Abstract
Avoiding capture from a fast-approaching predator is an important survival skill shared by many animals. Investigating the neural circuits that give rise to this escape behavior can provide a tractable demonstration of systems-level neuroscience research for undergraduate laboratories. In this paper, we describe three related hands-on exercises using the grasshopper and affordable technology to bring neurophysiology, neuroethology, and neural computation to life and enhance student understanding and interest. We simplified a looming stimuli procedure using the Backyard Brains SpikerBox bioamplifier, an open-source and low-cost electrophysiology rig, to extracellularly record activity of the descending contralateral movement detector (DCMD) neuron from the grasshopper’s neck. The DCMD activity underlies the grasshopper’s motor responses to looming monocular visual cues and can easily be recorded and analyzed on an open-source iOS oscilloscope app, Spike Recorder. Visual stimuli are presented to the grasshopper by this same mobile application allowing for synchronized recording of stimuli and neural activity. An in-app spike-sorting algorithm is described that allows a quick way for students to record, sort, and analyze their data at the bench. We also describe a way for students to export these data to other analysis tools. With the protocol described, students will be able to prepare the grasshopper, find and record from the DCMD neuron, and visualize the DCMD responses to quantitatively investigate the escape system by adjusting the speed and size of simulated approaching objects. We describe the results from 22 grasshoppers, where 50 of the 57 recording sessions (87.7%) had a reliable DCMD response. Finally, we field-tested our experiment in an undergraduate neuroscience laboratory and found that a majority of students (67%) could perform this exercise in one two-hour lab setting, and had an increase in interest for studying the neural systems that drive behavior.
Keywords: descending contralateral motion detector (DCMD), eye, electrophysiology, grasshopper vision, looming, escape response, DIY, mobile application, neuroethology
Undergraduate neuroscience programs often face the challenge of incorporating meaningful and engaging hands-on learning experiences into their curricula on limited budgets and resources. Many neuroscience educators have developed undergraduate-level teaching labs, particularly utilizing invertebrates as tractable models to teach neuroscience fundamentals. As demonstrated by Li et al. (2014), behavioral observations of well-studied neural systems, such as how a transection of the hindbrain or pharmacological factors can affect the swimming behavior of Xenopus laevis tadpoles, can provide an effective undergraduate lab exercise. For introducing basic electrophysiology concepts to students, the giant fibers in earthworms have been used in undergraduate classrooms to demonstrate how reflexes are elicited when the worms’ skin sensory cells are stimulated by touch (Kladt et al., 2010; Shannon et al., 2014). Cockroaches and crickets have also been a reliable model system to demonstrate fundamental concepts and techniques in neurophysiology, such as electrical stimulation and recording of axons in the cockroach’s leg, ventral nerve cord, and antenna (Ramos et al., 2007; Dagda et al., 2013; Marzullo and Gage, 2012). Instructors have also translated crustacean research into suitable undergraduate exercises, ranging from the behavioral and physiological experiments to computer simulations (Johnson et al., 2014). An online video-based lab manual for neurophysiology, called Crawdad, has been published to provide undergraduate instructors and students with various lab exercises that are inexpensive and engaging (Wyttenbach et al., 2014).
While recording action potentials from the peripheral nervous system of invertebrates demonstrates several core neuroscience concepts, often the students only record from neurons that reflect the encoding of sensory input, instead of the complete neural pathway, including the interneurons and output neurons that produce the neural signals that give rise to an observable animal behavior. An alternative approach to understanding neural activity is to capture the output that reflects this neural processing and to explore the controlled parameters that give rise to the observed responses. The grasshopper descending contralateral motion detector (DCMD) system is one way to introduce concepts relating input and output to behavior.
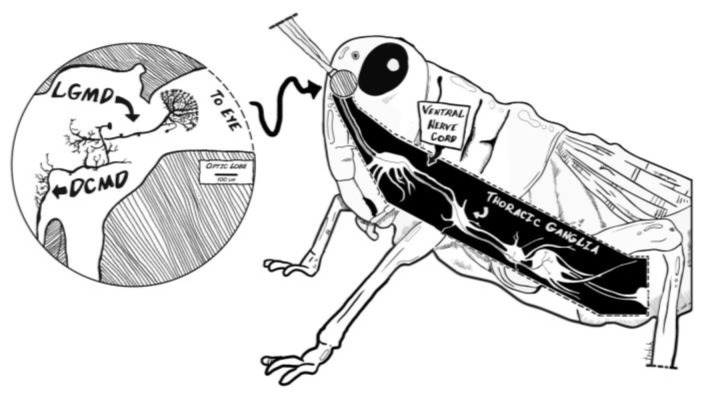
By studying the DCMD system, students also learn the role of natural selection in the evolution of the nervous system. The ability to avoid collision with surrounding objects or escape a predator is crucial to the survival of an animal and typically requires the visual detection of approaching stimuli and a motor response (Pearson and O’Shea, 1984; Card, 2012). When sensing a rapidly approaching object, grasshoppers escape by extending their powerful hind legs for a quick and strong jump. Flying locusts also respond to objects approaching one eye by steering and sweeping their wings into a gliding posture (Simmons and Young, 1999; Santer et al., 2005). In the grasshopper visual system (Fig. 1), two well-studied monocular motion-detecting neurons, the lobula giant movement detector (LGMD) and the descending contralateral movement detector (DCMD), underlie these characteristic escape responses (Rowell, 1971a; O’Shea et al., 1973; Rind and Simmons, 1999). The large fan-shaped LGMD resides in the optic lobe and is excited by the visual stimuli of approaching objects on a collision course with the animal. This neuron is highly sensitive to an object moving towards the eye by integrating two main features of the object’s appearance: whether the edges of this object increase or decrease in length, and if these edges are accelerating as they move across the retina (Simmons and Young, 1999). Then, the LGMD sends these neural signals to their postsynaptic targets in the hindbrain, the DCMD, which responds to object movements detected by the contralateral (opposite) eye. This in turn activates the appropriate motor neurons and interneurons in the thoracic ganglia to contribute to the hind leg flexion and preparation for the jumping response (Santer et al., 2008). The LGMD and DCMD together make up an early warning system to prepare the escape behavior in the face of possible collision with objects that exhibit small movements in the eye’s visual field (Rind and Simmons, 1992; Hatsopoulos et al., 1995; Gabbiani et al., 1999; Simmons and Young, 1999).
Figure 1.
Neuroanatomy of the grasshopper’s motion detector neurons. The grasshopper optic lobes (lamina, medulla and lobula) lie in the central brain beneath each of the compound eyes and processes visual information. The lobula giant motion detectors (LGMDs) receive visual signals from the lobula and pass these inputs to the descending contralateral motion detectors (DCMDs). The LGMDs respond to motion of an object seen moving across the insect eye as well as to the looming effect, where an object increases in size as it approaches the eye. The DCMD activates motor neurons in the thoracic ganglia. These primary motion detector neurons underpin the animal’s detector and motor response to a looming object, forming the see-and-jump escape mechanism.
