Abstract
Genotyping of Mycobacterium tuberculosis isolates is useful in tuberculosis control for confirming suspected transmission links, identifying unsuspected transmission, and detecting or confirming possible false-positive cultures. The value is greatly increased by reducing the turnaround time from positive culture to genotyping result and by increasing the proportion of cases for which results are available. Although IS6110 fingerprinting provides the highest discrimination, amplification-based methods allow rapid, high-throughput processing and yield digital results that can be readily analyzed and thus are better suited for large-scale genotyping. M. tuberculosis isolates (n = 259) representing 99% of culture-positive cases of tuberculosis diagnosed in Wisconsin in the years 2000 to 2003 were genotyped by using spoligotyping, mycobacterial interspersed repetitive unit (MIRU) typing, and IS6110 fingerprinting. Spoligotyping clustered 64.1% of the isolates, MIRU typing clustered 46.7% of the isolates, and IS6110 fingerprinting clustered 29.7% of the isolates. The combination of spoligotyping and MIRU typing yielded 184 unique isolates and 26 clusters containing 75 isolates (29.0%). The addition of IS6110 fingerprinting reduced the number of clustered isolates to 30 (11.6%) if an exact pattern match was required or to 44 (17.0%) if the definition of a matching IS6110 fingerprint was expanded to include patterns that differed by the addition of a single band. Regardless of the genotyping method chosen, the addition of a second or third method decreased clustering. Our results indicate that using spoligotyping and MIRU typing together provides adequate discrimination in most cases. IS6110 fingerprinting can then be used as a secondary typing method to type the clustered isolates when additional discrimination is needed.
The genotyping of Mycobacterium tuberculosis isolates plays a role in tuberculosis (TB) control by identifying possibly related cases not detected in routine contact investigation, detecting unsuspected false-positive cultures, and supporting investigations of suspected outbreaks and false-positive cultures. The utility of genotyping is greatly enhanced when all available isolates in a geographic area are typed. Such large-scale genotyping services have only been available to a limited number of TB control programs in the United States. In 1996, the Centers for Disease Control and Prevention established the National Tuberculosis Genotyping and Surveillance Network that provided universal genotyping services to five states and selected counties in two additional states. Over a 5-year period, 10,883 isolates were genotyped by using IS6110 fingerprinting as the primary typing method and spoligotyping for secondary typing of isolates containing low copy numbers (<7) of IS6110 (4). While there has been a desire to provide universal genotyping services to the entire country (∼12,000 isolates/year), the difficulties encountered with IS6110 fingerprinting as a primary typing method in the Network study clearly indicated that finding a new genotyping strategy would be necessary to meet this goal. The limitations of the previous approach included the technical difficulties associated with IS6110 fingerprinting—a lengthy turnaround time, a labor-intensive purification of genomic DNA, complex results that require sophisticated computer analysis, and problems with reproducibility—and the insufficient discrimination provided by the combination of IS6110 fingerprinting and spoligotyping for isolates containing low copy numbers of IS6110 (5). Ideally, the new strategy would include PCR-based methods that require minimal amounts of culture material, are conducive to high throughput analysis, have a fast turnaround time, and provide a high level of discrimination.
Of the PCR-based methods available for genotyping M. tuberculosis complex isolates, mycobacterial interspersed repetitive unit (MIRU) typing is the most promising. It is based on the variability found at 12 specific loci interspersed throughout the genome (16). Studies have shown that MIRU genotypes are stable over several years and are thus suitable for evaluating transmission links (8, 13), but they are also variable enough to generate a discriminatory power approaching that of the “gold standard, ” IS6110 fingerprinting (15). Only a limited number of studies using MIRU typing have been completed. These studies have included sets of isolates chosen for their diversity by IS6110 fingerprinting, for their lack of diversity by IS6110 fingerprinting, for their diversity by geographic origin, or for having low copy numbers of IS6110, serial isolates from chronically infected patients or patients with drug-resistant TB, and isolates from patients known to be epidemiologically related (3, 8, 10, 11, 13-15). However, the predictive value of genotyping to document relatedness of isolates and recent TB transmission is often considered stronger in studies where the links among patients are suspected or known. To date, no studies have described the discriminatory power of MIRU typing in a universal genotyping study and the epidemiological relatedness of the clustered isolates.
