA-DNA hybridization were subjected to porA sequencing (8).
TABLE 1.
Probes used to determine the meningococcal subtype by use of DNA-DNA hybridization
| Probe | Probe sequence | Source or reference |
|---|---|---|
| P1.2 | 5′-Biotin-CAT TTT GTT CAG CAG ACT CCT RAA AGT CAG CCT ACT CTC GTT CCG-3′ | 9 |
| P1.4 | 5′-Biotin-CAT GTT GTT GTG AAT AAC AAG GTT GCT ACT CAC GTT CCG-3′ | 8 |
| P1.7 | 5′-Biotin-GCA CAA GCC GCT AAC GGT GGA GCG AGC GGT CAG GTA AAA GTT-3′ | 8 |
| P1.13 | 5′-Biotin-TAT TGG ACT ACT GTG AAT ACC GGT AGT GCT ACT ACT ACT ACT-3′ | This study |
| P1.15 | 5′-Biotin-CAT TAT ACT AGG CAG AAC AAT GCT GAT GTT TTC GTT CCG-3′ | This study |
| P1.16 | 5′-Biotin-TAT TAT ACT AAG GAT ACA AAC AAT AAT CTT ACT CTC GTT-3′ | 12 |
| P1.19 | 5′-Biotin-CCG CCC TCA AAG AGT CAA CCT CAG GTA AAA GTT ACT AAG GCC-3′ | This study |
DNA extraction.
Genomic DNA was purified from other meningococcal cellular components by using cetyltrimethyl ammonium bromide (Sigma, St. Louis, Mo.) followed by phenol and chloroform extractions (15). DNA was quantitated using PicoGreen fluorescent dye (Molecular Probes, Eugene, Oreg.) in a fluorometer (BMG LabTechnologies, Offenburg, Germany). Stock DNA was stored at −70°C, and 5-ng/μl dilutions made in TE buffer (10 mM Tris, 1 mM EDTA) were stored at 4°C.
PCR-based analysis.
PCR Master Mix (Qiagen, Hilden, Germany) was used to amplify the PCR product. The porA products were amplified using primers PorA P1 and PorA P2 and sequenced using primers PorA P1 and PorA P4 (21). Multilocus restriction typing (MLRT) (2) and multilocus sequence typing (MLST) (7) were carried out as described previously, with the addition of primers to amplify fumC (fumC-A1, 5′-CAC CGA ACA CGA CAC GAT GG-3′, and fumC-A2, 5′-ACG ACC AGT TCG TCA AAC TC-3′). The porA promoter region was amplified and sequenced using primers PorA5 and P1-1 (28). Sequencing was carried out using a model 3100 genetic sequencer (Applied Biosystems, Foster City, Calif.). Sequence data were analyzed using sequence analysis programs (DNASTAR, Inc., Madison, Wis.). To assign porA VR sequences to families and multilocus sequence types, nucleotide sequences were submitted to the porA typing website (http://www.neisseria.org/nm/typing/pora) and MLST website (http://pubmlst.org/neisseria), respectively.
SDS-PAGE.
Whole-cell protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a Mini-Protean 3 electrophoresis cell (Bio-Rad, Hercules, Calif.). Samples were stacked in a 4% acrylamide gel and separated in a 12% acrylamide gel by using Tris-glycine buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Protein bands were visualized by staining for 1 h in Coomassie brilliant blue (0.1% [wt/vol] Coomassie brilliant blue in 50% [vol/vol] methanol-10% [vol/vol] acetic acid) and destaining overnight in 10% (vol/vol) methanol-10% (vol/vol) acetic acid. The sizes of the products were compared to that of the prestained SDS-PAGE broad-range standard (Bio-Rad) or the MagicMark molecular weight marker (Invitrogen, Carlsbad, Calif.).
Immunoblotting.
The method described by Wedege (34) was used for immunoblotting. The P1.7 (NIBSC 01/514) and P1.4 (NIBSC 02/148) monoclonal antibodies (NIBSC, Hertfordshire, England) were used for epitope detection. Bound antibodies were detected using protein A-peroxidase conjugate (Bio-Rad) and 4-chloro-1-naphthol for detection (Sigma).
Nucleotide sequence accession number.
The porA sequence for strain NZ02/203 was submitted to the GenBank database and has been assigned accession number AY653178.
