Abstract
More than 35 million people remain infected with HIV-1. Upon antiretroviral therapy cessation, HIV-1-positive individuals systematically fail to achieve sustained virological remission, revealing the presence of a reservoir. This reservoir takes into account anatomical sanctuaries where HIV-1 continues to replicate, and latently infected cells also known as the latent reservoir (LR). A better understanding of the nature and features of the LR and its quantification are crucial to evaluate the efficiency of therapeutic strategies aiming at purging HIV-1. Culture- and PCR-based assays have already been implemented to measure the LR, and new assays are continuously being developed. In this review, we will discuss these methods highlighting the difficulties to accurately measure the LR, one main obstacle in curing HIV-1.
KEYWORDS : assays, DNA, HIV-1, latent reservoir, quantification, RNA, size
30 years ago, it was unthinkable for HIV-1-positive individuals to have a life expectancy above a decade [1]. With the development of antiretroviral therapy (ART), the mortality and morbidity associated with HIV-1 infection have been successfully reduced although >35 million people remain infected with HIV-1 [2,3]. ART effectively reduces plasma viral loads to undetectable levels by targeting specific replication steps of HIV-1 thus allowing treated patients to live longer despite a lifelong treatment, high costs and severe adverse effects [4]. Indeed, upon ART interruption viremia rebounds typically within a few weeks suggesting the presence and release of virus from a stable reservoir [2]. This reservoir is established early during infection, providing the basis for treating HIV-1-positive individuals as soon as possible during acute infection [5,6]. To date, two main sources, which are not mutually exclusive, are thought to contribute to the viral reservoir: the persistence of productive infected cells in lymphoid tissues and anatomical sites that are inaccessible to ART; and the persistence of latently infected cells that are able to produce viral particles upon stimulation also known as the latent reservoir (LR), and which is the main focus of this review [7–9]. The existence of this HIV-1 reservoir is considered to be the major handicap to HIV-1 eradication and an increasing number of studies focus on finding new strategies to eliminate it [2,10–11].
HIV-1 primarily infects activated CD4+ T cells and macrophages. Macrophages can contribute to HIV-1 persistence mainly due to their presence in different anatomical sanctuaries such as the CNS and the gut [3,12]. Tissue-resident macrophages can live up to several months and can thus contribute to the maintenance of viral production, but their impact on the HIV-1 reservoir is still poorly understood [3]. Additional cell populations may affect the reservoir but their exact role remains to be elucidated [4]. During HIV-1 infection, the majority of activated CD4+ T cells usually die shortly after infection because of virus-mediated cytotoxicity or immune-mediated killing. However, some of these cells will return to a resting memory state and do not release viral particles, giving rise to latently infected cells [2,3]. As they do not express viral proteins, these latently infected cells are no longer targeted by the immune system, and ART is not effective. These resting memory CD4+ T cells have been identified as the main cellular reservoir harboring latent provirus [2–3,7,9]. The persistence of this LR is directly linked to key features of both the retrovirus and the resting memory of CD4+ T cells, which are the stable viral genome insertion in the host genome and the long half-life of infected cells [4,11,13].
Understanding the LR is key to design new approaches aiming at curing HIV-1. Cells harboring latent HIV-1 represent only 10–100 in 106 of resting CD4+ T cells; however, reactivation of these cells in HIV-1-positive individuals off ART is sufficient to viral rebound [11,13–14]. Some of the most attractive strategies, such as the ‘shock and kill’, aim at reactivating the latently infected CD4+ T cells using latency-reversing agents (LRA) to induce expression of viral proteins, which triggers the elimination of the infected cells by the immune system or by virus-induced cytotoxicity [2,3]. Although attractive, no LRA alone seemed potent enough to induce expression from all latently infected cells, suggesting that, as for drug treatment, combination of multiple LRA might represent an alternative strategy [14]. Thus, a better characterization of the LR and the multiple mechanisms leading to latency is required to better understand the ins and outs of the inducible LR.
