Abstract
Dental caries is a costly and prevalent disease characterized by the demineralization of the tooth’s enamel. Disease outcome is influenced by host factors, dietary intake, cariogenic bacteria, and other microbes. The cariogenic bacterial species Streptococcus mutans metabolizes sucrose to initiate biofilm formation on the tooth surface and consequently produces lactic acid to degrade the tooth’s enamel. Persistence of S. mutans biofilms in the oral cavity can lead to tooth decay. To date, no anticaries therapies that specifically target S. mutans biofilms but do not disturb the overall oral microbiome are available. We screened a library of 2-aminoimidazole antibiofilm compounds with a biofilm dispersion assay and identified a small molecule that specifically targets S. mutans biofilms. At 5 µM, the small molecule annotated 3F1 dispersed 50% of the established S. mutans biofilm but did not disperse biofilms formed by the commensal species Streptococcus sanguinis or Streptococcus gordonii. 3F1 dispersed S. mutans biofilms independently of biofilm-related factors such as antigen I/II and glucosyltransferases. 3F1 treatment effectively prevented dental caries by controlling S. mutans in a rat caries model without perturbing the oral microbiota. Our study demonstrates that selective targeting of S. mutans biofilms by 3F1 was able to effectively reduce dental caries in vivo without affecting the overall oral microbiota shaped by the intake of dietary sugars, suggesting that the pathogenic biofilm-specific treatment is a viable strategy for disease prevention.
Keywords: microbiota, rats, dental enamel, microscopy, bacteria, microbial ecology
Introduction
Dental caries is a detrimental disease characterized by the demineralization of the enamel tooth surface. Disease results from a susceptible tooth surface, frequent sucrose intake, poor dental hygiene, and the persistence of cariogenic bacteria. The cariogenic species Streptococcus mutans metabolizes dietary sucrose into glucans via glucosyltransferases (Gtfs). As a by-product of sucrose metabolism, S. mutans produces lactic acid, which leads to demineralization of the tooth’s enamel. Due to the importance of sucrose in both S. mutans persistence and disease progression, it is no surprise that the prevalence of S. mutans has been highly correlated with a sugary diet. The prevalence of S. mutans in plaque from ancient humans coincided with the introduction of processed sugar (Liu et al. 2011; Adler et al. 2013). However, selective targeting of S. mutans biofilms for disease prevention as well as the interactions between S. mutans biofilms and dietary sucrose on the oral microbiome has not been reported.
Extracts or derivatives of various natural products such as propolis and cranberries have been characterized and shown to prevent dental caries, although none have exhibited notable selectivity toward S. mutans (Koo et al. 2010; Falsetta et al. 2012). An early study designed an antimicrobial peptide C16G2 that selectively targeted S. mutans; however, C16G2 kills S. mutans and is not specific for biofilms, a key virulence factor of S. mutans (Eckert et al. 2006). In this study, we have identified a small molecule (denoted 3F1) that has a narrow spectrum activity against S. mutans biofilms. We used this small molecule as a chemical probe to investigate if it is efficient in preventing dental caries and preserving the oral microbiome in vivo. Through these studies, we have determined that 3F1 is capable of potently and selectively dispersing established S. mutans biofilms in vitro. Furthermore, we have demonstrated that 3F1 reduced dental caries in vivo without perturbing the overall composition of the oral microbiome. Moreover, probing with 3F1 revealed that daily dietary sucrose intake determined the microbiome community. Overall, we demonstrate that selective targeting of S. mutans biofilms is feasible and can effectively prevent dental caries without disturbing the overall oral microbiome, offering a new anticaries strategy.
Materials and Methods
Strains
S. mutans UA159 was used as the model organism. S. mutans ΔgtfB and ΔgtfC mutant strains were kindly provided by Dr. Robert Burne, and S. mutans ΔagI/II was kindly provided by Dr. Jeannine Brady (University of Florida, Gainesville, FL). All S. mutans strains, Streptococcus gordonii Challis, and Streptococcus sanguinis SK36 were grown in Todd-Hewitt broth (THB; BD Biosciences) at 37°C with 5% CO2. Colony-forming units (CFUs) were enumerated after plating 10-fold dilutions and analyzed by analysis of variance (ANOVA).
Biofilm Dispersion Assay
Single colonies of S. mutans were grown in THB overnight and diluted into fresh THB and grown to an OD470 of 0.5 to 0.6. Bacterial cultures were diluted into biofilm media (BM), a chemically defined media containing 1% sucrose (Liu et al. 2011). BM inoculated with bacteria was plated into 96-well polystyrene plates and sealed. Biofilms were formed statically at 37°C with 5% CO2 for 6 h. Biofilms were washed with phosphate-buffered saline (PBS) to remove planktonic cells. Small molecules from a 10-mM stock in dimethyl sulfoxide (DMSO) were diluted in BM, added to the biofilms, and incubated for 14 h. For comparison, identical volumes of DMSO were added to untreated wells. Briefly, biofilms were stained with crystal violet and biomass measured at OD562 (Garcia et al. 2016). Differences in biomass were analyzed by Student’s t test and ANOVA.