Here, we report the results of genotyping using three typing methods, IS6110 fingerprinting, spoligotyping, and MIRU typing, alone and in combination, for a set of isolates that included one isolate from each culture-positive patient diagnosed with TB in Wisconsin between 2000 and 2003. The clustering data are compared to previously collected patient demographic information and results of contact and cluster investigations.
MATERIALS AND METHODS
Study isolates.
One isolate from each culture-positive TB patient in Wisconsin diagnosed between January 2000 and October 2003 was included in the universal genotyping study (n = 259 isolates). These represent 99% of culture-positive cases (n = 262 cases) and 84% of all cases during that period (n = 310 cases). Of the 259 patients whose isolates were genotyped, 76 had a reported case of TB in 2000, 69 had a reported case in 2001, 68 had a reported case in 2002, and 46 had a reported case in 2003. All isolates were genotyped by using spoligotyping, MIRU typing, and IS6110 fingerprinting. The following demographic data was available for all of the patients in the study: city, county, and ZIP code of residence at the time of diagnosis, age, sex, race or ethnicity, HIV status, drug resistance of isolate, history of prior TB infection, drug use (injection and noninjection), alcohol abuse, homelessness, and residence in a long-term care facility or correctional institute. If the patient was born outside of the United States, the length of time residing in the United States prior to diagnosis was documented. The results of contact investigations and cluster investigations were also available for review.
Genotyping.
Crude DNA preparations suitable for PCR-based assays were prepared from cell suspensions by shaking with beads. Growth was washed from Lowenstein-Jensen slants with 1 ml of Tris-EDTA, and the suspension (at least 0.5 ml) was transferred to a 2.0-ml screw-top microtube containing a 200-μl aliquot of 0.1-mm zirconium beads (BioSpec Products, Inc., Bartlesville, Okla.) that had been treated with Sigmacote (Sigma Chemical, St. Louis, Mo.). The tube was centrifuged (Eppendorf 5415C microcentrifuge) at 14,000 rpm for 5 min, the supernatant was removed, and 300 μl of Tris-EDTA and 400 μl of chloroform were added to the tube. The tube was shaken in a Fastprep instrument (Qbiogene, Inc., Carlsbad, Calif.) at a setting of 4 for 20 s and then centrifuged (Eppendorf 5415C microcentrifuge) at 14,000 rpm for 5 min. The upper aqueous layer was transferred to a fresh tube and incubated at 90°C for 30 min. Samples were stored at −20°C. All manipulations were performed in a biosafety level 3 laboratory.
Spoligotyping was performed as previously described (12). Spoligotypes were reported by using an octal code (3, 6).
MIRU typing was performed by using the approach of Supply et al. (15) with modifications designed to facilitate analysis on a CEQ8000 capillary electrophoresis instrument (Beckman, Fullerton, Calif.). Each locus was amplified individually with primers (Table 1) that flanked the locus. The PCR mixtures consisted of 1× PCR buffer (QIAGEN, Valencia, Calif.), 0.25 U of HotStar Taq DNA polymerase, deoxynucleoside triphosphates (0.2 mM each), 1× Q buffer, 2.0 mM MgCl2, Beckman dye-labeled (D2, D3, or D4) forward primer and unlabeled reverse primer (0.2 μM each for loci 4, 16, and 20; 0.1 μM each for all other loci), and 2.5 μl of template DNA. The final volume of each reaction was 5 μl. Samples were amplified in a GeneAmp PCR System 9700 (Perkin Elmer Applied Biosystems, Foster City, Calif.) by using an amplification profile of 15 min at 95°C; 40 cycles of 30 s at 94°C, 10 s at 58°C, and 1 min at 72°C; and 7 min at 72°C. Following amplification, the 12 PCR products were combined into four pools (described in Table 1), where each pool contained 5 μl of D2-labeled amplicon, 1 μl of D3-labeled amplicon, and 1 μl of D4-labeled amplicon. Each pool was diluted by the addition of 20 μl of deionized formamide (Sigma). For analysis with the CEQ8000, 1 μl of the diluted pool was added to 40 μl of deionized formamide and 0.5 μl of 600-bp marker (Beckman). The number of MIRU copies at each locus was determined by using the convention described in Table 1. The final MIRU result is the concatenation of the individual results into a 12 character script (MIRU locus 2-4-10-16-20-23-24-26-27-31-39-40). To maintain a single-character result for each locus, the following convention was used for greater than 9 copies: the letter a indicated 10 copies, the letter b indicated 11 copies, the letter c indicated 12 copies, etc. A dash indicated deletion of the locus, i.e., when no product was obtained on amplification.