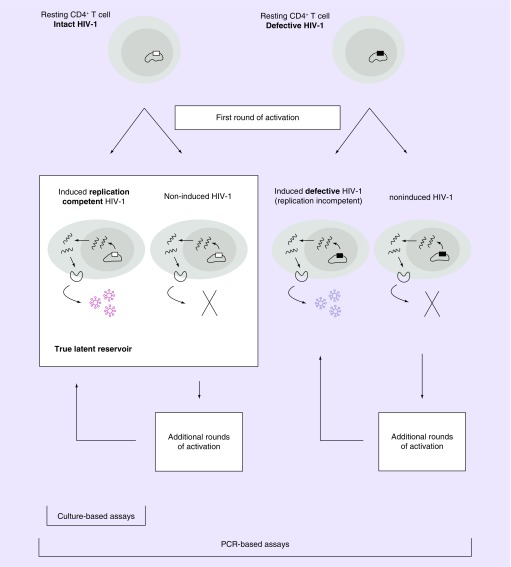
The accurate quantification of the LR in HIV-1-infected individuals is essential for evaluating HIV-1 eradication strategies and for deciding on whether and when ART interruption is legitimate [11]. Quantifying the true size of the LR is extremely challenging due to characteristics of both the viral genome and the latently infected cell. Indeed, as illustrated in Figure 1, many integrated proviral sequences are hypermutated, and thus defective and unable to produce replication-competent viral particles. A study by Ho et al. showed that about 88% of cells carrying HIV-1 DNA contained a large deletion rendering them defective [11]. Thus, only about 12% of infected cells carry a replication-competent viral genome and would represent the bona fide LR. Furthermore, in vitro, only a fraction of these cells was effectively inducible using one round of CD4+ T-cell stimulation [11,13]. Thus, these two features, replication-competent versus defective virus, and inducible versus noninducible virus, represent significant challenges for the development of reliable assays measuring the LR. The current review aims at providing an overview of assays currently used to evaluate the size of the ‘true’ LR as well as discussing their clinical relevance.
Figure 1. . Characterization of the latent reservoir.
Two major types of resting CD4+ T cells are shown, containing either intact or defective HIV-1. Defective HIV-1 contains large internal deletions, G→A hypermutations or other defects. In both cases, following a single round of T-cell activation, proviruses are either induced to produce virions that can infect other cells (induced, replication-competent HIV-1) or to produce defective virions unable to infect other cells (induced, replication noncompetent HIV-1). Proviruses that are not induced to produce virions following a single round of T-cell activation are termed noninduced HIV-1. The noninduced HIV-1 can still be induced after additional rounds of activation. On subsequent rounds of cellular activation, some noninduced proviruses can produce either competent or noncompetent virions. Culture-based assays detect only induced replication-competent HIV-1, while PCR-based assays detect all types of proviruses. The true latent reservoir includes only induced replication-competent HIV-1 and noninduced intact HIV-1.
Culture-based assays to measure the latent reservoir
Culture-based assays (Figure 2) aim at measuring the LR by assessing the number of cells that carry replication-competent viruses. These assays use pharmacological or immunological agents to reactivate the LR and induce replication-competent HIV-1 in resting CD4+ T cells. Additional CD4+ T cells are added to the primary cell culture to further promote HIV-1 replication and thereby allow HIV-1 amplification. These assays measure the frequency of cells producing viral particles, able to replicate and propagate and offer great potential to measure the LR. However, it is essential to note that these assays rely on the ability of cells to be stimulated and thus to induce the LR. To date, T-cell receptor-mediated stimulation is considered to be the most powerful stimuli to activate CD4+ T cells; however, they are not able to induce viral expression from all latently infected cells [11].
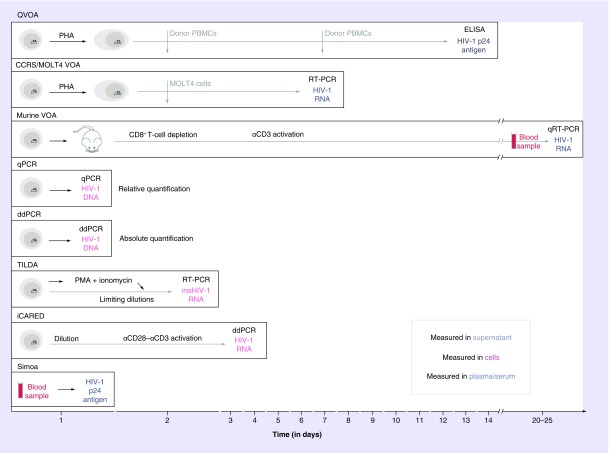
Figure 2. . Measurement of the latent reservoir by seven different assays.