Imaging Biofilms with Confocal Laser Scanning Microscopy
In total, 500 nM Cascade Blue–dextran conjugated dye (Molecular Probes) was added to BM prior to inoculation for visualization of the glucan matrix. Biofilms were grown on 8-well ibiTreat slides (ibidi, 80826) (Peng, Michalek, et al. 2016). Cells and extracellular DNA (eDNA) were stained with 1 µM Syto-9 (Molecular Probes) and 0.67 µM propidium iodide (Sigma-Aldrich), respectively. All images were taken on a Zeiss confocal microscope with a 63× oil immersion lens (University of Alabama [UAB] High Resolution Imaging Facility).
Rat Caries Model
The in vivo effect of 3F1 on S. mutans colonization and cariogenicity was assessed in specific pathogen-free rats using a modification of a previously described method (Crowley et al. 1999; Palmer et al. 2012). Fischer 344 rats were bred and maintained in trexler isolators. Eighteen 20-d-old pups were removed from isolators and randomly assigned into 3 groups of 6 animals each based on treatment regimen: small molecule 3F1, fluoride (positive control), or no treatment (negative control). All animals were provided sulfamethoxazole trimethoprim oral in their drinking water to suppress endogenous flora and facilitate S. mutans infection. Animals were then infected with S. mutans UA159 and provided a caries-promoting diet (Teklad Diet 305) containing 5% sucrose (Harlan Laboratories) and sterile drinking water ad libitum. Colonization of S. mutans UA159 was confirmed by plating. For treatment, a solution of fluoride (250 ppm) or 3F1 (100 µM) was topically applied to rat molars for 30 s with the aid of a camel hair brush, twice daily for 4 wk beginning 10 d postinfection. Brushes were cleaned between uses. Animals were weighed weekly to monitor for signs of toxicity. Following treatment, all animals were sacrificed and their mandibles excised for plaque analysis and scoring for caries by the method of Keyes (1958). Differences were compared statistically by ANOVA. Animal use was approved by the UAB Institutional Animal Care and Use Committee (IACUC-09771).This study was in accordance with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines for preclinical animal studies.
Microbiome Analysis
For microbiome analysis of the rat oral cavity throughout the study, the mouth of each rat was quickly and gently rubbed with a sterile swab so as not to disturb the plaque. Swab samples were stored at –80°C. DNA was extracted from swab samples with the ZR Fecal DNA Miniprep Kit (Zymo Research). The V4 region of the 16S ribosomal RNA (rRNA) gene was amplified and then sequenced on an Illumina Miseq as previously described (Kumar et al. 2014; Daft et al. 2015). Sequences were analyzed using the Quantitative Insight into Microbial Ecology (QIIME) suite v1.7 (Caporaso et al. 2010) and a QIIME wrapper called QWRAP (Kumar et al. 2014). Operational taxonomic units (OTUs) were assigned taxonomic groups using the Ribosomal Database Project (RDP) classifier (Wang et al. 2007) and the May 2013 Greengenes 16S rRNA sequence database (DeSantis et al. 2006). Distance matrices generated by clustering were used to generate principal components analysis (PCoA) plots. To determine whether samples clustered differently by treatment and/or time point, samples were grouped by treatment and/or time point and the distance matrices were tested using the permutational multivariate analysis of variance (PERMANOVA) test for significant differences in clustering (P < 0.05). To determine the differences in taxonomic frequency underlying differences in clustering, OTUs were grouped by phyla, classes, orders, families, and genera and tested for significant differences in frequency between groups using the Kurskal-Wallis test (P < 0.05 after false discovery rate [FDR] correction).
Quantitative Polymerase Chain Reaction
S. mutans UA159-specific primers (Table 1) were used to determine S. mutans CFUs in microbiome samples. SYBR Green Supermix (Bio-Rad) was used to perform real-time quantitative polymerase chain reaction (qPCR) as described before (Childers et al. 2011).
Table 1.
