Abstract
In a territory-wide surveillance study, Laribacter hongkongensis was isolated solely from freshwater fish (60% of grass carps, 53% of bighead carps, and 25% of mud carps). Comparing the pulsed-field gel electrophoresis patterns of fish and patient isolates revealed that most patient isolates were clustered together, suggesting that some clones could be more virulent.
Laribacter hongkongensis was first discovered in Hong Kong from the blood and empyema pus of a 54-year-old Chinese man with alcoholic cirrhosis (5). Subsequently, L. hongkongensis was discovered in three of our patients and three patients in Switzerland with community-acquired gastroenteritis (3). In a multicenter prospective study, we confirmed that L. hongkongensis is associated with community-acquired gastroenteritis and traveler's diarrhea (4). Freshwater fish were shown to be a reservoir of L. hongkongensis (4). The isolation of L. hongkongensis from patients who resided in or have recently traveled to Asia, Europe, America, and Africa implied that the bacterium is likely to be of global importance.
In this study, in order to determine the prevalence of L. hongkongensis in different animals commonly used for cooking in our locality, we carried out an ecoepidemiology study in Hong Kong. Furthermore, to determine the genetic diversity of L. hongkongensis, all L. hongkongensis isolates were typed by pulsed-field gel electrophoresis (PFGE). The PFGE patterns were compared to those of L. hongkongensis strains isolated from patients with gastroenteritis.
Fecal swabs were obtained from 350 pigs and 80 cows, and cloacal swabs were obtained from 400 chickens, 50 ducks, and 50 geese from slaughter houses and poultry farms in Hong Kong with assistance from Veterinary Public Health Section of the Food and Environmental Hygiene Department. Three hundred and sixty freshwater fish (mostly farmed) of six different species and 360 marine fish of six different species commonly purchased for cooking in Hong Kong were obtained from 10 retail food markets (six fish per species per market) located in different districts of Hong Kong. Samples were obtained from the midguts and hindguts of the fish by using sterile cotton wool swabs. All samples were plated onto cefoperazone MacConkey agar and incubated under aerobic conditions at 37°C for 48 h (1). All suspected isolates were identified phenotypically by standard conventional biochemical methods (2). Isolates suspected to be L. hongkongensis were subjected to 16S rRNA gene sequencing (3, 4). All L. hongkongensis isolates were subjected to PFGE (3, 4). Digital images were stored electronically as TIFF files and analyzed visually and with GelCompar II (version 3.0; Applied Maths, Kortrijk, Belgium) by using the Dice coefficient and represented by the unweighted pair-group method using average linkages with 1% tolerance and 0.5% optimization settings.
L. hongkongensis was isolated from the midguts and hindguts of 86 (24%) of 360 freshwater fish, including 36 (60%) grass carps, 32 (53%) bighead carps, 15 (25%) mud carps, and 3 (5%) largemouth bass (Table 1). L. hongkongensis was not isolated from samples of pigs, cows, chickens, ducks, geese, Chinese perch, tilapia, and marine fish.
TABLE 1.
L. hongkongensis isolation from ecoepidemiology study
| Animal group and type (generic name) | No. of samples obtained | No. (%) of samples positive for L. hongkongensis |
|---|---|---|
| Mammals | ||
| Pig | 350 | 0 (0) |
| Cow | 80 | 0 (0) |
| Fowl | ||
| Chicken | 400 | 0 (0) |
| Duck | 50 | 0 (0) |
| Goose | 50 | 0 (0) |
| Freshwater fish | 360 | 86 (24) |
| Grass carp(Ctenopharyngodon idella) | 60 | 36 (60) |
| Bighead carp (Aristichthys nobilis) | 60 | 32 (53) |
| Mud carp (Cirrhina molitorella) | 60 | 15 (25) |
| Largemouth bass (Micropterus salmoides) | 60 | 3 (5) |
| Chinese perch (Siniperca chuatsi) | 60 | 0 (0) |
| Tilapia (Oreochromis mossambicus) | 60 | 0 (0) |
| Marine fish | 360 | 0 (0) |
| Surf bream (Acanthopagrus australis) | 60 | 0 (0) |
| Golden thread fin bream (Nemipterus virgatus) | 60 | 0 (0) |
| Fourfinger threadfin (Eleutheronema tetradactylum) | 60 | 0 (0) |
| Yellow croaker (Pseudosciaena crocea) | 60 | 0 (0) |
| Green wrasse (Halichoeres chloropterus) | 60 | 0 (0) |
| Black pomfret (Parastromateus niger) | 60 | 0 (0) |
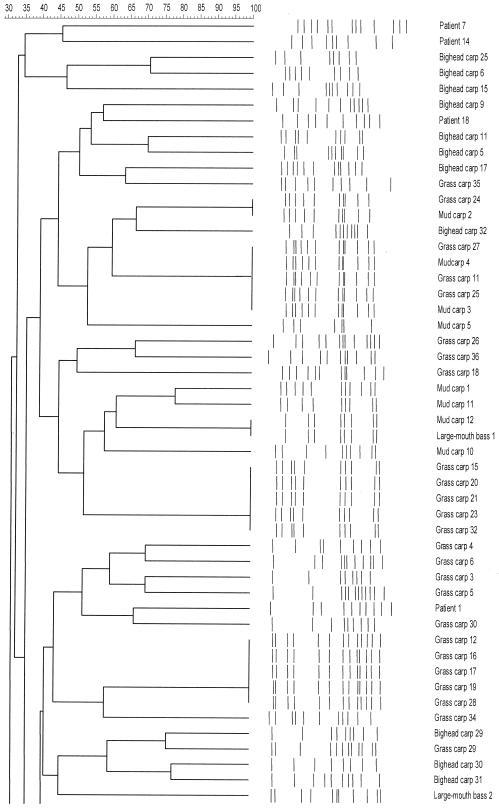
Sixty-seven different PFGE patterns were found in the 86 L. hongkongensis isolates from freshwater fish, including 25 in the 36 grass carp isolates, 28 in the 32 bighead carp isolates, 14 in the 15 mud carp isolates, and 3 in the 3 largemouth bass isolates (Fig. 1). Twenty different PFGE patterns were found in the 20 L. hongkongensis isolates from patients with gastroenteritis (Fig. 1).