TABLE 1.
Sequences of oligonucleotides used in MIRU typing
| Pool | Locus | Name (dye label) | Sequence | Apparent size of amplicon containing one MIRU copy + size of additional copies (bp)a |
|---|---|---|---|---|
| A | 04 | 4a(D2) | 5′GTCAAACAGGTCACAACGAGAGGAA | 187 + 72 |
| 4b | 5′CCTCCACAATCAACACACTGGTCAT | |||
| 02 | 2c(D3) | 5′CAGGTGCCCTATCTGCTGACG | 236 + 47 | |
| 2d | 5′GTTGCGTCCGGCATACCAAC | |||
| 10 | 10a(D4) | 5′ACCGTCTTATCGGACTGCACTATCAA | 148 + 52 | |
| 10d | 5′ATCCACCGCATCACGTTGGT | |||
| B | 16 | 16c(D2) | 5′CGAAGCCGCAGTACCACCTC | 118 + 51 |
| 16d | 5′CGACGCCTACGCTGATTCCA | |||
| 24 | 24a(D3) | 5′GAAGGCTATCCGTCGATCGGTT | 365 + 49 | |
| 24b | 5′GGGCGAGTTGAGCTCACAGAAC | |||
| 23 | 23a(D4) | 5′CGAATTCTTCGGTGGTCTCGAGT | 132 + 52 | |
| 23b | 5′ACCGTCTGACTCATGGTGTCCAA | |||
| C | 20 | 20a(D2) | 5′GCCCTTCGAGTTAGTATCGTCGGTT | 291 + 73 |
| 20b | 5′CAATCACCGTTACATCGACGTCATC | |||
| 40 | 40c(D3) | 5′ATACGGCAAGCGCAAGAGCAC | 172 + 47 | |
| 40b | 5′TCAGGTCTTTCTCTCACGCTCTCG | |||
| 26 | 26a(D4) | 5′GCGGATAGGTCTACCGTCGAAATC | 114 + 48 | |
| 26c | 5′CAACTGCCTCGCGGAATAGG | |||
| D | 39 | 39a(D2) | 5′CGGTCAAGTTCAGCACCTTCTACATC | 238 + 47 |
| 39c | 5′GCGTCCGTACTTCCGGTTCAG | |||
| 31 | 31a(D3) | 5′CGTCGAAGAGAGCCTCATCAATCAT | 160 + 52 | |
| 31b | 5′AACCTGCTGACCGATGGCAATATC | |||
| 27 | 27c | 5′CGGTGACCAACGTCAGATTCACT | 121 + 54 | |
| 27d | 5′TGACACGTGACGGGGGCATCTT |
A table listing the observed sizes for all alleles is available upon request.
IS6110 fingerprinting was performed by using previously described methods (17). The BioImage Whole Band Analyzer software package version 3.4 (Ann Arbor, Mich.) was used to analyze the fingerprint patterns. Patterns were considered identical only if the size of each of the matched bands differed by less than 2.5%. Patterns were considered a +1 band match if the patterns differed by the presence of one additional band.
All primers were synthesized by the Biotechnology Core Facility, Centers for Disease Control and Prevention, Atlanta, Ga.
The Hunter-Gaston discrimination index was calculated as originally described (9) by using the following equation: HGDI = 1 − ({1/[N × (N − 1)] } × {Σ(j = 1→s) [nj× (nj − 1)]}) where N is the total number of strains in the sample population, s is the total number of types described, and nj is the number of strains belonging to the jth type.