Quantitative Polymerase Chain Reaction Streptococcus mutans UA159-Specific Primers.
| Primer Set | Sequence | Source |
|---|---|---|
| S. mutans UA159 GtfB | Forward: 5′-CAAAATGGTATTATGGCTGTCG-3′ Reverse: 5′-GCTTAGATGTCACTTCGGTTG-3′ |
This study |
Results
Small Molecule 3F1 Disperses S. mutans Biofilm In Vitro
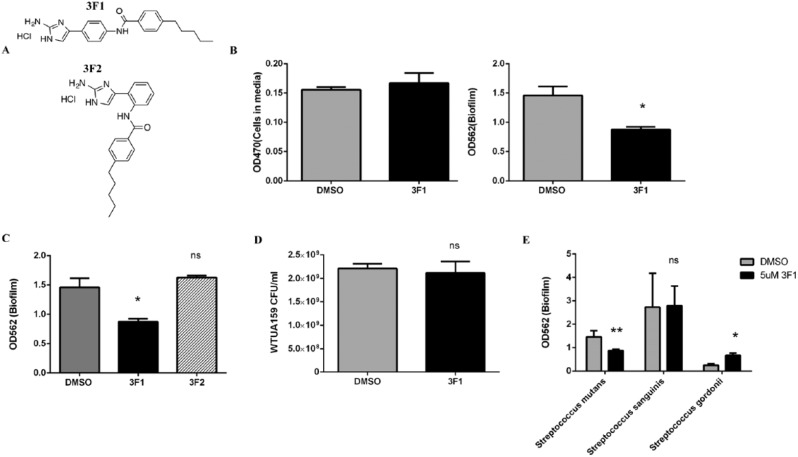
To determine whether selective targeting of S. mutans biofilms could effectively prevent dental caries, we first needed to identify a compound capable of selectively targeting S. mutans biofilms. Small molecules present ideal therapeutic candidates in comparison to antimicrobial peptides due to their stability, potential membrane permeability, activity at low concentrations, and low toxicity (Worthington et al. 2012; Pierce et al. 2015). 2-Aminoimidazole (2-AI) derivatives based on the marine sponge–derived natural product bromoageliferin have been shown to be potent and versatile antibiofilm agents across phylum and genera (Rogers and Melander 2008; Rogers et al. 2010). A previous study from our groups used a library of 2-AI derivatives and biofilm formation assays with S. mutans and successfully identified a small molecule inhibitor of S. mutans biofilm formation (Liu et al. 2011). In this study, we screened this library of 600 compounds using a biofilm dispersion assay to identify a small molecule specific for dispersing S. mutans biofilms. We only characterized small molecules that met the following criteria: low minimum dispersion concentration (MDC50, or concentration that a single dose of the small molecule needed to disperse 50% of the biofilm), little to no cytotoxicity, and selectivity toward S. mutans biofilms. We identified 1 small molecule capable of selectively dispersing S. mutans biofilms compared with biofilms treated with DMSO. Small molecule 3F1 (Fig. 1A) dispersed biofilms at an MDC50 of 5 µM (P < 0.05, 3F1, Fig. 1B). We investigated whether the structure of 3F1 was specific for the dispersal phenotype by characterizing the effects of structural analogue 3F2 (Fig. 1A) on biofilm dispersion. 3F2 did not disperse S. mutans biofilms (3F2, Fig. 1C), suggesting that dispersal activity was selective to the 3F1 structure. To determine whether 3F1 was inhibiting bacterial growth and thereby reducing biofilms, planktonic cells were treated with 5 µM 3F1 for 14 h and viable cells were enumerated. CFUs recovered from 3F1-treated cells were similar to CFUs recovered from cells treated with DMSO (Fig. 1D).
Figure 1.
Small molecule 3F1 selectively disperses Streptococcus mutans biofilms in vitro. (A) Chemical structure of small molecule 3F1 and its structural analogue, 3F2. 3F2 is a structural isoform of 3F1 and only varies in the side projection of the distal propyl oxy group from the center aromatic ring. (B) S. mutans UA159 biofilms were formed for 6 h. Biofilms were then washed and treated with dimethyl sulfoxide (DMSO), 5 µM 3F1, or 5 µM 3F2 for 14 h. After 14 h, cells released into the media were measured at OD470. Loosely adhered cells were then washed off and remaining biomass quantitated by crystal violet staining at OD562. (C) S. mutans UA159 biofilms were formed for 6 h, washed, and treated with DMSO, 5 µM 3F1, or 5 µM 3F2 for 14 h. Loosely adhered cells were then washed off and remaining biomass quantitated by crystal violet staining at OD562. (D) Planktonic S. mutans UA159 cells were treated for 14 h with DMSO or 5 µM 3F1. Viable cells were enumerated by colony-forming unit (CFU) plating. (E) Oral commensal species, Streptococcus sanguinis and Streptococcus gordonnii, were allowed to form biofilms for 6 h before treatment with DMSO or 5 µM 3F1 for 14 h. Bars represent the mean of 3 independent experiments. Differences were statistically compared by analysis of variance. Error bars represent standard error. *P < 0.05. **P < 0.01.
Next, we investigated whether 3F1 was selective for S. mutans biofilms by measuring its activity toward common commensal oral streptococci. To this end, biofilms formed by oral commensals S. sanguinis or S. gordonii were treated with 5 µM 3F1 or DMSO. 3F1 did not negatively affect biofilms formed by oral commensal species at this concentration (Fig. 1E). 3F1 had no effect on biofilms formed by the mutans streptococci member Streptococcus sobrinus (Appendix Fig. 1), implicating pathogen and species specificity of 3F1.