The DCMD underlies the grasshopper’s motor responses to looming monocular visual cues perceived by their eyes, and provides an excellent systems-level view of a decoding problem being computed by the grasshopper brain in an escape response. It is also easy to extracellularly record from this neuron by placing electrodes around a nerve cord, due to the relatively large spikes (large amplitude and high bursting firing rate) produced from the DCMD axons (Simmons and Young, 1999). In this paper, we present a tractable electrophysiology procedure using an Apple iPad and a low-cost, open-source electrophysiology kit called the SpikerBox (Marzullo and Gage, 2012) from Backyard Brains (BYB, backyardbrains.com) that extracellularly records and analyzes the firing rate of the grasshopper DCMD neuron. The SpikerBox provides connections to electrodes for recording electrophysiological responses from an invertebrate preparation, and includes a bioamplifier that connects to a smartphone, tablet, or computer. We also describe our software package, SpikeRecorder, which is currently available for iOS systems and generates visual looming stimuli while simultaneously recording neural activity. We provide analysis tools within the app, as well as separate MATLAB code, to perform spike-sorting of the DCMD unit and to analyze the neuronal activity. This software, along with the SpikerBox schematics, are open-source and available on the Backyard Brains website (backyardbrains.com). Alternative neurophysiology amplifiers can also be used, for example Land et al., (2001) describes a low-cost but high-performance amplifier and electrodes for extracellular recording. Also, high-precision micromanipulators for insect retinal physiology can be used for electrode placement (Krans et al., 2006).
We present three exercises for undergraduate labs: 1) Recording of DCMD signals when the grasshopper is exposed to approaching visual stimuli, 2) Finding the minimal inter-trial interval, meaning the minimum amount of time that is required between the presentation of the last visual stimuli trial and the subsequent trial (durations shorter than this minimum result in the grasshopper habituating to the repetition of presented stimuli and a substantial reduction in the DCMD responses), and 3) Testing the effect of visibility of the stimulus on DCMD response by adjusting screen brightness and therefore stimulus contrast against the background. These lab exercises are designed to guide students from an observational question like “why are grasshoppers hard to catch?” to quantifiable questions regarding the visual detection of dangerous objects using electrophysiology. Students can investigate the electrical transmission of highly processed visual information for this life-saving “see-and-jump” escape mechanism. Each exercise is estimated to take about two hours total, one for the animal preparation and another for the electrophysiological recording and data analysis, which can also be done as homework if necessary. We also provide results from our experiments to show the efficacy of the tools.
With these simple techniques and affordable equipment, students can observe neural responses to visual stimuli in a behavioral context in an invertebrate model and understand how such a behavioral output is produced by a network of interconnected neurons, from the visual to the motor systems.
MATERIALS AND METHODS
Animals & Electrophysiology Preparation
Many species of grasshoppers can be used, including various species of Schistocerca (e.g., Burrows and Rowell, 1973; Hatsopoulos et al., 1995) and Locusta (e.g., Rind and Simmons, 1992; Gray et al., 2001). In some locations outside the USA, grasshoppers are available for purchase as feeder insects and are inexpensive. Grasshoppers can also be found and caught in nearby grass fields by the students, which will provide context for the lab exercises as the activity allows for direct observation of the insects’ collision avoidance behavior in nature. Adult grasshoppers are preferred for their relative large size and ease of handling. However, we have had great success on small grasshoppers as well. The animals should be refrigerated (suggested temperature: 3°C) overnight until the exercises.
To prepare for extracellular recording of the DCMD neuron, further anesthetize the grasshopper in ice for 15 minutes or until the insect is inactive after storage in the fridge. Then, use masking tape to tape the animal ventral (belly) side up onto the corkboard on the SpikerBox apparatus with its head and part of the thorax exposed. Pull back its head using standard sewing thread and tape the thread to the SpikerBox to expose the neck connectives (Fig. 2). If the thread pulling back the grasshopper’s head does not stay in place, heat up a small mixture of beeswax and rosin (a teaspoon total) on a glass petri dish on a heat plate and use a sewing needle or similar to place a dab of the warm liquid on the thread, gluing it to the grasshopper’s neck. This mixture will dry and secure the thread restraint, and is quick and easy to remove from the animal after the exercise.
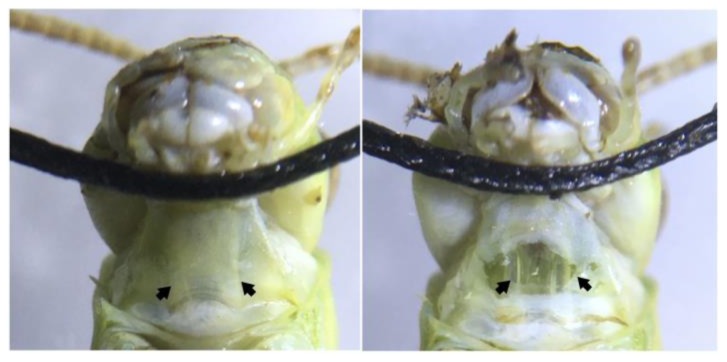
Figure 2.
Ventral view of the grasshopper neck showing the neck connectives from the head to the thoracic ganglia. (Left) Neck connectives (pointed to by arrows) are visible as white stripes under the neck skin. Sewing thread is used to hold the grasshopper’s head back and in place during the experiments. (Right) A simple neck cut exposes the connectives, two translucent tubes, where the part of the ventral nerve cord can be found. Placing a hook electrode around one connective and exposing the contralateral eye to looming visual stimuli allows DCMD activity to be recorded.
The neck connectives through which the DCMD axons pass can be seen with the naked eye, but a dissecting microscope (suggested magnification: 20X) should be used for more precise visualization and electrode placement. Using a 0.3mm insect pin, make a small incision in the middle of the neck. Modify the straight silver wire recording electrode (0.127mm) that comes with the BYB kit into a hook by bending the tip into a hook shape with tweezers. Then, guide the recording hook electrode with a micromanipulator (see 3D printed micromanipulator, Backyard Brains, Ann Arbor, MI) into the incision and around the neck connective contralateral to the eye that will be exposed to the visual stimuli (Fig. 3). When the electrode is in place, put a small dab of Vaseline on the incision to keep it from drying out. We fill a 1ml syringe with a small broken needle with Vaseline to make application to the recording site easier. Ground the reference electrode (sewing needle, 0.6mm diameter) in the grasshopper’s thorax. To check for proper electrode position, record DCMD responses (listen for popcorn-like pops on the speaker connected to the SpikerBox) when the grasshopper is exposed to an approaching object (e.g., your hand or a pen).
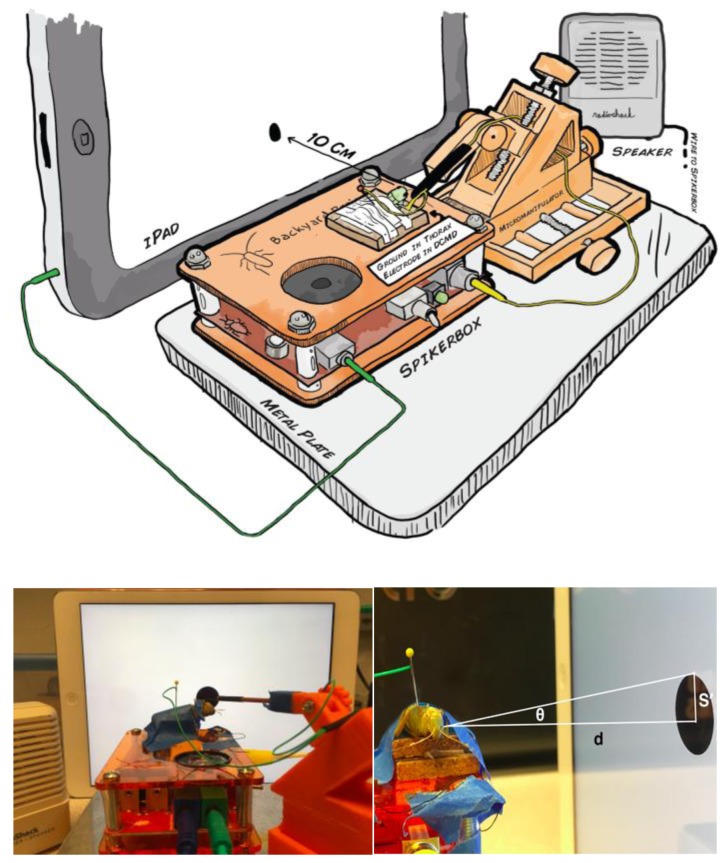
Figure 3.