RESULTS
From 1991 through 2003 inclusive, New Zealand's Meningococcal Reference Laboratory received 2,358 serogroup B case isolates, and all were subjected to serosubtyping. Of these, 85.7% (2,021 isolates) were determined to be type P1.4, 6.6% (156 isolates) were NST, and the remaining 7.7% (181 isolates) were other PorA serosubtypes. DNA-DNA hybridization and porA sequencing determined that, of the 156 B:NST isolates, 138 (88.5%) had porA subtypes distinct from that encoding the P1.7-2,4 PorA. The most common subtypes were P1.19, P1.22,14-6, P1.7,13, and P1.17,16. Of the remaining 18 isolates, 15 expressed variants of P1.7-2,4 PorA and three expressed P1.7-2,4 PorA without sequence variation although this was not identifiable using monoclonal antibodies.
Two meningococcal isolates that expressed a variant of P1.7-2,4 PorA epitope (strains NZ02/203 and NZ02/234) also expressed both the P1.7-2 and P1.4 PorA epitopes but had insertions in the region encoding PorA loop 4. Strain NZ02/203 had a 17-amino-acid insertion 11 amino acids away from PorA VR2-4 on the carboxy side. This porA sequence was submitted to the GenBank database under accession number AY653178. Strain NZ02/234 had an extra HV in the amino acid sequence at the start of the PorA VR2 (HVHVVVNNKVATHVP) compared to the PorA VR2-4 sequence (HVVVNNKVATHVP). The unique porA VR2 sequence found in strain NZ02/234 was submitted to the Neisseria meningitidis PorA VR database (http://neisseria.org/nm/typing/pora) and was assigned to VR2-4-12.
The remaining 13 meningococcal isolates that expressed a variant of P1.7-2,4 PorA showed variation in only porA VR2-4 (Table 2). Eleven had deletions of different sizes in the region encoding the P1.4 epitope, including three (strains NZ97/27, NZ99/226, and NZ02/68) with 39-bp deletions encompassing the entire porA VR2 region (Table 2). Two other meningococcal isolates (strains NZ98/214 and NZ01/56) had unique VR2 sequences involving a single-amino-acid change compared to the P1.4 epitope (Table 2). All unique porA VR2 sequences were searched against the PorA VR database and were given the P1.4-7 to P1.4-11 designations (Table 2). Immunoblotting showed that the P1.7 monoclonal antibody bound to a band of approximately 42 kDa in all isolates (Table 2). Binding of the P1.4 DNA probe was affected by the size and position of the deletion (Table 2).
TABLE 2.
Summary of serological typing, DNA-DNA hybridization, porA sequencing, and Western blotting results for all meningococci identified with variations in the porA VR2-4 sequence
| Strain | Phenotype |
porA VR2 sequence
|
PorA expressed | Outcome of Western blottingd
|
|||
|---|---|---|---|---|---|---|---|
| DNAc | Amino acid | Epitope code | P1.7 | P1.4 | |||
| NZ99/38 | B:4:P1.4 | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVVNNKVATHVP | P1.4 | Yes | + | + |
| NZ96/142 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVVNNKVATHVP | P1.4 | Weak | − | − |
| NZ01/278 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVVNNKVATHVP | P1.4 | Weak | − | − |
| NZ02/55 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVVNNKVATHVP | P1.4 | Weak | − | − |
| NZ98/214 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGTGAATAACCAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVVNNQVATHVP | P1.4-8 | Yes | + | − |
| NZ01/56 | B:14:NSTa | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCTCCTCACGTTCCGGCTGTTGTCGGC | HVVVNNKVAPHVP | P1.4-11 | Yes | + | + |
| NZ99/22 | B:4:NSTa | TATACGCC------------TGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | ---VNNKVATHVP | P1.4-9 | Yes | + | (+) |
| NZ02/68 | B:4:NSTb | TATACGCCGGCTCAT---------------------------------------GTTGTCGGC | HVV---------- | Deleted | Yes | + | − |
| NZ99/226 | B:4:NSTb | TATACGCCGGCTCAT---------------------------------------GTTGTCGGC | HVV---------- | Deleted | Weak | (+) | − |
| NZ97/27 | B:4:NSTb | TATACGCCGGCTCAT---------------------------------------GTTGTCGGC | HVV---------- | Deleted | Yes | + | (+) |
| NZ98/4 | B:4:NSTb | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTT---------------GCTGTTGTCGGC | HVVVNNKV----- | P1.4-5 | Yes | + | (+) |
| NZ03/43 | B:4:NSTb | TATACGCCGGCTCATGTTGTTGTGAATAACAAGGTTGCT------------GCTGTTGTCGGC | HVVVNNKVA---- | P1.4-5 | Yes | + | (+) |
| NZ94/124 | B:4:NSTb | TATACGCCGGCTCATGTTGTTGT------CAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVVV--KVATHVP | P1.4-7 | Yes | + | (+) |
| NZ00/202 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGT---TAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVV-NNKVATHVP | P1.4-10 | Yes | + | − |
| NZ01/199 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGT---TAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVV-NNKVATHVP | P1.4-10 | Yes | + | (+) |
| NZ01/224 | B:4:NSTa | TATACGCCGGCTCATGTTGTTGT---TAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVV-NNKVATHVP | P1.4-10 | Yes | + | (+) |
| NZ02/112 | B:4:NSTa | TATACGCCGGCTCAT---GTTGTGAATAACAAGGTTGCTACTCACGTTCCGGCTGTTGTCGGC | HVV-NNKVATHVP | P1.4-10 | Yes | + | (+) |
The porA P1.7 and P1.4 VR sequences identified by use of DNA-DNA hybridization.