From top to bottom: Culture-based assays, PCR-based assays and novel assays. For each assay, the methodology used to measure the latent reservoir is indicated proportionally to the time needed to obtain the results. Culture-based assays are very time-consuming compared with PCR-based assays, but neither offers an accurate estimation of the latent reservoir. Novel assays approach the ‘true’ size and are less time-consuming compared with culture-based assays.
PBMC: Peripheral blood mononuclear cell; PHA: Phytohemagglutinin; PMA: Phorbol myristate acetate; qRT: Quantitative real-time reverse transcription PCR.
• Quantitative viral outgrowth assay: ‘the gold standard’
Quantitative viral outgrowth assay (QVOA) was the first assay developed that permitted identification and measurement of the LR, and is currently considered as the gold standard [2,11].
QVOA measures the frequency of resting CD4+ T cells able to produce replication competent and therefore infectious virus upon stimulation. Resting CD4+ T cells are isolated from peripheral blood mononuclear cells (PBMCs) by density gradient centrifugation and further purified by negative selection. These cells are considered resting since they do not express classical activation markers such as CD69, CD25 or HLA-DR and in most cases do not produce virus without stimulation [2,13]. Fivefold dilutions are performed to distribute the cells into wells, such that the input number of patient cells ranges from 1,000,000 to 320 cells per well. To reverse latency, T-cell activation is carried out using mitogen phytohemagglutinin (PHA) and tenfold excess of γ-irradiated PBMCs from healthy donors. After 1 day of stimulation, CD4+ lymphoblasts from healthy donors are added at two different time points to allow HIV-1 replication and propagation. On day 14, supernatant of each well is analyzed by HIV-1 p24 antigen ELISA to identify wells positive for viral outgrowth. Based on this, the frequency of cells releasing virus can be determined using maximum likelihood method. This assay first indicated an average of one latently infected cell (able to produce replication-competent virus) in 1 million resting CD4+ T cells, thereby defining the first estimate of the LR size.
Although HIV-1 induction by PHA activation was thought to be uniform and maximal, studies by Ho et al. [11] followed by Cillo et al. [15] showed that one single round of activation was not sufficient to reactivate all intact proviruses. Indeed, 98.5% of the viral reservoir was not induced upon activation, which indicates that QVOA largely underestimates the size of the LR. Although the majority of these noninduced proviruses after one round of stimulation are defective, it has been shown that a significant proportion (11.7%) of noninduced proviruses has intact genomes and is potentially replication competent [13,15]. Additional rounds of stimulation could induce expression of infectious virus from additional latently infected cells, suggesting that the true LR size might rather be around 60–70 latently infected cells per million of resting CD4+ T cells. Thus, QVOA can only give a definitive minimal estimate of the frequency of latently infected CD4+ T cells and relies on the ability of compounds or stimuli to efficiently induce viral expression from the LR [2,13,15].
In addition to underestimating the LR size, QVOA presents multiple drawbacks. It is time-consuming, labor-intensive and expensive since it requires multiple weeks of cell culture with multiple media changes in a Biosafety Level 3 (BSL3) laboratory, and is therefore not suitable for large-scale studies. This assay also has a limited dynamic range and requires large sample volumes (120–180 ml) from HIV-1-positive individuals and healthy blood donors. Despite these limitations, it is considered as a gold standard since positive viral outgrowth is a direct proof of the persistence of the LR and thereby calls for caution for ART interruption. Attempts to improve this assay are still being pursued.