Experimental setup to measure descending contralateral movement detector (DCMD) activity during simulated stimulus presentation. (Top) Schematic representation of the experimental setup, grasshopper is taped down onto the corkboard on top of the SpikerBox. The reference electrode is grounded at the thorax and the recording hook electrode, guided by the 3D printed micromanipulator for precision, is placed around the neck connective where the ventral nerve cord and DCMD neuron’s axon is found. The iPad presenting the visual stimuli is placed 10cm from the grasshopper’s eye and the angle between the grasshopper’s eye and the center of the iPad is minimized. (Bottom left) Photograph of the apparatus. (Bottom right) Visual stimuli are displayed on the iPad to simulate the looming effect of an object approaching the grasshopper. The radius (S′) and subtending angle (θ) of a filled black dot can be manipulated to simulate a fixed sized object (S) approaching at a particular constant velocity (v) towards the eye of the insect a fixed distance (d) from the screen. DCMD recordings can be made during visual simulations of different sized objects moving at different velocities.
Finally, for the experiment to begin, the longitudinal axis of the grasshopper’s body should be oriented parallel to an Apple iPad 10cm away such that the angle between the eye and the center of the screen is as minimal as possible (Fig. 3 top). A spirit level can be used to easily minimize the angle by lining the level edges to the center of the iPad and the grasshopper’s eye and adjusting until the bubble in the horizontal tube is centered. To achieve the upright position of the iPad, we suggest simply taping it against a wall. If possible, recordings should be done in the dark for better contrast intensity of the experimental stimuli, and to isolate it from possible visual distractions. Placing the apparatus inside a cardboard box can easily achieve this. This setup (Fig. 3) is the initial step for all three exercises described below and materials are listed in Table 1. This preparation stage should take about one hour. See Supplementary Information at the end of this paper for a link to a video that describes these procedures.
Table 1.
A full list of materials for the animal preparation and spike recording and analysis for all three proposed lab exercises.
Materials
| Animal Preparation | Ice |
| Microscope (suggested: 20X or higher) | |
| Vaseline | |
| Sewing thread (or similar) | |
| Masking tape (or other types) | |
| Insect pin (0.3mm) | |
| Micromanipulator (BYB) | |
| Spirit level with horizontal tube | |
| Standard ruler | |
| Included with BYB Micromanipulator: | |
| Recording electrode (silver wire, 0.127mm, modify into hook with tweezers) |
|
| Reference electrode (sewing needle, 0.6mm diameter) |
|
| Optional: | |
| Beeswax | |
| Rosin | |
| Magnetic stirrer | |
|
| |
| Spike Recording & Analysis | iPad (Apple), iPhone, or laptop |
| Neuron SpikerBox (BYB) | |
| Green smartphone cable (BYB, SpikerBox to tablet) |
|
| SpikeRecorder application (BYB, currently available for iOS) |
|
| Speaker (suggested: RadioShack mini amplifier) |
|
| Blue audio cable (from SpikerBox to speaker) |
|
Visual Stimuli & Spike Recording, and Sorting
Electrophysiological measurements are carried out here using the Neuron SpikerBox (Backyard Brains, v.1.3c). The SpikerBox has a 4x gain instrumentation amplifier, 220x gain amplifier bandpass-filtered 340–1300Hz, and a 20x gain audio amplifier (Marzullo and Gage, 2012). It also contains an output port for a Radioshack mini speaker (blue port and cable) and an output port for laptops, tablets, and smartphones (green port and cable) to make neural activity visible and recordable. During all exercises, the blue and green cables from the SpikerBox should be connected to the speaker (optional) and iPad respectively.
The BYB SpikeRecorder iOS application provides both the monocular visual stimuli and recording of DCMD activity. Visual stimuli consist of expanding dark dots (of various object sizes, S, and velocities, v) on a white background to simulate an approaching and colliding object on a direct collision course. As S increases over time, so does the angle that subtends the grasshopper’s eye (Fig. 3 bottom right). This change in angular size of the dot describes the movement of the object towards the animal. Adjustable parameters in the application include: number of trials, distance between subject and screen (cm), an array of object sizes (S, cm) and velocities (v, m/s), inter-trial interval or ITI (s). The ITI is the amount of time between the end of the previous visual display and the commencement of the next looming trial. The iPad displays a blank white screen during this interval. During an experimental session, stimuli with parameters S and v are pseudo-randomly presented by the application such that each parameter pair is repeated an equal number of times.
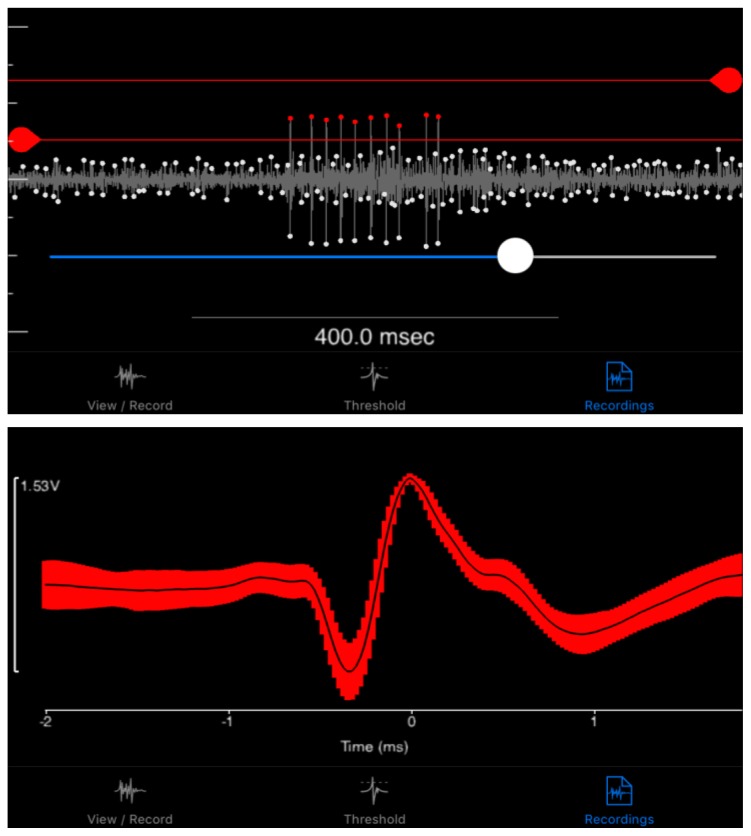
Spikes can be sorted from noise after recording in the iPad application (Fig. 4). Noise is identified as false spikes with constant and consistent amplitudes (mV) over time. These spikes have noticeably lower amplitude than the real DCMD spikes. The app allows the student to set minimum and maximum threshold values (mV), and spikes within the range are counted as spikes for further analysis. After sorting the spikes from one recording for one trial (one S, one v), the app automatically performs global sorting for the rest of the trials conducted during one experimental session using these threshold parameters.
Figure 4.