The porA P1.7 VR sequence identified by use of DNA-DNA hybridization.
The region in boldface identifies the porA VR2 sequence.
(+), weak reaction.
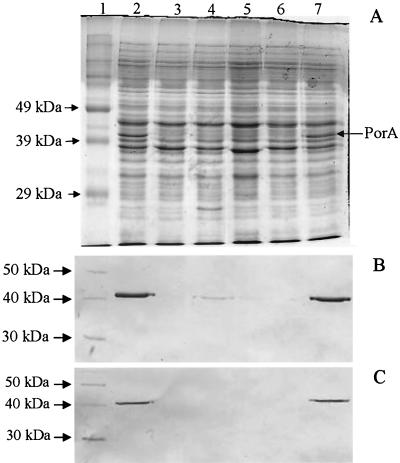
The three isolates (strains NZ96/142, NZ01/278, and NZ02/55) that expressed the P1.7-2,4 PorA epitope without sequence variation and yet were NST were examined for protein expression by SDS-PAGE analysis of whole-cell protein extracts. Strain NZ99/226 was also examined because, although it expressed the P1.7-2 epitope, it was recognized only weakly by the P1.7 monoclonal antibody during immunoblotting (Table 2 and Fig. 1B). Profiles for all four meningococci showed a weak band at the positions corresponding to 40 to 42 kDa on SDS-PAGE gels (Fig. 1A). Extracts from strains NZ96/142, NZ01/278, and NZ02/55 were not recognized by either the P1.7 or P1.4 monoclonal antibodies during immunoblotting (Fig. 1). To try to establish why strains NZ96/142, NZ99/226, NZ01/278, and NZ02/55 had reduced PorA expression, the porA promoter region was sequenced. Sequencing determined that there was variation in the polyguanidine track between the putative −35 and −10 regions of the porA promoter in New Zealand case isolates. Strains NZ96/142, NZ01/278, and NZ02/55 contained a string of 9 guanidine residues, whereas strain NZ99/226 contained a string of 10 guanidine residues. In contrast, four meningococcal isolates with a strong ∼42-kDa band (strains NZ03/43, NZ03/164, NZ99/38, and NZ99/109) contained either 11, 12, or 13 guanidine residues in the polyguanidine track.
FIG. 1.
SDS-PAGE profiles and immunoblotting results (B and C) for whole-cell extracts from four meningococcal isolates of phenotypes B:P1.4 (lanes 2 and 7) and B:NST (lanes 3 through 6). Strains assessed were NZ91/49 (lanes 2 and 7), NZ96/142 (lane 3), NZ99/226 (lane 4), NZ01/278 (lane 5), and NZ02/55 (lane 6). (A) SDS-PAGE gel profiles of strains stained using Coomassie blue. Lane 1 contains the prestained SDS-PAGE broad-range standard (Bio-Rad). (B) Immunoblot of gel with P1.7 monoclonal antibody (NIBSC 01/514). (C) Immunoblot of gel with P1.4 monoclonal antibody (NIBSC 02/148). For panels B and C, lane 1 contains the MagicMark molecular mass marker (Invitrogen).