• MOLT4/CCR5 viral outgrowth assay: a slight improvement of the gold standard
The limitations and drawbacks of QVOA are called for improvements and development of new assays to measure the LR. MOLT4/CCR5 viral outgrowth assay (VOA) allows a faster measurement of the number of latently infected CD4+ T cells [13]. To avoid the need of high amounts of donor cells, a MOLT4/CCR5 T-cell line is used to propagate the virus. These cells express both CXCR4 and CCR5 co-receptors, and are highly permissive to HIV-1 replication [2,13]. In addition, these MOLT4/CCR5 cells are added only once after activation. To replace the ELISA used in QVOA, a more sensitive reverse transcription (RT)-PCR assay is used only after 7 days to quantify viral RNA copies in the supernatant of each well, thereby reducing the time needed to perform the assay. The frequency of latent cells measured by MOLT4/CCR5 VOA correlates with QVOA and no significant difference was observed between the two methods [2,13].
MOLT4/CCR5 VOA offers great potential according to Laird et al. to replace the standard QVOA method, since it is more rapid and sensitive, cost-effective, less cell-consuming, less labor-intensive and therefore more suitable for large-scale clinical trials [13]. However, it is still quite demanding and, as for QVOA, the true size of the LR is likely to be underestimated [13].
• Murine VOA: the first in vivo measurement of the LR
To avoid the underestimation of the reservoir, Pate et al. proposed a novel VOA assay [16]. Instead of cell culture, it uses an immunocompromised murine model (NOD.Cg-PrkcscidIl2rgtm1Wjl/SzJ (NSG), Jackson Laboratories, Bar Harbor, ME, USA) in which purified PBMCs or resting CD4+ T cells from an HIV-1-positive individual are injected intraperitoneally to transfer the LR into the mouse. CD8+ T cells are then depleted using anti-human CD8 antibody to minimize the potential antiviral response and allow maximal viral amplification. T cells are then activated with anti-human CD3 antibody to induce viral replication from patient-derived latently infected cells. To quantify viral amplification in mice blood samples, HIV-1 RNA plasma levels are measured by quantitative RT-PCR. Detection of replication-competent HIV-1 occurred on average at day 20 with a median peak viral load of 4.6 × 103 copies/ml at an average of 25 days [16].
Pate et al. established a new in vivo model to detect residual virus, allowing prediction of viral rebound without endangering the patient [16]. The assay is relatively simple and although it is not quantitative, it is more sensitive than QVOA. However, murine VOA has some limitations. The main drawbacks are the requirements of a BSL3 animal facility, animal handling and specialized technicians, which may not always be applicable for clinics. In addition, the sensitivity of the method is highly related to the quality of the xenograft, and the time needed for viral detection varies between patients. The current model only measures the reservoir in the peripheral blood, but the authors estimate that it will be applicable to other tissues. The study was conducted on a limited number of patients and further investigation is required to determine whether murine VOA is really more sensitive than QVOA and whether this method will be suited for quantitation of the reservoir and for its future implementation in clinics.
PCR-based assays to measure the LR
PCR-based assays (Figure 2) are commonly used to quantify total HIV-1 provirus, as they usually require minimal amounts of specialized equipment. Furthermore, PCR can detect very small amounts of DNA/RNA, which is crucial due to the low copy numbers of HIV-1 in long-term ART-treated patients.
• Quantitative PCR: the silver standard
Nowadays, PCR-based assays are widely used to measure the LR size, quantitative PCR (qPCR) for proviral DNA being the most common one [2]. CD4+ T cells are first purified from peripheral blood. Afterward, DNA is extracted and amplified by qPCR using HIV-1-specific primers as well as a fluorescent probe. Primers and probe for an endogenous housekeeping gene are used as well to assess the equivalent number of cell copy. qPCR allows quantification of HIV-1-specific PCR products by following cycle by cycle the appearance and the accumulation of fluorescence released during amplification, which is directly proportional to the amount of amplified HIV-1 DNA [18,19]. The assay gives, therefore, a relative quantification of HIV-1 DNA from a standard curve, and estimates the amount of HIV-1 DNA to be at an average of 1000 copies per million cells. This frequency is at least two to three logs higher than measured by QVOA [2,20]. In fact, infected cells carry both intact and defective HIV-1 sequences, making this assay overestimating the LR size. Despite this overestimation, qPCR is less labor-intensive and faster than QVOA. Moreover, it does not require any extended culture time in BSL3 laboratory [2]. However, PCR efficiency can be biased due to viral sequence variability caused by mutations affecting primer binding, and to variable abundance of viral DNA between patients [19]. In summary, qPCR has been shown to be an effective surrogate to follow LR; and as HIV-1 DNA measured by qPCR is correlated to integrated HIV-1 DNA and to viral rebound in patients on suppressive ART, it can be used as a marker in cure research and patient monitoring to predict remission of HIV-1 [21,22]. Therefore, although DNA measurements of HIV-1 are likely to overestimate the LR, they are currently the best way to safely monitor the reservoir in clinical settings.