Spike sorting in the SpikeRecorder iPad app. SpikeRecorder provides both the looming visual stimuli and recording and analysis of DCMD activity. DCMD spikes can be sorted from noise after recording by providing minimum and maximum spike threshold values. These values can be applied to all recordings made within a single grasshopper’s experimental session. (Top) The app’s output of the DCMD recordings during a looming experiment (object size S = 6cm, velocity v = −2m/s) with a minimum spike amplitude of approximately 0.30mV, maximum of 0.65mV. Red dots are spikes recorded within the thresholds. (Bottom) The app can also analyze data after spike sorting. Shown here is an example graph showing the average waveform of DCMD activity over time, within 1 standard deviation.
Exercise 1: Looming Stimuli DCMD Experiments (1hr)
Objective: This exercise, coupled with instructors’ lectures and introduction to neuroanatomy of the grasshopper visual-motor system, demonstrates to students how sensory input might stimulate a dedicated neuronal system to drive a certain animal behavior. Students can follow the procedure below, with modifications appropriate for a classroom lab or other learning settings. Experiments can begin after students develop their own hypotheses, regarding the time in which the DCMD neuron would peak in the firing frequency when the object approaches the grasshopper’s eye, or how different object sizes and velocities may affect this DCMD firing response. For example, we might expect the DCMD firing to increase and peak before the virtual object would collide with the grasshopper’s eye. This would ensure a successful jump away from danger. What would we observe with objects of different sizes and approaching at different speeds?
Procedure: In the SpikeRecorder application, keep the following parameters constant: 10cm distance between the grasshopper’s eye and the iPad and inter-trial intervals, ITI, of 45s to reduce habituation of the DCMD response to visual stimuli. For the required values of choice for S (object radius) and v (object approaching velocity), we suggest choosing S of 6cm and a range of v values (−2, −4, −6, −8m/s) to begin. Each combination of S and v should be repeated for at least three simulation trials. Such parameters will create an experiment, approximately 10 minutes in duration, that allows students to observe the differences in DCMD response to objects of the same size but approaching at different velocities. Once the application is set up, check again for the correct iPad positioning and connected cables as described above and press ‘Start’ in the iPad app to begin. Observe the approach of the black dots and listen for DCMD spikes in form of popcorn popping sounds through the speakers.
Exercise 2: Inter-trial Interval Experiments (15min)
Objective: In this exercise, students will be exposed to the concept of inter-trial intervals (ITIs), the time between the stimuli presentations. ITI times are often important in behavior studies, as subjects may need a fixed amount of time to respond to the next stimuli. This is indeed the case in DCMD experiments (Rind and Simmons, 1992; Hatsopoulos et al., 1995). ITIs ensure that each trial’s stimulus is distinct and thus the animal would not develop habituation to the presented stimuli, preventing accurate DCMD responses. Instructors can discuss the use of ITIs of at least 40s used in literature (e.g., Rind and Simmons, 1992). Students can hypothesize the effect of different ITIs on peak DCMD activity, such as an marked decrease in bursting activity when there is no ITI, a short ITI (less than 10s) or a medium ITI (20s). Students can subsequently implement an experiment to investigate their predictions, and determine a sufficiently long interval to avoid habituation to the stimuli.
Procedure: With the same setup as Exercise 1, enter parameters to be kept constant for this experiment in the application: 10cm distance between eye and iPad, chosen number of trials per ITI, and one pair of S and v values. We recommend using three different ITIs, repeating each at least three times, to produce distinctly different DCMD responses: 45s, 22.5s, and 1s. This recording should take approximately 10–15 minutes.
Exercise 3: Screen Brightness Experiments (30min)
Objective: This exercise aims to engage students in further experimental inquiry after they have performed the previous two activities. From a neuronal perspective, contrast of the stimuli against the background is important for the DCMD response, as it shows that the neuron is encoded to respond to particular stimuli depending on the environmental conditions. Here, they can hypothesize and test the effect of screen brightness, which affects the contrast intensity and thus visibility of the black stimuli against the white background, on DCMD response. Can the grasshoppers effectively see a darken screen? Would there be a difference in DCMD activity and/or the time at which it peaks?
Procedure: Adjust brightness of the screen in the Settings of the iPad, whose brightness is approximately 425 cd/m2 at maximum and 5 cd/m2 at minimum. Constant parameters include 10cm distance between eye and iPad, 45s ITI, number of trials per brightness level, and one pair of S and v values. Students may choose different brightness levels, each is a separate experiment. We recommend initially observing the effects of the maximum and minimum screen brightness levels (the iPad screen will not be completely black at the minimum setting, but very dim). If ten stimulation trials are chosen for each brightness level, the recording for each brightness level should take about 10 minutes.
Data Analysis
The SpikeRecorder app includes spike sorting, data analysis and visualization functionality. DCMD activity and change in angular size of the visual stimulus over time can be visualized in perievent time histograms or PETHs (peri-stimulus time histograms, see Bretschneider and De Weille, 2006 for a review) and raster plots, with time of collision defined as the 0 second point. Students may save the data graphs directly from the app or engage in deeper analysis for a post-lab assignment.
For more in-depth analysis, data recorded and contained within the SpikeRecorder application can be exported to JSON files, which can then be imported into MATLAB (MathWorks, Inc.) using the open-source JSONlab toolbox. The MATLAB code files are available for download from the Backyard Brains GitHub repository (see Supplementary Information). A database of the experimental data (recording and spike timestamps, stimulus angles, time of collision) will be created by the provided code. Perievent time histograms and raster plots can then be plotted to visualize firing rate and timing of spikes in relation to stimulus. All datasets used in this paper as well as all analysis tools are provided in the supplementary repository link at the end of this paper.
Student Laboratory & Evaluation
During the Fall 2016 semester, students in the MCB81 course, Fundamentals of Neuroscience, taught by David Cox at Harvard University participated in a 2-hour classroom laboratory with a brief lecture and lab performing the first exercise described above. Data analysis to produce histograms and rasters was done strictly within the iOS App, and not via the advanced MATLAB tools provided. Students were sent an online survey a week after the lab to rate the effectiveness of the various aspects of the exercise, such as the ease of setting up the grasshopper preparation and whether the lab section improved the students’ understanding of the use of electrophysiology to study neural mechanisms of behavior. The answers were on the scale of ‘1=Strongly agree’ to ‘5=Strongly disagree.’ The overall rating at the end of the survey asked students to rate the whole lab as ‘1=Terrible’ to ‘10=Excellent.’ The last question asked students for ideas for improvements. Student learning assessment was not directly measured in this study.
RESULTS
We performed the three exercises described above to verify the efficacy of our equipment, to refine the protocol, as well as provide results as points of reference for instructors and students.
Animals
In our experiments, due to dependence on local availability and catching abilities, both adult and nymph, male and female, grasshoppers (Melanoplus femurrubrum and Melanoplus differentialis) were caught in Nichols Arboretum, MI, USA, in June and July 2016. The animals were refrigerated (average temperature 3°C) until the experiments (average 4 days).
Exercise 1: Looming Stimuli DCMD Experiments
In this replication of literature DCMD studies by Rind and Simmons, 1992 and Hatsopoulos et al., 1995, 22 grasshoppers were tested across 57 recording sessions. An average of 2.62 sessions was conducted on each subject (range = 1 to 5 sessions). Each grasshopper was individually exposed to visual stimuli on the iPad while the SpikeRecorder application recorded DCMD activity amplified by the SpikerBox. Stimuli virtually approached the grasshopper’s eye on a direct collision course. Session parameters kept constant across all DCMD experiments were: 10cm distance between the grasshopper’s eye and the iPad; 16 trials per pair of S and v; and inter-trial intervals, ITI, of 45s to reduce habituation of the DCMD response to visual stimuli. Each stimulus is a pair of S (6 and 8cm) and v (−2, −4, −6, −8m/s). Initially, two separate sessions of stimuli with constant S and varying v were performed on the same grasshopper. However, to prevent the effect of habituation of the DCMD to the stimuli presented later in a second session, the SpikeRecorder application was modified to pseudo-randomly present stimuli of varying S and v throughout a continuous session of approximately 2 hours, 128 trials. Because we were not limited to a classroom timescale, we allowed the experiments to run longer than the amount of trials suggested in Methods to obtain more data for analysis. Students may set up a similar, longer experimental session if desired to gather more data.