RESULTS
Genotyping was performed with isolates from the state of Wisconsin as part of a project designed to study the impact of genotyping in low-incidence areas. Between January 2000 and October 2003, one isolate from each culture-positive case identified in Wisconsin was submitted for genotyping. All isolates were spoligotyped, MIRU typed, and IS6110 fingerprinted. The results were analyzed retrospectively to evaluate the feasibility of a two-step approach for genotyping, the first step being the two PCR-based methods, spoligotyping and MIRU typing, that can be accomplished rapidly and directly from primary cultures, and the second step being traditional IS6110 fingerprinting for isolates that are clustered by the PCR methods. In this set of isolates, spoligotyping and MIRU typing clustered 64.1 and 46.7% of the isolates, respectively (Table 2). IS6110 fingerprinting was the most discriminating of the three methods, clustering only 29.7% of the isolates, with matches defined as identical patterns. However, seven clusters representing 45 isolates (17.3%) had low band number (less than seven), including 13 isolates with a common one-band pattern and 19 isolates with a common two-band pattern. The combination of the two PCR-based genotyping methods clustered 29.0% of the isolates and thus provided a level of discrimination comparable to IS6110 fingerprinting. The addition of IS6110 fingerprinting further reduced the number of clustered isolates from 75 to 30 (11.6%). If the definition of a matching IS6110 fingerprint is expanded to include patterns that differ by a single band, the number of clustered isolates is reduced only to 44 (17.0%)
TABLE 2.
Discriminatory power of genotyping methodsa
| Genotyping method | No. of distinct types | No. of unique isolates | No. of clustered isolates (%) | No. of clusters (no. of isolates per cluster) | HGDI |
|---|---|---|---|---|---|
| Spoligotyping | 128 | 93 | 166 (64.1) | 35 (2-23) | 0.979 |
| MIRU | 177 | 138 | 121 (46.7) | 39 (2-8) | 0.995 |
| IS6110-RFLP | 201 | 182 | 77 (29.7) | 19 (2-19) | 0.991 |
| low copy number isolatesb | 21 | 14 | 45 | 7 (2-19) | NR |
| high copy number isolates | 180 | 168 | 32 | 12 (2-4) | NR |
| Spoligotyping and MIRU | 210 | 184 | 75 (29.0) | 26 (2-7) | 0.997 |
| Spoligotyping and IS6110-RFLP | 223 | 203 | 56 (21.6) | 20 (2-4) | 0.998 |
| MIRU and IS6110-RFLP | 237 | 223 | 36 (13.9) | 14 (2-4) | 0.999 |
| All three methods | 241 | 229 | 30 (11.6) | 12 (2-4) | 0.999 |
HGDI, Hunter Gaston discrimination index (9); NR, not relevant; RFLP, restriction fragment length polymorphism.
Low copy number isolates had less than seven bands.
Only 12 of the 26 clusters defined by spoligotyping and MIRU typing contained isolates with exact matches by IS6110 fingerprinting (Table 3). In 9 of the 12 clusters, all of the isolates in the cluster had identical IS6110 fingerprints. For the other three clusters, there were multiple isolates in each cluster with identical IS6110 fingerprints and an additional isolate whose IS6110 fingerprint differed from the fingerprint of the clustered isolates by the presence of one additional band. One of these clusters also contained an isolate with an IS6110 fingerprint that did not match that of the clustered isolates, i.e., it differed by more than two bands. In 5 of the 12 clusters, probable transmission links were identified for nearly all (17 out of 18) of the patients. The remaining 14 clusters defined by spoligotype and MIRU type did not contain any isolates with identical IS6110 fingerprint patterns. In five of these clusters, the IS6110 fingerprint patterns differed by the presence of an additional band. The remaining nine clusters contained isolates whose IS6110 fingerprint patterns differed by two or more bands.
TABLE 3.