• Droplet digital PCR or the digital revolution
Droplet digital PCR (ddPCR) is an emerging technology enabling high precision and reproducibility at low DNA copy number [2,18–19,23]. ddPCR was initially developed to reduce noise observed in qPCR and to better detect low amounts of HIV-1 nucleic acids in patient samples. Following DNA extraction, ddPCR systems separate the sample into compartments, or so-called drops, by emulsification of the aqueous PCR reaction mixture in thermostable oil. Thus, only few molecules are present in each PCR partition so that rare targeted viral DNA will stand out after PCR amplification reactions. Poisson statistics are used to predict how much starting material was present before amplification [24]:
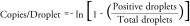 |
Unlike qPCR, ddPCR is a highly precise and sensitive method that allows absolute quantification of HIV-1 DNA. ddPCR can be achieved in 1 day and is well appropriate to measure the LR size. By increasing the number of replicates we can decrease the LOD. However, the rate of false positives, whose source is still unclear, remains a real challenge in measuring the LR with ddPCR; and like qPCR, it overestimates the LR size [23]. Only few laboratories use ddPCR since the instrumentation is expensive and requires specific expertise. Nonetheless, by performing simultaneously multiple qPCR reactions, ddPCR is an elegant adaptation of the current qPCR assay [2,19].
The next-generation assays
We have described above two main categories of assays measuring LR that both face major drawbacks: culture-based assays underestimate the size of the reservoir while PCR-based assays tend to overestimate it. The ‘true’ size is defined to be in-between, and new methods are needed to approach reality. Recently, three assays have been proposed to offer an alternative to estimate the LR: the tat/rev-induced limiting dilution assay (TILDA), the inducible cell-associated RNA expression in dilution (iCARED) assay and single molecule arrays (Simoa™, Quanterix Corporation, Lexington, MA, USA) (Figure 2).
• Should TILDA, iCARED & Simoa be considered as alternatives to the gold standard?
TILDA measures the frequency of total CD4+ T cells that produce viral tat/rev HIV-1 msRNA upon maximal cell activation [25]. The production of these cell-associated viral transcripts is used as a marker of the cell's ability to produce replication-competent virus [11,25]. Isolated total CD4+ T cells are stimulated for 12 h with phorbol myristate acetate/Ionomycin, a presumably stronger activator than PHA [25]. Cells, serially diluted 1:3 from 18,000 to 1000 cells, and in 22–24 replicates, are then directly added to the one-step RT-PCR mix to perform the reverse transcription reaction and amplify tat/rev sequence. A fraction of the first PCR is then used to perform a nested qPCR with HIV-1-specific primers and probe to quantify tat/rev transcripts. To calculate the frequency of cells containing inducible HIV-1, a maximum likelihood method that takes into account the number of cell input as well as the number of positive detections is used. This assay was used to measure the LR size of ART-suppressed individuals and estimated a median of 24 out of 1 million latently infected CD4+ T cells producing replication-competent virus upon stimulation. By comparison, the frequency of latently infected cells measured by TILDA was 48-times larger than QVOA, and 6–27 lower than by qPCR on total viral DNA.