We show results of data analysis in both the SpikeRecorder app (Fig. 5) and MATLAB (Fig. 6) for comparison. Students may simply use the data analyses function of the app, or use the export function in the app to use the MATLAB code to gain experience in more sophisticated data analysis and visualization. The SpikeRecorder app (Fig. 5) generates analysis of an extracellular recording of DCMD response for dots 6cm and 8cm in radius approaching at four velocities (−2, −4, −6, −8 m/s) in a perievent time histogram (PETH) and raster plot. The MATLAB code we developed allows data analyses similar to the SpikeRecorder app, with a wider range of options. As shown in Fig. 6, compared to the overall graph from the app (Fig. 5), DCMD activity patterns of a dot of the same size (S: 6cm) approaching at various velocities (v: −2, −4, −6 m/s) can be better visualized in various figure types. Both the SpikeRecorder app and MATLAB analyses provide the following information about the grasshopper DCMD and its role in the insect’s escape mechanism: As shown in Fig. 5 and 6B, baseline DCMD firing rate was silent or very low (below 20Hz) for more than 1s after the onset of the stimulus, then increased approximately 0.2s before the time of collision (0s on all graphs) and peaked around the collision between the object and the eye, when the object’s image on the grasshopper’s retina reached a certain angular size (Fig. 6A & 6B). Note that there are various factors affecting the observed baseline activity, such as different grasshopper preparations (i.e., how clean a contact was made between the electrode and the DCMD axon), and biological differences between individual animals. In addition, the baseline firing rate is also subject to the minimum voltage selected during spike sorting, which is at the discretion of the human analyst.
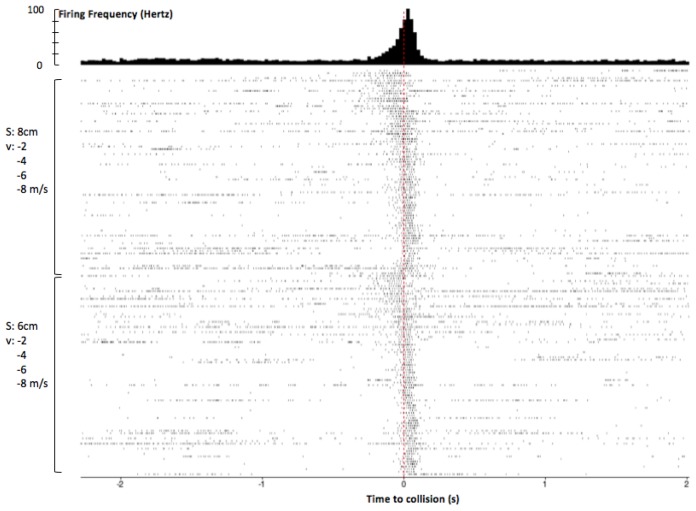
Figure 5.
Mobile app analysis of DCMD response to approaching objects, with additional annotations. After spike sorting, the SpikeRecorder app produces a perievent time histogram (PETH) and raster plot to show DCMD firing activity 2s before and after collision. Peak firing was 92Hz around collision. Rasters are ordered top to bottom by large to small S, then slow to fast v. Similar patterns of DCMD activity are seen for dots 6cm and 8cm in size, with peak firing frequency before collision when the dots are approaching at slower velocities (−2 and −4m/s) compared to those at faster velocities (−6 and −8m/s). Session: G17-071416-01 (see Supplementary Information). Parameters: d: 10cm; S: 6, 8cm; v: −2, −4, −6, −8m/s; trials per pair of S and v: 30; ITI: 45s. Collision at 0s.
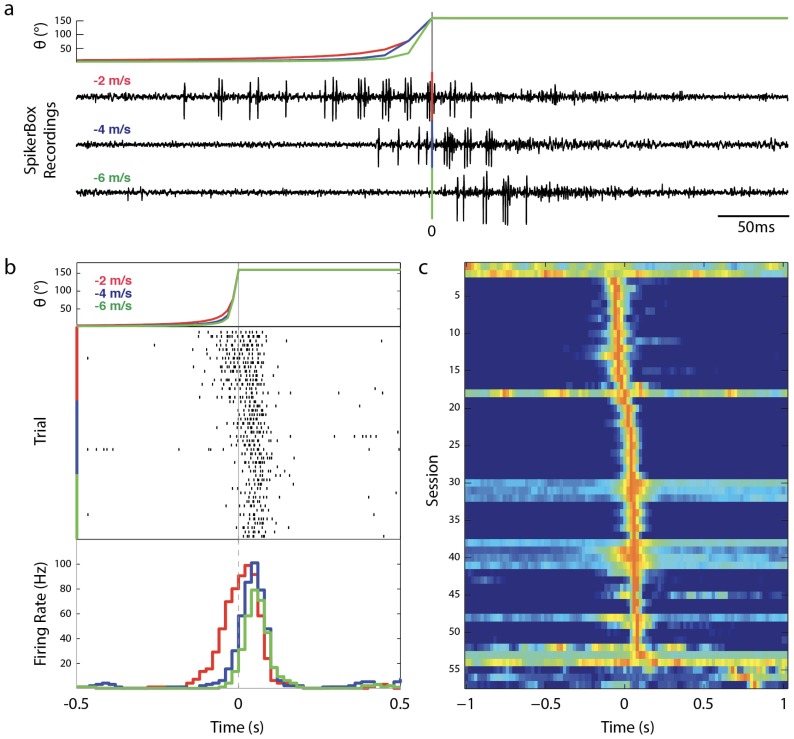
Figure 6.
Analysis of DCMD response to approaching objects in MATLAB. Importing data from the SpikeRecorder app into MATLAB allows further data analysis and visualizations. (a) Comparison of DCMD activity in SpikerBox traces from iPad across 3 values of v: −2, −4, −6 m/s for a fixed S: 6 cm. Theta (θ) is the angle of the approaching object (See Fig 3). DCMD firing rate increases and peaks as the angle of the object subtended on the grasshopper eye reaches maximum. 0s indicates computed collision with grasshopper. Session: G17-071416-01 (see Supplementary Information). Parameters: d: 10cm; trials per S/v pair: 30; ITI: 45s. Collision at 0s. Timescale: 0.5s (b) Spike rasters (center) show DCMD spiking activity (tick marks) across trials within a session and are sorted by velocity. Note delay in DCMD onset and decrease in spike timing variance as velocity increases. Session: G15-071316-01(see Supplementary Information). Parameters: d: 10cm; trials per S/v pair: 30; ITI: 45s. Collision at 0s. Timescale: 1s (c) Session analysis of DCMD responses. Each row shows a false-color representation of DCMD peri-collision firing rate each of the total 57 sessions in our database (bin size = 0.2s). Red indicates peak firing rate. Sessions are sorted by time of peak firing rate. 50 of the 57 sessions (87.7%) had a peak in firing within +/− 0.1 s of collision. Timescale: 2s.