MLRT was used to determine the genetic relatedness of B:NST and B:P1.7-2,4 meningococcal isolates. All 18 B:NST meningococcal isolates expressing P1.7-2,4 PorA or a variant of P1.7-2,4 PorA belonged to one of the ST-41/ST-44 complex, lineage III, subclones causing New Zealand's epidemic: RT-41 (n = 2), RT-42 (n = 11), or RT-154 (n = 5). In contrast, most of the 60 NST meningococcal isolates with porA types distinct from that encoding P1.7-2,4 PorA belonged to a diverse range of multilocus restriction types unrelated to the ST-41/ST-44 complex, lineage III (Table 3). The P1.19 and/or P1.15 porA type was the most prevalent porA type carried by these NST meningococci. A number of different porA VR2 types were found among the 50 meningococcal isolates expressing the P1.19 VR1 epitope, including P1.19,15 (29 isolates), P1.19,26 (7 isolates), P1.19-3,15 (5 isolates), and P1.19,15-1 (4 isolates). Meningococcal isolates with the P1.19,15 porA type were mostly unrelated to New Zealand's epidemic strain and shared a common MLRT profile with a case isolate from Cuba, which belonged to ST-33. The few that did belong to the ST-41/ST-44 complex, lineage III, included three meningococcal isolates expressing the P1.19,15 PorA proteins that were typed as RT-42 (n = 2) and RT-154 (n = 1) and three meningococcal isolates expressing the P1.19-3,15 PorA proteins that were typed as RT-42 (n = 2) and RT-154 (n = 1). The only other meningococcal isolates that expressed PorA types distinct from P1.7-2,4 PorA, yet belonged to the ST-41/ST-44 complex, lineage III, had porA types P1.7-2,25 (n = 1) and P1.7,16 (n = 1) and were ST-2763 (which differs from ST-42 by the presence of aroE15 and RT-154).
TABLE 3.
Summary of porA subtyping results with MLRT results for all nst meningococci derived from disease cases in New Zealand between 1991 and 2003
| porA type | Total no. of isolates | No. of isolates
|
|||
|---|---|---|---|---|---|
| Assessed by MLRT | Of other complexes | Of the ST-41/ST-44 complex, lineage III
|
|||
| Total | ST-41/ST-42/ ST-154 | ||||
| P1.19 | 50 | 41 | 23 | 18 | 6 |
| P1.22, 14-6 | 16 | 0e | |||
| P1.7-2,4 varianta | 15 | 15 | 0 | 15 | 15 |
| P1.7,13 | 12 | 5 | 5 | 0 | 0 |
| P1.17,16 | 10 | 3 | 3 | 0 | 0 |
| P1.7-2,4b | 3 | 3 | 0 | 3 | 3 |
| Other definedc | 20 | 8 | 3 | 6 | 2 |
| Other undefinedd | 30 | 0 | 0 | 0 | 0 |
| Total | 156 | 75 | 29 | 42 | 26 |
Expressed P1.7-2,4 PorA with the exception of deletions, insertions, or single-nucleotide substitutions around the VR2-4 sequence.
NST but determined to express P1.7-2,4 PorA by DNA sequencing.
Includes porA types P1.5,10, P1.21, 16, P1,18, 34, P1.7,16, P1.7-2,25, P1.21,15, P1.7-1,14, P1.22,28, and P1.21-2,28.
Isolates were NST and negative for all DNA probes used in DNA-DNA hybridization.
All 18 serosubtype P1.14 meningococcal isolates tested had restriction profiles identical to those for ST-457 meningococci.
Between 1991 and 2003 inclusive, five New Zealand case isolates expressing P1.7-2,4 PorA that were not serogroup B were identified. These five isolates were typed as C:2a:P1.7-2,4 (strains NZ96/59 and NZ03/236), C:2b:P1.7-2,4 (strain NZ96/211), W135:2a:P1.7-2,4 (strain NZ03/243), and W135:2a:P1.4 (strain NZ03/266). One further meningococcal strain (strain NZ04/8), type W135:2a:P1.4, was identified in early 2004. MLST showed that strain NZ96/59 had a multilocus sequence type dissimilar to that of any meningococcus deposited in the MLST database (http://pubmlst.org/neisseria), and sequence type ST-2344 was assigned to this combination of alleles. Strain NZ96/211 was type ST-66, which belongs to the ST-8/A4 complex. MLST showed that NZ03/266 belonged to the ST-11/ET-37 complex. Strains NZ03/236, NZ03/243, and NZ04/8 had the same restriction profile as strain NZ03/266 and were assumed therefore to belong to the ST-11 complex.