TILDA is highly attractive since it requires only 10 ml of blood, does not rely on cell culture nor on RNA extraction, and the result is obtained within 2 days. In addition, this first study shows that TILDA is highly sensitive, reproducible, has a wide dynamic range and is suitable for clinical trials. However, the authors report a potential limitation since, even though all cells that do produce virus give a positive signal, some cells might produce tat/rev even without viral production. The resulting overestimation of the reservoir seems lower than DNA-based qPCR assays, and the frequency of latently infected cells is closer to the prediction by Ho et al. [11]. Further studies are needed to validate TILDA to all quasispecies, as tat/rev region is variable and may introduce biases in viral detection. However, TILDA appears as an appealing alternative to VOA- and PCR-based assays.
iCARED is also a promising method, based on measurement of viral RNA [17]. Similarly to QVOA, CD4+ T cells are purified and used in threefold serial dilutions from 1,500,000 to 686 cells in six to eight replicates [17]. Cells are activated to induce viral particle production using anti-CD3/anti-CD28 monoclonal antibodies that mimic antigen presentation, and in presence of the HIV-1 integrase inhibitor raltegravir to block viral propagation. After 3-day culture, RNA (from both virion-containing supernatant and cells) is extracted using magnetic beads to enrich for HIV-1-specific RNA. Primers and probe targeting either the gag region or the tat/rev region were used to quantify cell-free and/or cell-associated viral RNA by RT-qPCR using ddPCR. The size of HIV-1 LR as measured by iCARED was estimated to be 51 latently infected cells per million of CD4+ T cells, which is about 7–13-times more than by QVOA and 19-times lower than total viral DNA by ddPCR. iCARED correlated well with QVOA at day 7 and with measurements of total viral DNA [17]. iCARED is easy to perform, fast and highly sensitive. Moreover, it is less cell demanding and less expensive than QVOA, and allows testing large volumes of cells and culture supernatants [17]. Thus, iCARED appears as a simple and innovative method to quantify the HIV-1 LR, which can be useful for clinical investigations.
In addition to nucleic acids, proteins are also considered as reliable biomarkers for disease progression. However, relevant biomarkers are often present in very low abundance and conventional immunoassays lack in sensitivity [26]. Similarly to TILDA and iCARED, Simoa digital ELISA from Quanterix Corporation represents a novel promising assay for ultrasensitive detection of proteins at femtogram per milliliter levels [26–28]. This represents a >1:200-fold sensitivity improvement compared with classical ELISA [26]. Simoa technology uses standard paramagnetic beads that are coupled with antibodies designed to bind the protein of interest in the sample. At low-protein abundance, each bead will only bind to one target protein or none. A biotin-labeled detection antibody is added into the reaction with its substrate streptavidin-β-galactosidase. The beads are then loaded onto microarrays that consists of >200,000 femtoliter-sized wells. A single target molecule generates enough fluorescence to be detected by a camera. The number of positive wells will allow calculating protein quantification in the whole sample.
The Simoa platform is highly sensitive, allows fast single-molecule diagnostics and requires only 100 μl reaction volume [30]. Additionally, the assay is fully automated and is already implemented in different fields such as inflammation, oncology and neurology [28–30]. Although it has not yet been implemented in HIV-1 diagnostics, Simoa offers great potential in quantifying HIV-1 p24 capsid protein. In a study by Wilson et al., 24 blood samples from individuals with early HIV-1 infection were assayed for p24 antigen detection [29]. Although most samples were negative by conventional p24 immunoassay, they were all detected using Simoa assay, demonstrating its superior sensitivity. The LOD was estimated to be 2.5 fg/ml, which corresponds approximately to 60 RNA copies/ml. The assay was as sensitive as PCR for detecting HIV-1 in early infection and may thus offer a new opportunity for HIV-1 screening during acute infection and therapy follow-up. Although this assay needs some specialized equipment, it is simple, low cost and holds great potential for measuring the true size of the LR.
Conclusion & future perspective
Herein we reviewed the complexity of measuring the HIV-1 LR, whose exact nature and size is still not precisely determined. Currently, LR lies somewhere between 1 and 1000 latently infected cells per million CD4+ T cells, and estimated to be more likely around 10–100. Current methods fail to measure accurately the LR, even if TILDA, iCARED and Simoa are probably closer to reality. Additional effort is required to develop low-cost assays that will capture only intact and thus replication-competent viral sequences with high accuracy, sensitivity and specificity. Moreover, these assays should be optimized for routine use in clinical settings.