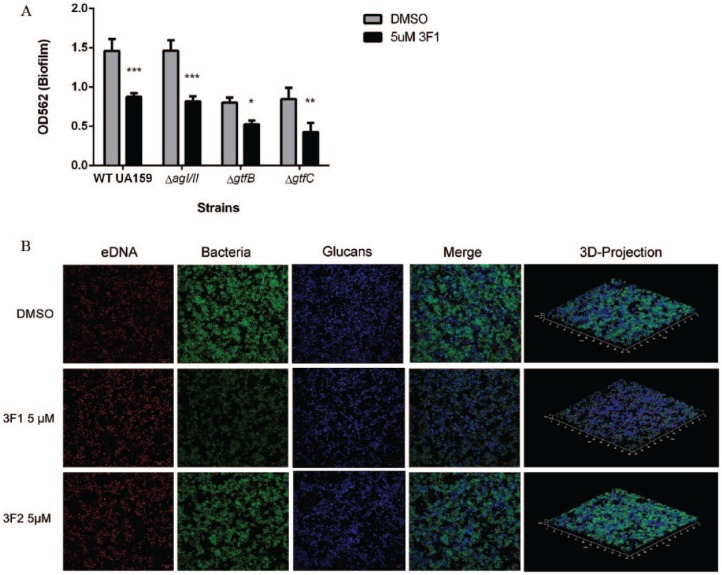
Antigen I/II (AgI/II) and glucans produced by Gtfs, particularly GtfB and GtfC and to a lesser extent GtfD, play a role in initial bacterial attachment and biofilm development (Ahn et al. 2008; Xiao et al. 2012). Therefore, we reasoned 3F1 could be targeting AgI/II, GtfB, or GtfC. Biofilms of single-gene mutants of agI/II, gtfB, or gtfC were treated with 5 µM 3F1. All of the mutants were similarly sensitive to 3F1 treatment as the parent strain (Fig. 2A). So was the GtfD mutant (Appendix Fig. 2). We further investigated the mechanism of 3F1 by examining the effect of 3F1 on the S. mutans biofilm structure and composition. The S. mutans biofilm is a 3-dimensional complex composed mainly of bacterial microcolonies, glucans, eDNA, and proteins (Klein et al. 2012; Peng, Zhang, et al. 2016). DMSO-, 3F1-, and 3F2-treated biofilms exhibited similar fluorescent levels of glucans and eDNA (glucans, eDNA, Fig. 2B), suggesting 3F1 treatment did not affect glucans, cell death, or eDNA scaffolding. Minimal to no reduction of glucans or eDNA corroborated the GtfBCD mutant and CFU studies, respectively, presented above (Fig. 2A, Fig. 1D, respectively). In contrast, the fluorescence of bacterial microcolonies was visibly decreased in the 3F1-treated biofilm compared with DMSO or 3F2 (3F1 bacteria, Fig. 2A), suggesting that the decrease in biomass (Fig. 1B) is due to the dispersion of bacteria from biofilms.
Figure 2.
Small molecule 3F1 disperses Streptococcus mutans biofilm independent of known biofilm-related factors. (A) S. mutans single-gene knockout mutants lacking antigen I/II (ΔagI/II), GtfB (ΔgtfB), or GtfC (ΔgtfC) and the parental strain (wild-type [WT] UA159) were allowed to form biofilms for 6 h. Biofilms were then washed and treated with dimethyl sulfoxide (DMSO) or 5 µM 3F1 for 14 h. Loosely adhered cells were then washed off and remaining biomass quantitated by crystal violet staining at OD562. Bars represent the mean of 3 independent experiments. Differences were statistically compared by analysis of variance. Error bars represent standard error. *P < 0.05. **P < 0.01. ***P < 0.001. (B) Confocal laser scanning microscopy (CLSM) images of wild-type (WT) UA159 biofilm treated with DMSO, 5 µM 3F1, or 5 µM 3F2. Bacterial cells (green), extracellular DNA (eDNA; red), and glucans (blue) were visualized by CLSM after the dispersion assay. Representative images are shown.