DISCUSSION
Throughout New Zealand's meningococcal disease epidemic, the P1.7-2,4 PorA epitope expressed by clinical isolates has been easily identifiable by using the P1.4 subtyping antibody. The presence of 156 B:NST case isolates presented the possibility that the ST-41/ST-44 complex, lineage III, meningococci expressing the P1.7-2,4 PorA epitope were responsible for a larger proportion of epidemic disease than serological typing had indicated. This was determined not to be the case, as the majority (138 of 156, 88.5%) of the B:NST isolates expressed PorA epitopes distinct from P1.7-2,4 PorA. Most of these meningococci belonged to clonal complexes other than the ST-41/ST-44 complex, lineage III, and are therefore not related to the meningococcal isolates causing the epidemic. All 18 NST case isolates that expressed a PorA related to P1.7-2,4 PorA belonged to the ST-41/ST-44 complex, lineage III. These meningococci are genetic variants of the epidemic strain that have arisen during the epidemic.
Four meningococcal isolates that did not express a detectable level of PorA on an SDS-PAGE gel were identified. Three of these contained a porA encoding the P1.7-2,4 PorA epitope but were NST, while the fourth had a 39-bp deletion in porA VR2. The observation that all four meningococcal isolates with reduced PorA expression contained a polyguanidine tract with 9 or 10 guanine residues between the putative −35 and −10 regions of the porA promoter has previously been reported (22, 27). It has been suggested that the presence of 11, 10, or 9 contiguous guanidine nucleotides in the promoter region is associated with high, medium, or no expression of PorA mRNA, respectively (28). It has also been suggested that the level of PorA expression is affected by substitutions in the polyguanidine tract (22, 27, 28). No New Zealand case isolate was identified in which a guanidine nucleotide in the polyguanidine track was replaced by an alternative nucleotide has been identified.
The inability to serosubtype isolates expressing variant P1.7-2,4 PorA was due mostly to the presence of mutations in porA VR2-4. Both deletions and single-nucleotide substitutions in porA VR2-4 compromised the ability of the subtype P1.4 monoclonal antibody to recognize the VR2 epitope in a whole-cell enzyme-linked immunosorbent assay. The binding of the P1.4 DNA probe and the P1.4 monoclonal antibody to porA and the PorA protein, respectively, was affected by the size and position of the VR2-4 mutation. The insertion of 2 amino acids on the amino side of the P1.4 epitope, as well as the insertion of 17 amino acids on the carboxy side of the P1.4 epitope, resulted in the inability of the P1.4 monoclonal antibody to bind. The insertions appear to have modified the position of the epitope recognized by the P1.4 antibody, although the epitopes were recognized during immunoblotting. This result creates a situation similar to that for the “hidden” P1.7-2 epitope described by Wedege (35) which resulted from a 3-amino-acid deletion on the carboxy side of the epitope.
NST isolates were relatively uncommon, accounting for only 156 (6.6%) of all serogroup B case isolates. This study identified three major reasons for the failure of monoclonal antibodies to recognize the PorA subtype in New Zealand's serogroup B case isolates. First, the range of monoclonal antibodies available for typing in our laboratory did not cover all of the possible PorA epitopes identified in the population. This deficiency accounted for most of the NST results. Second, sequence variations, including insertions or deletions around the VR2-4 region of PorA, prevented recognition by the P1.4 subtyping monoclonal antibody. Third, reduced PorA expression appears to have resulted in NST meningococci. The inclusion of additional serosubtyping antibodies in the panel, such as the P1.19 antibody, would have reduced the number of isolates for which the subtype could not be determined serologically. However, with the low numbers of NST isolates, genetic characterization with porA sequencing remains a suitable method to determine their porA types.
Gorla and coworkers (4) reported that case isolates from vaccine failures expressed low levels of PorA protein. However, these researchers showed that there was no association between a low level of PorA expression and vaccination status, as there were equal numbers of meningococci expressing low levels of PorA in immunized and nonimmunized children. Vermont and colleagues (33) showed that immune responses directed against meningococci with significantly different porA VR2 sequences were reduced, whereas responses against isolates with porA variants that had most of the VR2-4 sequence conserved were less affected. Whether people challenged with meningococci demonstrating the P1.7-2,4 deletions or the transcriptional variations detected in this study would be protected by anti-P1.7-2,4 antibodies induced by a strain-specific vaccine cannot be determined from this study but is currently being investigated. It is possible that alternative epitopes on the PorA protein, which are not recognized by anti-VR1 and anti-VR2 monoclonal antibodies, may induce functional responses that are normally overshadowed by antibodies against the VR1 and VR2 targets. Rouppe van der Voort and coworkers (18) observed that the serum bactericidal antibody responses in some vaccinees were dependent on the presence of loops 1 and/or 4, whereas some volunteers developed PorA-specific bactericidal antibodies which did not depend on loops 1 and/or 4.