Isolates clustered by the combination of spoligotyping and MIRU typing
| Spoligotype | MIRU pattern | IS6110 fingerprint result
|
Patient birthplacec | Length of residency prior to diagnosis (yr)d | Transmission links identified? | ||
|---|---|---|---|---|---|---|---|
| No. of exact-match isolates (no. of bands)a | No. of + 1 band isolates (no. of bands)a | No. of no-match isolates (no. of bands)b | |||||
| 777760077760771 | 124326153224 | 4 (9) | 1 (10) | United States | Yes | ||
| 777777607760071 | 214327143228 | 4 (10) | United States | Yes | |||
| 717737777760771 | 223225153322 | 3 (8) | 1 (9) | United States | Yes | ||
| 407777777760771 | 223326153321 | 3 (11, 12, 12) | United States | ||||
| 737776777760601 | 223325153324 | 3 (2) | United States | ||||
| 776177607760771 | 224326133323 | 2 (15) | United States | ||||
| 777777777720771 | 225313153323 | 2 (9, 10) | United States | ||||
| 777776777760771 | 125325143225 | 2 (2) | United States | ||||
| 777776777760601 | 224325153323 | 2 (2, 3) | United States | ||||
| 677737607760771 | 224225163321 | 2 (10) | United States | ||||
| 777777777760771 | 223425153322 | 2 (9) | United States | ||||
| 777777777760771 | 223125153324 | 2 (9, 9) | United States | ||||
| 777777777760731 | 223225153423 | 2 (9, 11) | United States | ||||
| 677777477413771 | 254326223432 | 7 (10-13)e | Philippines | 1.9 | |||
| 777777777760771 | 222315153324 | 2 (9, 10) | Laos | 20.5 | |||
| 000000000000371 | 223325143535 | 2 (15, 16) | Laos | 14.5 | |||
| 703707740003771 | 226425173533 | 2 (13) | India | 0.5 | |||
| 737777777760031 | 223326143321 | 2 (13) | Mexico | 34.5 | Yes | ||
| 000000000003771 | 223325173523 | 2 (13, 14) | China, Korea | 3 | |||
| 703777740003771 | 225425173533 | 2 (10, 13) | India, Nepal | 2 | |||
| 000000000003771 | 223325173533 | 6 (15-22)e | Burma (1), China (2), Laos (2), United States (1) | 4.8f | |||
| 000000000003771 | 223425173533 | 2 (14) | 1 (15) | 1 (12) | Laos (3), United States (1) | 16.3 | Yes |
| 777777774020771 | 225323153323 | 3 (9, 9, 10) | Mexico, Morocco, United States | 3.0 | |||
| 777777777760771 | 223125143324 | 3 (7, 9, 14) | United States (2), Ethiopia | 16 | |||
| 777777774020771 | 225325153323 | 3 (7, 8, 10) | Gambia, Puerto Rico, United States | 12.5 | |||
| 777777777760771 | 222325153324 | 2 (8) | Japan, United States | 43 | |||
Matching IS6110 fingerprints are defined as two patterns that are identical. + 1 band IS6110 fingerprints are defined as a pattern that is an exact match with the exception of having one additional band. Patterns that do not match differ by at least two bands.
The number of copies of IS6110 for each of the isolates is listed in parentheses. If a cluster contained isolates with different IS6110 pattern with the same number of copies of IS6110, then the copy number is repeated.
The number of patients from each country is listed only for those clusters that contain patients from multiple countries.
Length of residency in the United States prior to diagnosis was calculated only for patients born outside of the United States. The average length of residency for clusters containing patients from both the United States and foreign countries does not include the U.S.-born patient.
Only the range of IS6110 copy numbers is listed. The IS6110 fingerprints for these isolates are shown in Figure 1.
Length of residency for the second patient from Laos was unknown. The average was calculated for the remaining four patients.
Transmission links were identified for five clusters. Three of the clusters with transmission links contained solely U.S.-born patients; the transmission links between many of the patients were unknown prior to an epidemiological investigation initiated during the course of this study. Transmission links between the patients in the two other clusters had been previously identified through traditional contact investigations; the patients in these clusters were primarily foreign-born family members who had resided in the United States for over 20 years prior to diagnosis.
Of the 259 patients included in the universal genotyping study, 122 (47%) were born in the United States and 136 were born in one of 30 foreign countries (India, 25 patients; Mexico, 24 patients; Laos, 17 patients, and the Philippines, 13 patients); the origin of 1 patient is unknown. For the foreign-born patients, the average length of residency in the United States before diagnosis was 8.2 years. Clustering by combined spoligotype and MIRU pattern was higher among the U.S.-born patients (34%) than among the foreign-born patients (24%). The addition of IS6110 fingerprinting reduced the number of clustered isolates from 42 to 24 among U.S.-born patients and from 33 to 6 among the foreign-born patients.