Selective Targeting of S. mutans by Small Molecule 3F1 Treatment Prevents Dental Caries and Does Not Perturb the Oral Microbiome
The oral cavity is a unique environment of saliva, teeth, the occasional intake of food and drink, and hundreds of bacterial species. To examine the activity of our small molecule in vivo, we used the well-established rat caries model for the characterization of anticaries efficacy in a controlled experimental environment with a caries-promoting diet (CPD) (Larson et al. 1977). In the model, rapid caries progression can be assessed with an established caries scoring system for rats’ molars. However, the oral microbiome throughout the study has yet to be investigated. Rats were infected with S. mutans UA159, fed a CPD (5% sucrose), and then treated with 3F1 or fluoride. A third group received no treatment. After 4 wk of treatment, the severity of enamel lesions was significantly reduced in all locations for the 3F1 and fluoride groups compared with the no-treatment group (caries, P < 0.001; Table 2). Repeated topical application of treatment did not account for these differences (Appendix Table 1). S. mutans CFUs recovered from the dental plaque of animals treated with 3F1 or fluoride were decreased compared with the no-treatment group (CFUs; Table 2).
Table 2.
Summary of Results from the Rat Caries Study.
| No Treatment, Mean ± SD | Small Molecule 3F1, Mean ± SD | Fluoride, Mean ± SD | |
|---|---|---|---|
| Enamel caries score | |||
| Buccal | 13.0 ± 0.9 | 6.0 ± 0.4* | 6.5 ± 0.7* |
| Sulcal | 20.3 ± 0.7 | 12.0 ± 0.4* | 11.5 ± 0.7* |
| Proximal | 3.5 ± 0.4 | 0* | 0* |
| Weight, g | 171 ± 14 | 158 ± 16ns | 157 ± 11ns |
| CFUs recovered Streptococcus mutans UA159 (×106) | 8.2 ± 1.2 | 6.1 ± 1.1ns | 6.8 ± 1.1ns |
Rats were separated by treatment and grouped by sex. Samples were collected at the end of the study. n = 6 per group. Differences between groups compared by analysis of variance. CFUs, colony-forming units.
P < 0.001 compared with no treatment, ns = not significant compared with untreated.
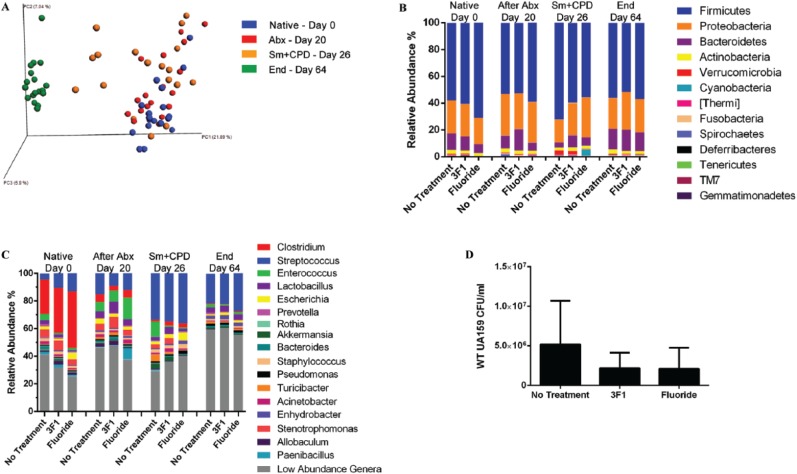
We next investigated whether selective targeting of S. mutans by small molecule 3F1 disturbed the overall oral microbiota. Saliva samples were taken from all rats at different time points: initial removal from isolators (Native [day 0]), after antibiotic treatment (After Abx [day 20]), S. mutans UA159 inoculation and concurrent start of a caries-promoting diet (Sm + CPD [day 26]), and end of study (End [day 64]). Saliva samples are ideal because they are noninvasive and the microbiomes between caries-active and healthy humans are distinguishable (Yang et al. 2012). Beta-diversity of groups over time was compared by PCoA plots (Fig. 3A). Notably, groups clustered together by time rather than by treatment (P < 0.001). At the end of the study, all animal microbiomes were closely clustered together regardless of treatment (P > 0.05). To investigate whether treatment perturbed the composition of the oral microbiome, we first compared bacterial composition at the phylum level. Firmicutes were consistently the most represented. TM7 and Gemmatimonadetes were only significantly enriched at the end of the study in the no-treatment group (P < 0.031 and P < 0.042) and 3F1-treated group (P < 0.031 and P < 0.042), albeit their relative abundance was low (Appendix Table 2). At the genus level, there was a shift after antibiotic treatment from predominantly Clostridium to Enterococcus and commensal Streptococci (After Abx, Fig. 3C). Inoculation with S. mutans and the concurrent start of CPD undoubtedly led to the dramatic increase in the Streptococcus genus (Sm + CPD, Fig. 3C). After 4 wk of treatment and CPD, all microbiomes despite treatment regimen were composed mainly of Streptococcus and low-abundance genera (End, Fig. 3C). In our “end” saliva microbiome samples, we used qPCR to assess S. mutans (Childers et al. 2011). There were fewer S. mutans CFUs detected in 3F1- and fluoride-treated rats compared with the no-treatment control (Fig. 3D), validating our microbiome data.
Figure 3.