Nowadays, a proper understanding of the in vivo correlation between LR size and time to rebound or cure remains crucial. It is becoming increasingly urgent to identify reliable biomarkers able to quantify the in vivo reservoir over time and predict time to viral rebound. Recent promising studies by Kiselinova et al. [31] and Williams et al. [32] suggest that the pool of HIV-1 DNA (total or integrated) reflects the size of the replication-competent virus in patients receiving ART and also better predict disease progression and time for viremia to rebound upon treatment interruption than plasma viral load. Hence, PCR-based assays quantifying HIV-1 DNA likely represent the best surrogate marker to date to evaluate the size of the LR and thus to assess the efficiency of cure strategies. This would be key to identify HIV-1 infected individuals able to safely interrupt ART in future clinical HIV-1 eradication trials. The clinical benefit of novel assays such as iCARED, TILDA and Simoa has not yet been investigated in vivo but may contribute to the determination of LR size in the future.
Finally, although the simplest way to measure the LR remains blood sampling, this does not reflect anatomical sites harboring stably integrated proviruses [2,15]. It remains thus essential in the coming years to adequately define if all potential reservoirs must be analyzed and quantified, and how to be confident that these reservoirs have reached a threshold sufficient for remission or cure. Certainly, size matters.
EXECUTIVE SUMMARY.
What is the HIV-1 reservoir?
Two main sources are thought to contribute to the viral reservoir: the persistence of productive infected cells in lymphoid tissues and anatomical sites that are inaccessible to antiretroviral therapy (ART); and the persistence of latently infected cells that are able to produce viral particles upon stimulation.
The reservoir is established early during infection and remains difficult to apprehend.
Current methods only quantify the latent reservoir (LR).
Culture-based assays to measure the LR
Culture-based assays aim at measuring the LR by assessing the number of cells that carry replication-competent viruses.
Quantitative viral outgrowth assay (QVOA) is the current gold standard to measure the LR.
Improvements of QVOA include MOLT4/CCR5 viral outgrowth assay and murine viral outgrowth assay.
Culture-based assays tend to underestimate the bona fide size of the LR as they are depending on the ex vivo ability of cells to be stimulated and produce infectious viral particles. Therefore, these measurements may lead to premature discontinuation of ART, where patients may experience viremia rebounds.
PCR-based assays to measure the LR
PCR-based assays are commonly used methods that detect all intracellular proviruses regardless of their ability to be replication competent or not, and overestimating consequently the total LR.
Quantitative PCR (qPCR) is used to assess HIV-1 proviruses. HIV-1 DNA measured by qPCR is correlated to integrated HIV-1 DNA and to viral rebound in patients on suppressive ART.
Improvements of qPCR include droplet digital PCR.
PCR-based assays tend to overestimate the bona fide size of the LR as they quantify both competent and defective viral sequences. Therefore, these measurements may represent the current best option to help deciding the legitimacy of a safe ART cessation.
The next-generation assays
Next-generation assays, such as tat/rev-induced limiting dilution assay, inducible cell-associated RNA expression in dilution and single molecule arrays, offer an alternative to better estimate the LR, based on viral RNA or protein.
These assays have not yet been investigated in vivo but may contribute to the determination of LR size in the future.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Open Access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.AIDS epidemic update: December 2007. UNAIDS, Geneva. http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf
- 2.Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 2015;23(4):192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar A, Darcis G, Van Lint C, Herbein G. Epigenetic control of HIV-1 post integration latency: implications for therapy. Clin. Epigenetics. 2015;7:103. doi: 10.1186/s13148-015-0137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melkova Z, Shankaran P, Madlenakova M, Bodor J. Current views on HIV-1 latency, persistence, and cure. Folia Microbiol. 2016 doi: 10.1007/s12223-016-0474-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]; •• Describes mechanisms leading to HIV-1 latency.
- 5.Rothenberger MK, Keele BF, Wietgrefe SW, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc. Natl Acad. Sci. USA. 2015;112(10):e1126–e1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitney JB, Hill AL, Sanisetty S, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512(7512):74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates the early establishment of the viral reservoir.