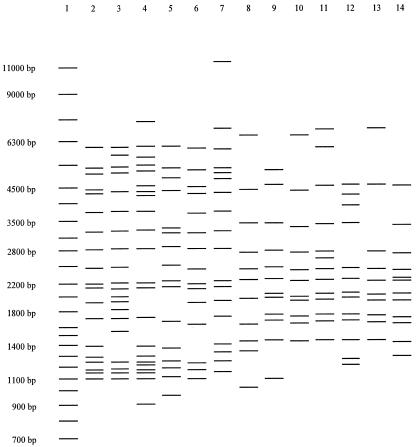
Thirteen of the 33 foreign-born clustered patients belong to one of two large clusters. The first cluster is defined by the spoligotype 000000000003771 and MIRU pattern 223325173533; however, the six isolates had distinct IS6110 fingerprints (Fig. 1). These isolates belong to the Beijing genotype family, as defined by spoligotyping (18), which is not only widely found in Eastern Asia and Russia but also prevalent in the United States. The patients in this cluster were from Burma, China, Laos, and the United States. Of the 63 different MIRU patterns that we have observed for Beijing isolates in this study and in our other U.S. genotyping studies (unpublished data), this is the most frequently observed pattern (80 out of 316 Beijing isolates); isolates with this pattern have many different IS6110 fingerprints. The second cluster is defined by the spoligotype 677777477413771 and MIRU pattern 254326223432, and again the seven isolates had distinct IS6110 fingerprints (Fig. 1). These isolates belong to the recently described Manila genotype family that predominates in the Philippines (7); the seven patients in this cluster were from the Philippines. Of the 22 different MIRU patterns that we have observed for isolates belonging to the Manila family, this is the most frequently observed MIRU pattern (50 out of 92 Manila isolates); isolates with this pattern have many different IS6110 fingerprints.
FIG. 1.
Emergence of common spoligotype-MIRU patterns. Lane 1, size markers; lanes 2 to 7, IS6110 fingerprints for isolates with the spoligotype 000000000003771 and MIRU pattern 223325173533 (Beijing genotype family); lanes 8 to 14, IS6110 fingerprints for isolates with the spoligotype 677777477413771 and MIRU pattern 254326223432 (Manila genotype family).
MIRU typing was able to discriminate between isolates clustered by both spoligotype and IS6110 fingerprint in 11 of 20 clusters. These 11 clusters (32 isolates) were reduced to 3 clusters (7 isolates) and 25 unique isolates by the addition of the MIRU results (Table 4). Seven of the clusters contained isolates with one or two copies of IS6110. The one-band pattern is common among patients from East Africa and India, and the two-band pattern is common among U.S.-born patients. A more unexpected result was that MIRU typing discriminated between isolates with identical high copy number IS6110 fingerprints (clusters I to K). The patient demographics and contact investigations were carefully reviewed for each cluster to ensure that MIRU typing was not discriminating between epidemiologically related isolates, but no transmission links were suspected. Only one of these clusters contained solely foreign-born patients; the remaining clusters contained either U.S.-born patients or a combination of both U.S.- and foreign-born patients.
TABLE 4.
Isolates clustered by the combination of spoligotyping and IS6110 fingerprinting but discriminated by MIRU typing
| Cluster | Spoligotype | IS6110 band no.a | MIRU pattern | No. of patients | Birthplace | Length of residency prior to diagnosis (yr) |
|---|---|---|---|---|---|---|
| A | 777777777413731 | 1 | 254326223513 | 1 | Somalia | 2 |
| 264226223513 | 1 | Somalia | 1 | |||
| 264225223523 | 1 | Cambodia | 21 | |||
| B | 477777777413071 | 1 | 253126223534 | 1 | India | 1 |
| 254326223434 | 1 | India | 0 | |||
| 254326223634 | 1 | India | 5 | |||
| C | 474377777413771 | 1 | 24432a223533 | 1 | India | 1 |
| 25432a223533 | 1 | India | 4 | |||
| D | 777777777760601 | 2 | 224325153323 | 1 | United States | |
| 22432-151223 | 1 | United States | ||||
| E | 777776777760771 | 2 | 125325143225 | 2 | United States | |
| 225325153323 | 1 | United States | ||||
| 222325153314 | 1 | United States | ||||
| F | 777776777760601 | 2 | 224325143323 | 1 | United States | |
| 224325153223 | 1 | United States | ||||
| 224325153324 | 1 | United States | ||||
| 224325151223 | 1 | United States | ||||
| 22432a151223 | 1 | United States | ||||
| G | 737776777760601 | 2 | 223325153324 | 3 | United States | |
| 224325153323 | 1 | United States | ||||
| H | 700076777760700 | 5 | 224225153324 | 1 | United States | |
| 224325153324 | 1 | United States | ||||
| I | 7777777777760771 | 9 | 223125143324 | 1 | United States | |
| 223125153324 | 1 | United States | ||||
| J | 703707740003771 | 13 | 226425173533 | 2 | India | 0.1 |
| 225425173533 | 1 | India | 3 | |||
| K | 000000000000371 | 15 | 223325143535 | 1 | Laos | 13 |
| 223326143535 | 1 | United States |
All isolates in clusters A to C had the same one-band IS6110 fingerprint (1.4 kb), and all isolates in clusters D to G had the same two-band IS6110 fingerprint (1.4 and 4.5 kb).