3F1 treatment does not perturb the microbiome. Oral microbiome samples were obtained from individual rats at the following time points: native (Native), after antibiotics (After Abx), after inoculation of Streptococcus mutans and start of a caries-promoting diet (Sm + CPD), and at the end of the study (End). The microbiota between groups at different time points was analyzed for diversity and composition. (A) Principal components analysis plots of beta-diversity of all animals in groups over time: native (blue), after antibiotics (red), infect with S. mutans and start CPD (orange), and end (green). Each dot represents 1 rat. Clusters did not change depending on sex or caging (data not shown). (B) Phyla composition in all groups. Each color represents 1 phylum, and length of bar reflects relative abundance. The major phyla detected throughout the study were Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, Thermi, Cyanobacteria, Deferribacteres, Tenericutes, and Verrucomicrobia. n = 6 per group. (C) Genus-level composition of all groups. Clostridium dominated in the native microbiome and, to a lesser extent, Enterococcus and Stenotrophomonas. Infection with S. mutans and daily sucrose intake shifted the composition to a Streptococcus-dominated community. n = 6. (D) Quantitative polymerase chain reaction of wild-type (WT) UA159 recovered from microbiome samples at the end of the study.
Discussion
Overall, our studies suggest that selective targeting of S. mutans biofilms by a noncytotoxic small molecule 3F1 can effectively prevent dental caries and preserve the overall microbiome. This small molecule selectively dispersed S. mutans biofilms in vitro, independent of major biofilm-related factors, as determined by biofilm dispersion assays and confocal laser scanning microscopy (CLSM) (Figs. 1 and 2). In contrast, structural isoform 3F2 was inactive (Fig. 1C). In an in vivo rat caries study, 3F1 reduced dental caries (Table 2) while preserving the dynamics of the microbiome (Fig. 3). S. mutans colonization was reduced in 3F1-treated rats, albeit modestly (CFUs Table 2, Fig. 3D). The observation that 3F1 treatment increased S. gordonii biofilms in vitro may contribute to a positive outcome in vivo since S. gordonii antagonizes S. mutans through production of hydrogen peroxide (Kreth et al. 2008). Unsurprisingly, the mechanism of 3F1 is not dependent on well-characterized biofilm factors, including AgI/II, Gtfs, glucans, and eDNA, since they are often associated with initial development of S. mutans biofilms, while proteins and factors involved in maintenance and maturation are largely unknown (Crowley et al. 1999; Bowen and Koo 2011). Slight changes in 2-AI compounds can significantly affect biofilm dispersal properties (Ballard et al. 2008). This structure-specific activity is consistent with most small molecule/protein interactions since compound geometry and architecture play key roles in protein binding (Bunders et al. 2010). Indeed, small molecules from this library have been found to target biofilm-essential proteins (Liu et al. 2011). Thus, 3F1 may target a novel biofilm-related protein potentially inhibiting a crucial interaction between S. mutans and the biofilm matrix in vitro. Reduction in S. mutans CFUs after treatment with 3F1 in vivo (Table 2) corroborates with results from other rat caries studies where a slight reduction in S. mutans CFUs resulted in a significant decrease in dental caries (Branco-de-Almeida et al. 2011; Falsetta et al. 2012). Under our in vivo conditions, 3F1 treatment may have an impact on S. mutans virulence, such as reducing the production of lactic acid while only modestly affecting bacterial colonization.
One of the novel aspects of our study is the full characterization of the oral microbiome throughout the rat caries model. Phylum-level comparison of groups from initial infection of S. mutans to after 4 wk of treatment showed that selective control of S. mutans by 3F1 did not perturb the overall rat oral microbiome (Fig. 3A–C). Together, our data suggest that the S. mutans biofilm-specific therapy we reported here is a novel viable approach for preventing caries and preserving the oral microbiome. A therapeutic combining both species-specific selectivity toward S. mutans and preservation of the oral microbiome in vivo has not been reported. Derivatives or extracts from natural products such as cranberries, green tea, tea tree oil, garlic, or hop polyphenols are able to reduce or inhibit the oral biofilm but do not specifically target S. mutans (Tagashira et al. 1997; Houshmand et al. 2013; Thomas et al. 2015). Killing and elimination of S. mutans by synthetic antimicrobial peptide C16G2 in an in vitro oral biofilm model in saliva also nonspecifically eliminated noncariogenic species, leading to a drastic shift of the structure of the microbiota (Guo et al. 2015). We speculate that under our conditions, the combination of a sugary diet and the presence of S. mutans biofilm shape the microbiota diversity and composition, while control of S. mutans colonization is sufficient to decrease the incidence of dental caries. The minimal changes in the microbiome after fluoride treatment are corroborated by studies in humans (Koopman et al. 2015; Reilly et al. 2016). While these studies do not take diet into consideration; we show that sugary intake can significantly affect the composition of the microbiome despite the treatment and colonization of S. mutans. The influence of the frequent ingestion of sucrose, rather than the prevalence of S. mutans, has been shown to correlate with the dysbiosis of the oral microbiome (Rudney et al. 2015; Tian et al. 2015; Bernabe et al. 2016). Such a dysbiotic community may precede the development of caries lesions, which can be employed as a microbial biomarker to predict the disease pathogenesis.