Given that PorA is an important target for immune recognition and that meningococci have a number of ways to facilitate variation in the porA gene, it is surprising that the P1.7-2,4 PorA epitope has remained so stable over the 13 years of New Zealand's epidemic. Meningococci expressing P1.7-2,4 PorA have continuously dominated group B isolates, accounting for 85.7% of all group B case isolates from 1991 through 2003. The observation that some meningococci belonging to the ST-41/ST-44 complex, lineage III, have a porA other than that encoding the P1.7-2,4 PorA epitope suggests that these meningococci have at some stage acquired an alternative porA. Twenty-nine such meningococcal isolates (1.2%, 29 of 2,358) have been identified. Increasing variability in PorA subtype among isolates of ST-41/ST-44 complex, lineage III, was reported during a period of heightened disease rates in both The Netherlands and Belgium in the 1980s (24, 32). The greater population density and the greater diversity of strains in these countries (23, 31) than those in New Zealand may have contributed to greater mixing of meningococcal DNA, allowing for the increased variety of PorA types.
The expression of P1.7-2,4 PorA by meningococci of serogroups other than serogroup B could not be explained by capsular switching following exchange of the gene encoding the polysialyltransferase, as was shown by Swartley and coworkers (25). Instead, the PorA gene appears to have been transferred from meningococci belonging to the ST-41/ST-44 complex, lineage III, to meningococci from other clonal complexes. Group W135 and group C isolates with the P1.7-2,4 PorA epitope have also been obtained among isolates recovered from a throat carriage study in university students in Dunedin, New Zealand (unpublished data). It is likely that anti-P1.7-2,4 PorA antibodies induced by the strain-specific vaccine confers some protection against such meningococci. We have demonstrated that antibodies induced by the New Zealand strain-specific vaccine (MeNZB) prepared from NZ98/254 (B:4:P1.7-2,4) are bactericidal against the C:2b:P1.7-2,4 strain NZ96/211 (unpublished results).
Identification of New Zealand case isolates with sequence variation in the P1.4 epitope prior to a population-based introduction of a strain-specific vaccine is important. The fact that PorA variants to the epidemic type were rare and apparently unconnected suggests that they have not become established in the community. However, it is possible that if such variants are present within the population of meningococci and are being carried asymptomatically, immune selection could occur. In Norway, increased resistance to bactericidal antibodies induced by the Norwegian B OMV vaccine were associated with point mutations in VR2 (16). However, Gorla et al. (4), reported that point mutations were not a strategy used by meningococci in Brazil to escape antibody pressure. No variation in the P1.19 or P1.15 epitopes was identified in case isolates following immunization with the Cuban VA-MENINGOC-BC vaccine (4). Continuing intensive surveillance of New Zealand case isolates will ensure detection and monitoring of any variants that occur following vaccine delivery to the broader community.
Acknowledgments
We acknowledge the receipt of strains from laboratories throughout New Zealand as a part of the surveillance of meningococcal disease on behalf of the Ministry of Health of New Zealand. We thank the staff of the Invasive Pathogens Laboratory, ESR, for the phenotyping of isolates received. This research made use of the Neisseria MLST website (http://pubmlst.org/neisseria) maintained by the University of Oxford and developed by M.-S. Chan and K. Jolley.
This work was supported in part by a grant from Lotteries Health Research. Kristin Dyet was an ESR Ph.D. scholar.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb. Pathog. 4:27-32. [DOI] [PubMed] [Google Scholar]
- 2.Dyet, K. H., R. S. Simmonds, and D. R. Martin. 2004. Multilocus restriction typing method to predict the sequence type of meningococci. J. Clin. Microbiol. 42:1742-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyet, K. H., R. S. Simmonds, and D. R. Martin. 2002. New Zealand's meningococcal disease epidemic caused by two subclones of the hypervirulent ST-44 complex. In Thirteenth International Pathogenic Neisseria Conference. Nordberg Aksidenstrykkeri AS, Oslo, Norway.
- 4.Gorla, M. C., A. P. S. Lemos, C. T. Sacchi, J. Cássio de Moraes, and L. G. Milagres. 2003. Comparison of PorA VR types and porA promoter sequence from Neisseria meningitidis B isolated from non-immunised children and vaccine failures immunised with a serogroup B outer membrane protein vaccine. Vaccine 21:2871-2876. [DOI] [PubMed] [Google Scholar]
- 5.Holst, J., I. S. Aaberge, P. Oster, D. Lennon, D. Martin, J. O'Hallahan, K. Nord, H. Nøkelby, L. M. Næss, K. Møyner, P. Kristiansen, A. G. Skryten, K. Byrn, A. Aase, R. Rappuoli, and E. Rosenqvist. 24 July 2003, posting date. A ‘tailor-made’ vaccine trialled as part of public health response to group B meningococcal disease epidemic in New Zealand. Eurosurveillance Wkly. 7:5. [Online.] http://www.eurosurveillance.org/ew/2003/030724.asp.