DISCUSSION
IS6110 fingerprinting and spoligotyping, individually and together, have been evaluated extensively for genotyping M. tuberculosis isolates to detect recent transmission. A newer method, MIRU typing, has not been evaluated in conjunction with the other methods on a large set of consecutive isolates from a single site. The results presented here show that combining multiple genotyping methods provides the greatest level of discrimination and therefore increases the probability that the clustered isolates are epidemiologically related.
While the authors of previous studies concluded that MIRU typing generated results similar to those obtained by IS6110 and suggested that MIRU typing is an efficient alternative to IS6110 fingerprinting (13), our studies have consistently shown that the addition of any secondary typing method to MIRU typing reduces clustering. In a study of 180 isolates from Michigan with low copy numbers of IS6110 collected over a 5-year period, MIRU typing clustered 120 isolates; the addition of spoligotyping or IS6110 fingerprinting reduced the number of clustered isolates by 14.2 or 19.2%, respectively (3). Even though IS6110 fingerprinting provides the greatest levels of discrimination, MIRU typing can differentiate between some isolates with matching IS6110 patterns. A recent study reported that a set of 12 Beijing isolates with the same nine-band fingerprint had two MIRU patterns; this finding contradicted the notion that isolates with matching high copy number fingerprint patterns were clonal (10). Unfortunately, there was insufficient patient information available to support the conclusion that these isolates were truly from epidemiologically unrelated patients as opposed to isolates from related patients with a change in one MIRU locus. In the present study, there were three clusters of isolates with matching spoligotypes and IS6110 fingerprint patterns with seven or more bands that each contained two MIRU patterns. All demographic and epidemiological data indicated that these isolates were from epidemiologically unrelated patients.
Spoligotyping and, to a lesser extent, MIRU typing and IS6110 fingerprinting have been useful in characterizing genotype families of M. tuberculosis (e.g., Beijing and Manila). Although MIRU typing can discriminate members of these two families, certain MIRU types are more common. IS6110 fingerprinting is more discriminatory in these two families. Thus, it is difficult to predict the utility of the combination of spoligotype and MIRU type in regions where one of these genotype families predominates. In the United States, half of the TB patients are foreign born and there is a pool of diverse M. tuberculosis strains from around the world. However, endemic strains exist even in the United States, and some common IS6110 fingerprint patterns, e.g., the common two-band pattern (1), are not predictive of transmission. It is likely that common spoligotype and MIRU patterns that define large clusters of possibly unrelated isolates will emerge.
Due to differences in the capillary electrophoresis systems, our MIRU typing protocol differs from previously published studies (15). Because the CEQ8000 system allows sizing of fragments only up to 640 bp, new primers were designed to produce smaller amplicons. Primarily due to the fluorescent dyes available for analysis in the CEQ8000, the system does not easily accommodate multiplexed amplifications, but instead requires that individual amplification reactions be pooled for analysis. Although the raw data generated is different from the published multiplex method, the two methods generate the same final result, as shown by a set of isolates that were MIRU typed by using both methods (13).
While MIRU patterns can be determined by using conventional agarose gel analysis, the capillary electrophoresis systems provide automated analysis and increased throughput. Although all spoligotyping data in this paper were generated by using the standard membrane hybridization method, we are now employing a high-throughput spoligotyping assay performed by using microsphere-based multianalyte profiling technology (2). These high-throughput genotyping techniques have many important advantages. Both the technical skill required and the time involved in preparing the DNA samples for genotyping are dramatically reduced compared to IS6110 fingerprinting. Primary cultures, grown either on Lowenstein-Jensen slants or in liquid medium, can be used. Since the assays are highly automated, use mostly commercially provided reagents, and require fewer manipulations from the user, genotyping is easier to perform and the results are more reproducible. Reproducibility is also increased due to the nearly automated analysis of the data. A simple spreadsheet program was used in this study to automatically convert the size of each amplified locus to the number of MIRU copies at that locus and then to a final 12-character MIRU pattern without requiring any manual recording of results. A similar spreadsheet program is available for automated spoligotyping. The final results obtained for both spoligotyping and MIRU typing are digital and much easier for the end user in the TB control program to receive and compare. Automated analysis also helps to reduce the turnaround time. It is our experience that both spoligotyping and MIRU results can be obtained within 48 h of receiving the isolate.