As shown in both Fig. 5 and 6A & 6B, with the looming objects at the slowest velocity (−2 m/s), peak firing occurred approximately before the time to collision between the eye and the stimuli. With increasingly faster velocities (−4, −6, −8m/s), DCMD began firing before the collision, but peaked up to approximately 0.05s later. The greater the velocity of the stimulus, the less spikes were observed with less variance in the spiking activity, as shown in Fig. 6A and 6B comparing stimuli of a fixed S (6cm) approaching at −2, −4, −6m/s. In Fig. 6C, we show a false-color representation graph of DCMD peri-collision firing rate of all 57 sessions from 22 grasshoppers. As seen, 50 of the 57 sessions (87.7%) had a clear peak in firing (red) within +/− 0.1 s of collision.
From a neuroethological perspective, instructors can discuss these results as explaining how the DCMD neuron contributes to its function in the grasshopper’s escape behavior. The more rapid the approaching object (seen in our stimuli at v of −6 and −8m/s), the fewer and later the DCMD spikes. Does this imply a lower chance of successful avoidance of collision, or does a higher firing rate of slower objects inform the grasshopper it has more time to make a final decision?
Exercise 2: Inter-trial Interval Experiments
ITI experiments were carried out with 2 grasshoppers. The setup was the same as Experiment 1. Constant parameters included 10cm distance between eye and iPad, 30 trials per ITI, and one pair of S (6cm) and v (−2m/s). Three ITIs were chosen: 45s, 22.5s, and 1s and the total experiment consisted of 90 trials.
An ITI of 1s yields low and late firing frequency (approximately 20Hz), compared to the frequencies of 22.5s (45Hz) and 45s (75Hz), shown in Fig. 7. The 45s TI showed the most consistent and frequent firing, and is the suggested minimum for the other experiments.
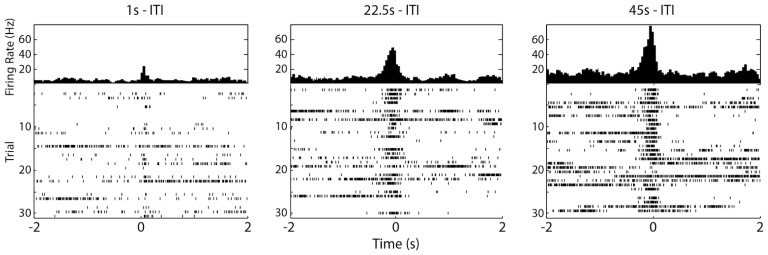
Figure 7.
Varying the inter-trial interval reveals DCMD response needs time to recover. Peak firing frequency is around 20, 45, and 75Hz for 1, 22.5, 45s ITI, respectively. The 45s ITI shows the most consistent and frequent firing of the DCMD, and is the ITI of choice for other experiments. Sessions: G25-072416-01 through G25-072416-03 (see Supplementary Information). Parameters: d: 10cm; S: 6cm; v: −2m/s; trials per ITI: 30; ITIs: 45, 22.5, 1s. Collision at 0s.
Exercise 3: Screen Brightness Experiments
Maximum and minimum brightness levels were set for two separate experiments. Constant parameters included 10cm distance between eye and iPad, 45s ITI, 20 trials per brightness level for 40 trials total, and one pair of S (6cm) and v (−2m/s). As shown in Fig. 8, the DCMD activity profiles at maximum and minimum iPad screen brightness are not substantially different, with 99.2Hz and 96.7Hz peak frequency, respectively. These results suggest that light level of the iPad does not affect how visual stimuli evoke DCMD responses. In dim background lighting, the grasshopper’s eyes could still detect the edges of the expanding dots and react to avoid the looming collision– an advantageous escape behavior in dark, natural settings. Many students may have predicted (as did we) a difference between the brightness extremes, but these results demonstrate that the neurons are encoded to respond to particular stimuli invariant of the environmental conditions. We provide this as a learning example of a negative result in hypothesis testing, but instructors may wish to eliminate this exercise.
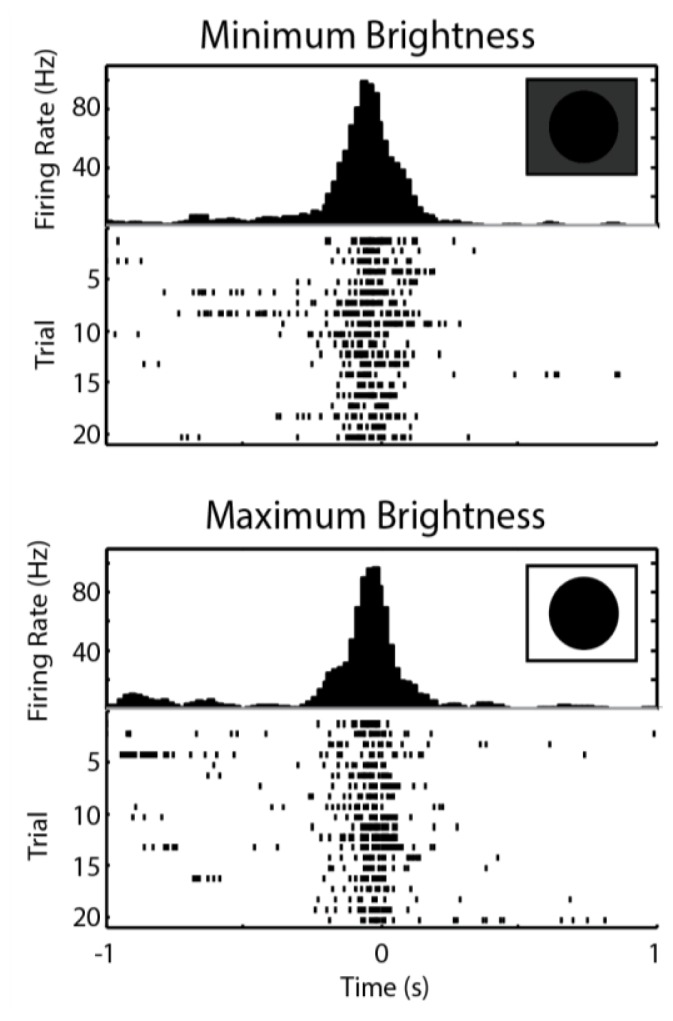
Figure 8.
DCMD shows little difference in response to varying iPad screen brightness. At minimum brightness (top), DCMD firing rate is consistent and peaks at −0.02s with 96.7Hz. At maximum brightness (bottom), firing rate also peaks at approximately the same rate (99.2Hz) and time (−0.06s). There was not a significant difference detected from spiking activity between the dimmest and brightest screen levels (Paired t-test of number of spikes in trials between −0.1s and 0.1s; P = 0.320). Sessions: G26-072516-02 and G26-072516-03 (see Supplementary Information). Parameters: d: 10cm; S: 6cm; v: −2m/s; trials per brightness level: 20; ITIs: 45s. Collision at 0s.