- 7.Ciuffi A, Mohammadi P, Golumbeanu M, di Iulio J, Telenti A. Bioinformatics and HIV latency. Curr. HIV/AIDS Rep. 2015;12(1):97–106. doi: 10.1007/s11904-014-0240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzo-Redondo R, Fryer HR, Bedford T, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530(7588):51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates the persistence of productively infected cells in tissues.
- 9.Fletcher CV, Staskus K, Wietgrefe SW, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl Acad. Sci. USA. 2014;111(6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates the persistence of productively infected cells in tissues.
- 10.Chun T-W, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat. Immunol. 2015;16(6):584–589. doi: 10.1038/ni.3152. [DOI] [PubMed] [Google Scholar]
- 11.Ho Y-C, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sattentau QJ, Stevenson M. Macrophages and HIV-1: an unhealthy constellation. Cell Host Microbe. 2016;19(3):304–310. doi: 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird GM, Eisele EE, Rabi SA, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimata JT, Rice AP, Wang J. Challenges and strategies for the eradication of the HIV reservoir. Curr. Opin. Immunol. 2016;42:65–70. doi: 10.1016/j.coi.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Recent updated review covering the challenges for HIV reservoir eradication.
- 15.Cillo AR, Sobolewski MD, Bosch RJ, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc. Natl Acad. Sci. USA. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf Pate KA, Pohlmeyer CW, Walker-Sperling VE, et al. A murine viral outgrowth assay to detect residual HIV type 1 in patients with undetectable viral loads. J. Infect. Dis. 2015;212(9):1387–1396. doi: 10.1093/infdis/jiv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massanella M, Yek C, Lada SM, Strain MC, Richman DD. Improvedassays to measure the inducible latent HIV reservoir. Towards an HIV CureSymposium Abstract PE18; Vancouver, Canada: July 2015. [Google Scholar]
- 18.Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS ONE. 2014;9(1):e85999. doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosman KJ, Nijhuis M, van Ham PM, et al. Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir. Sci. Rep. 2015;5:13811. doi: 10.1038/srep13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson S, Graf EH, Dahl V, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avettand-Fènoël V, Hocqueloux L, Ghosn J, et al. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin. Microbiol. Rev. 2016;29(4):859–880. doi: 10.1128/CMR.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Covers the use of HIV-1 DNA PCR as a valuable surrogate marker for measuring the size of reservoir.
- 22.Mexas AM, Graf EH, Pace MJ, et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26(18):2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strain MC, Lada SM, Luong T, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE. 2013;8(4):e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Droplet Digital PCR Applications Guide; Bio-Rad Laboratories, Inc. Bulletin officiel Bio-Rad 6407 Rev A. www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf
- 25.Procopio FA, Fromentin R, Kulpa DA, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2(8):874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson DH, Rissin DM, Kan CW, et al. The Simoa HD-1 analyzer: a novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J. Lab. Autom. 2016;21(4):533–547. doi: 10.1177/2211068215589580. [DOI] [PubMed] [Google Scholar]
- 27.Fischer SK, Joyce A, Spengler M, et al. Emerging technologies to increase ligand binding assay sensitivity. AAPS J. 2015;17(1):93–101. doi: 10.1208/s12248-014-9682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung D, Ciotti S, Purushothama S, et al. Evaluation of highly sensitive immunoassay technologies for quantitative measurements of sub-pg/ml levels of cytokines in human serum. J. Immunol. Methods. 2016;437:53–63. doi: 10.1016/j.jim.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Wilson DH, Hanlon DW, Provuncher GK, et al. Fifth-generation digital immunoassay for prostate-specific antigen by single molecule array technology. Clin. Chem. 2011;57(12):1712–1721. doi: 10.1373/clinchem.2011.169540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogoslovsky T, Wilson D, Chen Y, et al. Increases of plasma levels of glial fibrillary acidic protein, tau, and amyloid β up to 90 days after traumatic brain injury. J. Neurotrauma. 2016 doi: 10.1089/neu.2015.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. Integrated and total HIV-1 DNA predict ex vivo viral outgrowth. PLoS Pathog. 2016;12(3):e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams JP, Hurst J, Stöhr W, et al. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife. 2014;3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]