- 6.Jódar, L., I. M. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359:1499-1508. [DOI] [PubMed] [Google Scholar]
- 7.Maiden, M. C. 1998. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin. Infect. Dis. 27:S12-S20. [DOI] [PubMed] [Google Scholar]
- 8.Maiden, M. C., J. A. Bygraves, J. McCarvil, and I. M. Feavers. 1992. Identification of meningococcal serosubtypes by polymerase chain reaction. J. Clin. Microbiol. 30:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiden, M. C., J. Suker, A. J. McKenna, J. A. Bygraves, and I. M. Feavers. 1991. Comparison of the class 1 outer membrane proteins of eight serological reference strains of Neisseria meningitidis. Mol. Microbiol. 5:727-736. [DOI] [PubMed] [Google Scholar]
- 10.Martin, D., R. McDowell, E. Sneyd, and M. Baker. 2004. The epidemiology of meningococcal disease in New Zealand in 2003. A report to the Ministry of Health of New Zealand. [Online.] http://www.moh.govt.nz.
- 11.Martin, D. R., S. J. Walker, M. G. Baker, and D. R. Lennon. 1998. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J. Infect. Dis. 177:497-500. [DOI] [PubMed] [Google Scholar]
- 12.McGuinness, B., A. K. Barlow, I. N. Clarke, J. E. Farley, A. Anilionis, J. T. Poolman, and J. E. Heckels. 1990. Deduced amino acid sequences of class 1 protein (PorA) from three strains of Neisseria meningitidis. Synthetic peptides define the epitopes responsible for serosubtype specificity. J. Exp. Med. 171:1871-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuinness, B. T., I. N. Clarke, P. R. Lambden, A. K. Barlow, J. T. Poolman, D. M. Jones, and J. E. Heckels. 1991. Point mutation in meningococcal porA gene associated with increased endemic disease. Lancet 337:514-517. [DOI] [PubMed] [Google Scholar]
- 14.McGuinness, B. T., P. R. Lambden, and J. E. Heckels. 1993. Class 1 outer membrane protein of Neisseria meningitidis: epitope analysis of the antigenic diversity between strains, implications for subtype definition and molecular epidemiology. Mol. Microbiol. 7:505-514. [DOI] [PubMed] [Google Scholar]
- 15.Moore, E., A. Arnscheidt, A. Krüger, C. Strömpl, and M. Mau. 1999. Simplified protocols for the preparation of genomic DNA from bacterial cultures, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 1.6.1. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 16.Rosenqvist, E., E. A. Høiby, E. Wedege, D. A. Caugant, L. O. Frøholm, B. T. McGuinness, J. Brooks, P. R. Lambden, and J. E. Heckels. 1993. A new variant of serosubtype P1.16 in Neisseria meningitidis from Norway, associated with increased resistance to bactericidal antibodies induced by a serogroup B outer membrane protein vaccine. Microb. Pathog. 15:197-205. [DOI] [PubMed] [Google Scholar]
- 17.Rouppe van der Voort, E., M. Schuller, J. Holst, P. de Vries, P. van der Ley, G. van den Dobbelsteen, and J. Poolman. 2000. Immunogenicity studies with a genetically engineered hexavalent PorA and a wild-type meningococcal group B outer membrane vesicle vaccine in infant cynomolgus monkeys. Vaccine 18:1334-1343. [DOI] [PubMed] [Google Scholar]
- 18.Rouppe van der Voort, E. M., B. Kuipers, H. F. Brugghe, L. M. A. van Unen, H. A. M. Timmermans, P. Hoogerhout, and J. T. Poolman. 1997. Epitope specificity of murine and human bactericidal antibodies against PorA P1.7,16 induced with experimental meningococcal group B vaccines. FEMS Immunol. Med. Microbiol. 17:139-148. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Saukkonen, K., H. Abdillahi, J. T. Poolman, and M. Leinonen. 1987. Protective efficacy of monoclonal antibodies to class 1 and class 3 outer membrane proteins of Neisseria meningitidis B:15:P1.16 in infant rat infection model: new prospects for vaccine development. Microb. Pathog. 3:261-267. [DOI] [PubMed] [Google Scholar]
- 21.Saunders, N. B., W. D. Zollinger, and V. B. Rao. 1993. A rapid and sensitive PCR strategy employed for amplification and sequencing of porA from a single colony-forming unit of Neisseria meningitidis. Gene 137:153-162. [DOI] [PubMed] [Google Scholar]