Student Evaluation
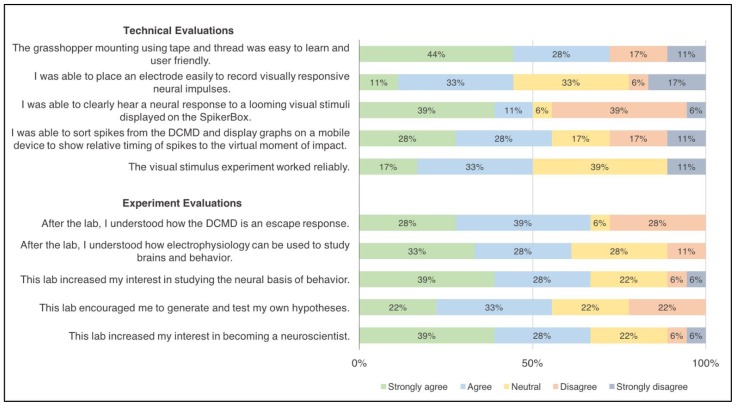
Students generally enjoyed the DCMD exercise and responded positively. Post-lab survey data were collected from 18 undergraduates. For the technical aspects of the experiment, 72% ‘strongly agreed’ or ‘agreed’ that the grasshopper preparation mounting using tape and thread was easy to learn and user friendly (Fig. 9). 50% or more students ‘strongly agreed’ or ‘agreed’ that the visual stimulus worked reliably, that they were able to clearly hear a neural response to the looming stimuli displayed on the SpikerBox, and that they were able to sort spikes from the recordings and display graphs on a mobile device to show relative timing of spikes to the virtual moment of collision. The most challenging part of the protocol might be electrode placement, which only 44% of the students thought was easy.
Figure 9.
Student evaluation of the Looming Stimuli DCMD Experiment. Survey data from 18 undergraduates in the MCB 81: Fundamentals of Neuroscience course taught by David Cox at Harvard University. Most students were able to perform the protocol, including grasshopper preparation and data analysis on a tablet or mobile device, with electrode placement possibly being the most difficult step. The lab enhanced student understanding and interest in electrophysiology as a tool and in neuroscience overall.
As for the evaluation of the lab’s effect on student understanding and interest in neurophysiology, at least 60% of the students ‘strongly agreed’ or ‘agreed’ that the lab increased their interest in studying the neural basis of animal behavior, that they were able to understand how electrophysiology can be used to study neural correlates of behavior, that they understood how the DCMD works as an escape response, that the lab encouraged them to generate and test their own hypotheses and increased their interest in becoming a neuroscientist. Overall, 16 out of the 18 students rated the lab 7 or higher out of 10, with 10 being ‘Excellent.’
DISCUSSION
The three exercises presented in this paper aim to provide classroom laboratories to introduce undergraduates to concepts of neurophysiology using a manageable grasshopper model and a simplified essential tool of neuroscience, electrophysiology. The affordable and simple exercises discussed in this paper could be implemented in undergraduate laboratories to provide students with hands-on activities to better understand the neurophysiology concepts of neuroanatomy, sensory responses, and the electrical properties of the brain in the context of a natural escape behavior grasshoppers exhibit to avoid collisions with predators or avoid other individuals during swarming (Simmons and Young, 1999).
Our results demonstrate the efficacy of the tools; the SpikerBox and SpikeRecorder app successfully reproduced grasshopper DCMD responses seen in the current literature (Rind and Simmons, 1992; Hatsopoulos et al., 1995; Gabbiani et al., 1999). Similar to Hatsopoulos et al.’s 1995 and Rind and Simmons’ 1992 results that show silence in DCMD activity during the first 2s of a 2.75s approach, our results find that the DCMD begins to fire rapidly approximately 1.8s after the stimulus begins the simulated collision course with the grasshopper’s eye. The exact timing of increased DCMD activity depends on stimulus size and velocity, but a general trend is established: silence in or low DCMD activity before and immediately after the stimulus onset and a rapid increase to peak firing frequency around the time of collision between the stimulus and the animal. Our results also show that with stimuli of faster velocities, DCMD firing frequency is relatively lower, as Rind and Simmons found in their 1992 paper. Additionally, cited literature studies (Rind and Simmons, 1992; Hatsopoulos et al., 1995; Gabbiani et al., 1999; Rowell, 1971b) use ITIs of at least 40s and our experiments have demonstrated the importance of using an ITI sufficiently long (at least 40s) to consistently obtain DCMD signals. As Gabbiani et al. (1999) found, this LGMD-DCMD motion-detection neuronal system is also unaffected by changes in contrast between the background and the stimulus.
These exercises demonstrate to students how recording the neural activity of a complete sensory-motor neural pathway- from the grasshopper’s eyes to its hind legs. The DCMD response produces an evolutionarily advantageous behavior for the grasshoppers. The start of DCMD firing and time of peak firing frequency are important, as the timing (before or after the expected collision with an object) determines the success of the animal avoiding danger. Students can also observe the importance of an interval stimuli presentation (useful for many electrophysiological preparations) by optimizing for consistent DCMD responses. The students also discover that neuronal escape response is surprisingly consistent across changes in the contrast between the stimulus and the background. Beyond the biological implications, students learn about negative results in hypothesis testing which occurs commonly in research labs, but not in student teaching labs.
While the iOS app was designed for the iPad, the students who performed the visual looming stimuli DCMD exercise at Harvard University during the Fall of 2016 replicated this investigation mostly using iPhone and iPod devices. This produced similar results to those presented here using the iPad, even with the smaller screen size.
Effectiveness of the exercises
Overall, the Harvard students provided enthusiastic feedback and evaluated the hands-on activity as contributing to their understanding of electrophysiology as a tool to investigate the neural basis of a particular animal behavior, as well as to their general interest in neuroscience as a potential academic pursuit. The solicited responses were similarly positive to similar surveys performed in teaching neuroscience laboratories (Pulver et al., 2011, Titlow et al., 2015). The students also made valuable suggestions for improvements. They asked to provide a clearer diagram for electrode placement in the grasshopper’s neck (now incorporated into Fig 2 and Supplementary Video) and improving the user interface of the SpikeRecorder app by framing the data analysis steps as a sequence (e.g., spike sorting before graph viewing). This suggestion has been addressed in the current version of our app.
Procedural challenges
Our results serve as points of reference for students to compare their results against, though variations are expected. Possible procedural challenges may arise, especially given the limited time in a classroom lab, and here we discuss several potential problems and solutions. First, noise signals are likely to be present during neural recordings and may obscure the desired signals. Since a noise-free environment is not realistic in a classroom, simple Faraday cages may be used. This provides the instructor with an opportunity to briefly explain how a closed conductive container, like a wire mesh box, can shield the materials inside from outside electromagnetic interference. If desired, this can also be an additional experiment, in which students can observe the differences in noise and neural signals during various levels of interference. Another challenge students may encounter is the animal preparation. If the grasshopper wakes up early from the anesthesia and becomes active during the surgery, it should be replaced into ice until it is inactive once more. Electrode placement, especially the hook recording electrode around the neck connective, is relatively simple but does demand using a microscope and micromanipulator for precision. Adult grasshoppers should be used if possible, as their neck connectives are larger and more visible. If the electrode placement around a neck connective proves too difficult, students may modify the hook electrode into a simple straight wire and place it between the two neck connectives. This is too imprecise for full DCMD analysis, but the amplification would be sufficient for DCMD spikes to be visualized and recorded. Determining the threshold for spike sorting can be difficult due to drift in the recording for long periods of time and movement-related EMG noise. However, we have shown that even in these cases (e.g., note the presumably false pre-stimulus spikes in Fig 5 & 7) that a rough placement of thresholds around visually-determined DCMD spikes can still show the time of peak firing.
Further exercises
We encourage modifications and experiments that may arise from our procedures. Example research ideas using the SpikerBox amplifier and grasshoppers or similar invertebrates include: somatotopy with grasshopper legs, the effect of temperature on neuronal responses or spikes, and the effect of drugs (e.g., nicotine) on the nervous system. Detailed exercises can be found on the Backyard Brains website.