In summary, we identified novel small molecule 3F1 capable of selectively dispersing S. mutans biofilms independent of well-studied biofilm-related factors. 3F1 treatment controlled S. mutans in a rat caries model and prevented dental caries. The dysbiosis of the oral microbiome is mainly attributed to daily sugar intake and not the abundance of S. mutans, as the reduction of S. mutans colonization by 3F1 did not significantly alter overall the microbiome at the phylum and genus levels. The necessity of sucrose intake in the maintenance of the oral microbiome suggests that environmental factors such as diet are necessary to properly assess anticaries therapies. To our knowledge, this is the first report to demonstrate selective targeting of the cariogenic biofilms to prevent dental caries and characterize the oral microbiome throughout the rat caries model. Our studies demonstrate that a therapy aimed at S. mutans can effectively prevent dental caries and preserve the microbiome. This report provides a scientific basis for developing a complete anticaries therapy that is selective for key pathogen S. mutans despite the presence of a dysbiotic microbiome fueled by dietary intake.
Author Contributions
S.S. Garcia, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; H. Wu, contributed to conception, design, data analysis, and interpretation, critically revised the manuscript; S. Michalek, contributed to conception, design, data acquisition, and analysis, critically revised the manuscript; M.S. Blackledge, contributed to design and data interpretation, critically revised the manuscript; L. Su, C. Morrow, contributed to design and data acquisition, critically revised the manuscript; T. Ptacek, contributed to data analysis and interpretation, critically revised the manuscript; E.J. Lefkowitz, contributed to data interpretation, critically revised the manuscript; C. Melander, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; P. Eipers, contributed to design, data acquisition, and analysis, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We thank Katherine Taylor for designing the qPCR primers for GtfB and Qiong Zhang for constructing the ΔgtfD mutant.
Footnotes
A supplemental appendix to this article is available online.
This work was supported by National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) F31 DE024041, NIH/NIDCR T90 DE022736, NIH/NIDCR RO1DE022350, and NIH CTSA award UL1TR001417. The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165), and Heflin Center.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJ, Townsend G, Soltysiak A, Alt KW, et al. 2013. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the neolithic and industrial revolutions. Nat Genet. 45(4):450–455, 455e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. 2008. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun. 76(9):4259–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard TE, Richards JJ, Wolfe AL, Melander C. 2008. Synthesis and antibiofilm activity of a second-generation reverse-amide oroidin library: a structure-activity relationship study. Chemistry. 14(34):10745–10761. [DOI] [PubMed] [Google Scholar]
- Bernabe E, Vehkalahti MM, Sheiham A, Lundqvist A, Suominen AL. 2016. The shape of the dose-response relationship between sugars and caries in adults. J Dent Res. 95(2):167–172. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. 2011. Biology of Streptococcus mutans–derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45(1):69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-de-Almeida LS, Murata RM, Franco EM, dos Santos MH, de Alencar SM, Koo H, Rosalen PL. 2011. Effects of 7-epiclusianone on Streptococcus mutans and caries development in rats. Planta Med. 77(1):40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunders CA, Richards JJ, Melander C. 2010. Identification of aryl 2-aminoimidazoles as biofilm inhibitors in gram-negative bacteria. Bioorg Med Chem Lett. 20(12):3797–3800. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. 2010. Qiime allows analysis of high-throughput community sequencing data. Nat Methods. 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers NK, Osgood RC, Hsu KL, Manmontri C, Momeni SS, Mahtani HK, Cutter GR, Ruby JD. 2011. Real-time quantitative polymerase chain reaction for enumeration of Streptococcus mutans from oral samples. Eur J Oral Sci. 119(6):447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 67(3):1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG. 2015. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome. 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16s rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. 2006. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother. 50(11):3651–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Lemos JA, Silva BB, Agidi S, Scott-Anne KK, Koo H. 2012. Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo. Antimicrob Agents Chemother. 56(12):6201–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SS, Du Q, Wu H. 2016. Streptococcus mutans copper chaperone, CopZ, is critical for biofilm formation and competitiveness. Mol Oral Microbiol. 31(6):515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, McLean JS, Yang Y, Eckert R, Kaplan CW, Kyme P, Sheikh O, Varnum B, Lux R, Shi W, et al. 2015. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc Natl Acad Sci USA. 112(24):7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshmand B, Mahjour F, Dianat O. 2013. Antibacterial effect of different concentrations of garlic (Allium sativum) extract on dental plaque bacteria. Indian J Dent Res. 24(1):71–75. [DOI] [PubMed] [Google Scholar]