Choosing the optimal approach for genotyping is dependent upon the needs of the program. To provide universal genotyping services in the United States, an approach is needed that will allow a small number of laboratories (currently two) to cost-effectively genotype a large number of isolates in a timely manner (<2 weeks). Although IS6110 fingerprinting had the lowest clustering rate (29.7%) as a single typing method, both spoligotyping and MIRU typing would still be required to provide the greatest levels of discrimination. In this study, IS6110 fingerprinting clustered 77 isolates, of which 45 had low copy numbers of IS6110 (less than seven bands). Experience has shown that such clusters do not reliably predict epidemiological relationships, and secondary typing is required to increase discrimination. Also, our data show that some isolates with matching high copy number IS6110 fingerprints can be discriminated by MIRU typing. In addition, the clustering rate of 29.7% was based on exact matches for IS6110 fingerprints; because IS6110 fingerprints do change with time, the use of exact matches only will exclude some cases that are part of transmission chains. IS6110 fingerprinting is not conducive to high-throughput studies nor does it have a rapid turnaround time, and the resulting patterns must be analyzed and arbitrarily named by the laboratory. Neither spoligotyping nor MIRU typing alone provides suitable discrimination, but when the two methods are combined, the clustering rate is similar to that of IS6110 fingerprinting. The rates of change of spoligotypes and MIRU patterns appear to be less than that for IS6110 fingerprints, implying that a smaller percentage of related isolates will be missed because of changes in one of these two markers if exact matches are used to define clusters.
These results suggest that the two-step approach will be both feasible and useful for universal genotyping in the United States. While IS6110 fingerprinting will continue to be necessary to provide the greatest level of discrimination, the number of isolates requiring this lengthy and laborious technique can be greatly reduced by the two-step genotyping approach. To maximize efficiency for the TB control programs, the results of primary typing will be immediately reported to the program for evaluation in conjunction with patient data. For routine purposes, clustering of isolates based on the spoligotype and MIRU type may be sufficient to confirm suspected links based on contact investigations, further reducing the need for IS6110 fingerprinting. Generally, IS6110 fingerprinting will not be performed when contact investigations have already identified clear links between clustered cases or when it is unlikely that clustered cases are linked due to disparate geography, patient demographics, or risk factors. This approach will be most effective when applied to limited geographic areas, such as single states or metropolitan areas. Comparing isolates from larger geographic areas or the entire country will likely increase the number of clusters based on spoligotype and MIRU type and decrease the significance of the clusters for TB control. However, because the spoligotype and MIRU patterns are digital, it will be easy to monitor the national data set for specific patterns.
REFERENCES
- 1.Cowan, L. S., and J. T. Crawford. 2002. Genotype analysis of Mycobacterium tuberculosis isolates from a sentinel surveillance population. Emerg. Infect. Dis. 8:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowan, L. S., L. Diem, M. C. Brake, and J. T. Crawford. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford, J. T., C. R. Braden, B. A. Schable, and I. M. Onorato. 2002. National Tuberculosis Genotyping and Surveillance Network: design and methods. Emerg. Infect. Dis. 8:1192-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronin, W. A., J. E. Golub, L. S. Magder, N. G. Baruch, M. J. Lathan, L. N. Mukasa, N. Hooper, J. H. Razeq, D. Mulcahy, W. H. Benjamin, and W. R. Bishai. 2001. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis with low copy numbers of IS6110. J. Clin. Microbiol. 39:3709-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standard nomenclature. Int. J. Tuber. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 7.Douglas J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. A. van Embden, D. van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwara, A., Schiro, R., L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. R. Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazars, E., S. Lesjean, A.-L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molhuizen, H. O. F., A. E. Bunschoten, L. M. Schouls, and J. D. A. van Embden. 1998. Rapid detection and simultaneous strain differentiation of Mycobacterium tuberculosis complex bacteria by spoligotyping. Methods Mol. Biol. 101:381-394. [DOI] [PubMed] [Google Scholar]
- 13.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 15.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 35:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 17.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Soolingen D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]