Footnotes
Supplementary Information:
Video S1: We have prepared a short video that describes the procedure and shows the preparation of the grasshopper: how to tape the grasshopper, how to place the thread around the neck, how to prepare and position the electrode for DCMD recording, how to align the grasshopper to the iPad, and finally a quick demonstration of the looming stimuli activating the DCMD Response. See: https://youtu.be/CEy67YGRmQk
Online Repository: All the data and analysis tools discussed in this paper are available on an online GitHub repository: https://github.com/BackyardBrains/Publications. Instructions are available to generate your own database from SpikerBox iOS recordings, as well scripts to generate the figures in this paper. Session numbers (e.g., G25-072416-01) are included in figure descriptions to allow you to reproduce these figures from this online repository. Scripts can be modified for your own needs.
Schematics, parts and building instructions for the SpikerBox are on the BYB website: https://backyardbrains.com. The iOS application GitHub repository is at: https://github.com/BackyardBrains/.
Funding for this project was supported by the National Institute of Mental Health Small Business Innovation Research grant #2R44MH093334: “Backyard Brains: Bringing Neurophysiology into Secondary Schools.” This project was initially developed in the Methods of Computational Neuroscience course at the Marine Biological Labs, which is supported by an NIH grant #2R25MH062204. MR was funded by a Queen Mary University of London PhD scholarship. We thank David Cox and his students for their constructive feedback. We thank Stefan Pulver for his guidance on developing our questionnaire survey. GJG is a co-founder and co-owner of Backyard Brains, Inc., a company that sells the open-source hardware described in this paper. SM is a paid employee of Backyard Brains, Inc. MR declares he is a paid employee of Ben Thorns Ltd. The authors thank Tanner Petch for his beautiful illustrations in Figures 1 and 3.
REFERENCES
- Bretschneider F, De Weille JR. Introduction to electrophysiological methods and instrumentation. London: Elsevier Ltd; 2006. Stimulus-response characteristics: The PSTH; pp. 188–190. [Google Scholar]
- Burrows M, Rowell CHF. Connections between descending visual interneurons and metathoracic motoneurons in the locust. J Comp Physiol A. 1973;85:221–234. [Google Scholar]
- Card GM. Escape behaviors in insects. Curr Opin Neurobiol. 2012;22:180–186. doi: 10.1016/j.conb.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Thalhauser RM, Dagda R, Marzullo TC, Gage GJ. Using crickets to introduce neurophysiology to early undergraduate students. J Undergrad Neurosci Educ. 2013;12:A66–A74. [PMC free article] [PubMed] [Google Scholar]
- Gabbiani F, Krapp HG, Laurent G. Computation of object approach by a wide-field, motion-sensitive neuron. J Neurosci. 1999;19:1122–1141. doi: 10.1523/JNEUROSCI.19-03-01122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JR, Lee JK, Robertson RM. Activity of descending contralateral movement detector neurons and collision avoidance behaviour in response to head-on visual stimuli in locusts. J Comp Physiol A. 2001;187:115–219. doi: 10.1007/s003590100182. [DOI] [PubMed] [Google Scholar]
- Hatsopoulos N, Gabbiani F, Laurent G. Elementary computation of object approach by a wide-field visual neuron. Science. 1995;270:1000–1003. doi: 10.1126/science.270.5238.1000. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Wyttenbach RA, Hoy RR. Crustacean as model systems for teaching neuroscience: Past, present, and future. In: Derby C, Thiel M, editors. The natural history of crustacea: Vol 3, Nervous systems and control of behavior. New York: Oxford UP; 2014. pp. 535–553. [Google Scholar]
- Kladt N, Hanslik U, Heinzel HG. Teaching basic neurophysiology using intact earthworms. J Undergrad Neurosci Edu. 2010;9:A20–A35. [PMC free article] [PubMed] [Google Scholar]
- Krans J, Gilbert C, Hoy R. Teaching insect retinal physiology with newly designed, inexpensive micromanipulators. Adv Physiol Educ. 2006;30:254–261. doi: 10.1152/advan.00029.2006. [DOI] [PubMed] [Google Scholar]
- Land BR, Wyttenbach RA, Johnson BR. Tools for physiology labs: an inexpensive high-performance amplifier and electrode for extracellular recording. J Neurosci Methods. 2001;106:47–55. doi: 10.1016/s0165-0270(01)00328-4. [DOI] [PubMed] [Google Scholar]
- Li WC, Wagner M, Porter NJ. Behavioral observation of Xenopus tadpole swimming for neuroscience labs. J Undergrad Neurosci Educ. 2014;12:A107–A113. [PMC free article] [PubMed] [Google Scholar]
- Marzullo TC, Gage GJ. The SpikerBox: a low cost, open-source bioamplifier for increasing public participation in neuroscience inquiry. PLoS One. 2012;7:e30837. doi: 10.1371/journal.pone.0030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea M, Rowell CHF, Williams JLD. The anatomy of a locust visual interneurone; The descending contralateral movement detector. J Exp Biol. 1973;60:1–12. [Google Scholar]
- Pearson KG, O’Shea M. Escape behavior of the locust. In: Eaton RC, editor. Neural mechanisms of startle behavior. New York: Springer US; 1984. pp. 163–178. [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv Physiol Educ. 2011;35:82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RL, Moiseff A, Brumberg JC. Utility and versatility of extracellular recordings from the cockroach for neurophysiological instruction and demonstration. J Undergrad Neurosci Educ. 2007;5:A28–A34. [PMC free article] [PubMed] [Google Scholar]
- Rind FC, Simmons PJ. Orthopteran DCMD neuron: a reevaluation of responses to moving objects. I. Selective responses to approaching objects. J Neurophysiol. 1992;68:1654–1666. doi: 10.1152/jn.1992.68.5.1654. [DOI] [PubMed] [Google Scholar]
- Rind FC, Simmons PJ. Seeing what is coming: building collision-sensitive neurones. Trends Neurosci. 1999;22:215–220. doi: 10.1016/s0166-2236(98)01332-0. [DOI] [PubMed] [Google Scholar]
- Rowell CHF. The orthopteran descending movement detector (DMD) neurones: a characterisation and review. Z Vergl Physiol. 1971a;73:167–194. [Google Scholar]
- Rowell CHF. Variable responsiveness of a visual interneurone in the free-moving locust, and its relation to behavior and arousal. J Exp Biol. 1971b;55:727–747. [Google Scholar]
- Santer RD, Simmons PJ, Rind FC. Gliding behaviour elicited by lateral looming stimuli in flying locusts. J Comp Physiol A. 2005;191:61–73. doi: 10.1007/s00359-004-0572-x. [DOI] [PubMed] [Google Scholar]
- Santer RD, Yamawaki Y, Rind FC, Simmons PJ. Preparing for escape: an examination of the role of the DCMD neuron in locust escape jumps. J Comp Physiol A. 2008;194:69–77. doi: 10.1007/s00359-007-0289-8. [DOI] [PubMed] [Google Scholar]
- Shannon KM, Gage GJ, Jankovic A, Wilson WJ, Marzullo TC. Portable conduction velocity experiments using earthworms for the college and high school neuroscience teaching laboratory. Adv Physiol Educ. 2014;38:62–70. doi: 10.1152/advan.00088.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons PJ, Young D. Nerve cells and animal behaviour. Cambridge, UK: Cambridge UP; 1999. pp. 122–128. [Google Scholar]
- Titlow JS, Johnson BR, Pulver SR. Light activated escape circuits: a behavior and neurophysiology lab module using Drosophila optogenetics. J Undergrad Neurosci Educ. 2015;13:A166–A173. [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach RA, Johnson BR, Hoy RR. Crawdad: an online lab manual for neurophysiology. Sunderland, MA: Sinauer Associates, Inc; 2014. [Google Scholar]