- 22.Sawaya, R., F. F. Arhin, F. Moreau, J. W. Coultin, and E. L. Mills. 1999. Mutational analysis of the promoter region of the porA gene of Neisseria meningitidis. Gene 233:49-57. [DOI] [PubMed] [Google Scholar]
- 23.Scholten, R. J., H. A. Bijlmer, J. T. Poolman, B. Kuipers, D. A. Caugant, L. Van Alphen, J. Dankert, and H. A. Valkenburg. 1993. Meningococcal disease in The Netherlands, 1958-1990: a steady increase in the incidence since 1982 partially caused by new serotypes and subtypes of Neisseria meningitidis. Clin. Infect. Dis. 16:237-246. [DOI] [PubMed] [Google Scholar]
- 24.Scholten, R. J., J. T. Poolman, H. A. Valkenburg, H. A. Bijlmer, J. Dankert, and D. A. Caugant. 1994. Phenotypic and genotypic changes in a new clone complex of Neisseria meningitidis causing disease in The Netherlands, 1958-1990. J. Infect. Dis. 169:673-676. [DOI] [PubMed] [Google Scholar]
- 25.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tappero, J. W., R. Lagos, A. M. Ballesteros, B. Plikaytis, D. Williams, J. Dykes, L. L. Gheesling, G. M. Carlone, E. A. Høiby, J. Holst, H. Nøkleby, E. Rosenqvist, G. Sierra, C. Campa, F. Sotolongo, J. Vega, J. Garcia, P. Herrera, J. T. Poolman, and B. A. Perkins. 1999. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA 281:1520-1527. [DOI] [PubMed] [Google Scholar]
- 27.van der Ende, A., C. T. P. Hopman, and J. Dankert. 2000. Multiple mechanisms of phase variation of PorA in Neisseria meningitidis. Infect. Immun. 68:6685-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Ende, A., C. T. Hopman, S. Zaat, B. B. Essink, B. Berkhout, and J. Dankert. 1995. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J. Bacteriol. 177:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Ley, P., J. E. Heckels, M. Virji, P. Hoogerhout, and J. T. Poolman. 1991. Topology of outer membrane porins in pathogenic Neisseria spp. Infect. Immun. 59:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Ley, P., J. van der Biezen, and J. T. Poolman. 1995. Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine 13:401-407. [DOI] [PubMed] [Google Scholar]
- 31.Van Looveren, M., D. A. Caugant, S. Chapelle, F. Carion, and H. Goossens. 2001. Interpreting the rising incidence of meningococcal disease in Belgium: the contribution of molecular typing. J. Med. Microbiol. 50:986-990. [DOI] [PubMed] [Google Scholar]
- 32.Van Looveren, M., P. Vandamme, M. Hauchecorne, M. Wijdooghe, F. Carion, D. A. Caugant, and H. Goossens. 1998. Molecular epidemiology of recent Belgian isolates of Neisseria meningitidis serogroup B. J. Clin. Microbiol. 36:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermont, C. L., H. H. van Dijken, A. J. Kuipers, C. P. J. van Limpt, W. C. M. Keijzers, A. van der Ende, R. de Groot, L. van Alphen, and G. P. J. M. van den Dobbelsteen. 2003. Cross-reactivity of antibodies against PorA after vaccination with a meningococcal B outer membrane vesicle vaccine. Infect. Immun. 71:1650-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wedege, E. 2001. Immunoblot analysis of sera from patients and vaccinees, p. 275-288. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal vaccines: methods and protocols, vol. 66. Humana Press, Totowa, N. J. [DOI] [PubMed] [Google Scholar]
- 35.Wedege, E., R. Dalseg, D. A. Caugant, J. T. Poolman, and L. O. Frøholm. 1993. Expression of an inaccessible P1.7 subtype epitope on meningococcal class 1 proteins. J. Med. Microbiol. 38:23-28. [DOI] [PubMed] [Google Scholar]
- 36.Wedege, E., B. Kuipers, K. Bolstad, H. van Dijken, L. O. Frøholm, C. Vermont, D. A. Caugant, and G. van den Dobbelsteen. 2003. Antibody specificities and effect of meningococcal carriage in Icelandic teenagers receiving the Norwegian serogroup B outer membrane vesicle vaccine. Infect. Immun. 71:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]