- Keyes PH. 1958. Dental caries in the molar teeth of rats. II. A method for diagnosing and scoring several types of lesions simultaneously. J Dent Res. 37(6):1088–1099. [DOI] [PubMed] [Google Scholar]
- Klein MI, Xiao J, Lu B, Delahunty CM, Yates JR, III, Koo H. 2012. Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics. PLoS One. 7(9):e45795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Duarte S, Murata RM, Scott-Anne K, Gregoire S, Watson GE, Singh AP, Vorsa N. 2010. Influence of cranberry proanthocyanidins on formation of biofilms by Streptococcus mutans on saliva-coated apatitic surface and on dental caries development in vivo. Caries Res. 44(2):116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JE, van der Kaaij NC, Buijs MJ, Elyassi Y, van der Veen MH, Crielaard W, Ten Cate JM, Zaura E. 2015. The effect of fixed orthodontic appliances and fluoride mouthwash on the oral microbiome of adolescents—a randomized controlled clinical trial. PLoS One. 10(9):e0137318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 190(13):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, Morrow CD. 2014. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet. 82:18.8.1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RH, Amsbaugh SM, Navia JM, Rosen S, Schuster GS, Shaw JH. 1977. Collaborative evaluation of a rat caries model in six laboratories. J Dent Res. 56(8):1007–1012. [DOI] [PubMed] [Google Scholar]
- Liu C, Worthington RJ, Melander C, Wu H. 2011. A new small molecule specifically inhibits the cariogenic bacterium Streptococcus mutans in multispecies biofilms. Antimicrob Agents Chemother. 55(6):2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SR, Crowley PJ, Oli MW, Ruelf MA, Michalek SM, Brady LJ. 2012. Yidc1 and Yidc2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology. 158(Pt 7):1702–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Michalek S, Wu H. 2016. Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of Streptococcus mutans revisit. Environ Microbiol. 18(11):3612–3619. [DOI] [PubMed] [Google Scholar]
- Peng X, Zhang Y, Bai G, Zhou X, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol. 99(5):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CG, Chaturvedi AK, Lazzell AL, Powell AT, Saville SP, McHardy SF, Lopez-Ribot JL. 2015. A novel small molecule inhibitor of Candida albicans biofilm formation, filamentation and virulence with low potential for the development of resistance. NPJ Biofilms Microbiomes. 1:15012. doi: 10.1038/npjbiofilms.2015.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly C, Goettl M, Steinmetz M, Nikrad J, Jones RS. 2016. Short-term effects of povidone iodine and sodium fluoride therapy on plaque levels and microbiome diversity. Oral Dis. 22(2):155–161. [DOI] [PubMed] [Google Scholar]
- Rogers SA, Huigens RW, III, Cavanagh J, Melander C. 2010. Synergistic effects between conventional antibiotics and 2-aminoimidazole-derived antibiofilm agents. Antimicrob Agents Chemother. 54(5):2112–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SA, Melander C. 2008. Construction and screening of a 2-aminoimidazole library identifies a small molecule capable of inhibiting and dispersing bacterial biofilms across order, class, and phylum. Angew Chem Int Ed Engl. 47(28):5229–5231. [DOI] [PubMed] [Google Scholar]
- Rudney JD, Jagtap PD, Reilly CS, Chen R, Markowski TW, Higgins L, Johnson JE, Griffin TJ. 2015. Protein relative abundance patterns associated with sucrose-induced dysbiosis are conserved across taxonomically diverse oral microcosm biofilm models of dental caries. Microbiome. 3:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagashira M, Uchiyama K, Yoshimura T, Shirota M, Uemitsu N. 1997. Inhibition by hop bract polyphenols of cellular adherence and water-insoluble glucan synthesis of mutans streptococci. Biosci Biotechnol Biochem. 61(2):332–335. [DOI] [PubMed] [Google Scholar]
- Thomas A, Thakur S, Mhambrey S. 2015. Comparison of the antimicrobial efficacy of chlorhexidine, sodium fluoride, fluoride with essential oils, alum, green tea, and garlic with lime mouth rinses on cariogenic microbes. J Int Soc Prev Community Dent. 5(4):302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Qin M, Ma W, Xia B, Xu H, Zhang Q, Chen F. 2015. Microbiome interaction with sugar plays an important role in relapse of childhood caries. Biochem Biophys Res Commun. 468(1–2):294–299. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington RJ, Richards JJ, Melander C. 2012. Small molecule control of bacterial biofilms. Org Biomol Chem. 10(37):7457–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8(4):e1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Zeng X, Ning K, Liu KL, Lo CC, Wang W, Chen J, Wang D, Huang R, Chang X, et al. 2012. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 6(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.