FIG. 1.
Relationships among SpeI profiles of 86 L. hongkongensis isolates in freshwater fish and 20 isolates from patients with gastroenteritis. The dendrogram was constructed with PFGE data by similarity and clustering analysis using the Dice coefficient and unweighted pair-group method using average linkages with GelCompar II. The patient numbers corresponded to those in Table 2 of reference 4.
A heterogeneous population of L. hongkongensis was recovered from freshwater fish commonly consumed in Hong Kong. Overall, L. hongkongensis was recovered from intestines of 24% of freshwater fish examined in this study. Although L. hongkongensis was recovered from over 50% of grass carp and bighead carp but not from tilapia and Chinese perch, there was no relationship between the rate of recovery of L. hongkongensis and individual retail market locations. This is in contrast to infectious disease outbreaks which originated from a single location, where a gradient of recovery of the pathogen can be observed. Furthermore, there was no relationship between the type of freshwater fish and the location of the retail markets. Our previous studies showed that all L. hongkongensis strains had the same morphotype and biochemical profile (3-5). Furthermore, almost all L. hongkongensis strains had the same antimicrobial susceptibility pattern. All 20 strains of L. hongkongensis recovered from our patients with gastroenteritis were sensitive to amoxicillin-clavulanate, meropenem, levofloxacin, and gentamicin but resistant to ampicillin, cefuroxime, ceftriaxone, and erythromycin. The only discriminatory characteristic was that 16 of the 20 strains were sensitive and 4 were resistant to tetracycline (4). Due to the difficulty in distinguishing different strains by phenotypic tests, we performed SpeI digestion and PFGE to examine the relatedness of the different isolates of L. hongkongensis. Overall, heterogeneous PFGE patterns were observed in different strains of L. hongkongensis, although the same PFGE patterns can be observed in some strains. These results show that L. hongkongensis has been endemic in our locality, and probably also southern China, where over half of the patients reported in our previous study had recently visited prior to development of gastroenteritis. Interestingly, when the PFGE patterns of L. hongkongensis isolated from fish and those from patients with gastroenteritis were compared, it revealed that most patient isolates were clustered together, suggesting that some clones of L. hongkongensis could be more virulent than others (Fig. 1). Further studies on the seasonal variation in the recovery of L. hongkongensis in freshwater fish, the pathogenic potential of L. hongkongensis on freshwater fish, and the presence of L. hongkongensis in other freshwater living organisms and different water bodies should be performed to determine the ecology and life cycle of the bacterium and the possibilities of other modalities of disease transmission. L. hongkongensis could be included in the surveillance of food-borne infectious disease agents in Hong Kong.
The methods by which freshwater fish are prepared and consumed in southern China are related to Laribacter gastroenteritis. In southern China, steamed, freshly prepared freshwater fish is one of the most popular dishes. Steamed whole freshwater fish is considered delicious only if the fish is just done, limiting the time of steaming to a few minutes. Consumption of undercooked fish, most often the “belly” part that is easily contaminated with the gastrointestinal contents, is not uncommon and could lead to Laribacter gastroenteritis. Careful handling of fish, proper cooking of the fish and related products, and prevention of cross-contamination at the processing, food preparation, and service steps are crucial in preventing infections associated with L. hongkongensis.
Acknowledgments
This work is partly supported by a Research Grant Council grant (7357/04 M); the University Development Fund, The University of Hong Kong; the Research Fund for the Control of Infectious Diseases (02040212); and the William Benter Infectious Disease Fund.
We thank King-man Chan for his technical help.
REFERENCES
- 1.Lau, S. K. P., P. C. Y. Woo, W. T. Hui, M. W. S. Li, J. L. L. Teng, T. L. Que, R. W. H. Yung, W. K. Luk, R. W. M. Lai, and K. Y. Yuen. 2003. Use of cefoperazone MacConkey agar for selective isolation of Laribacter hongkongensis. J. Clin. Microbiol. 41:4839-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray, P. R., E. J. Baro, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 3.Woo, P. C. Y., P. Kuhnert, A. P. Burnens, J. L. L. Teng, S. K. P. Lau, T. L. Que, H. H. Yau, and K. Y. Yuen. 2003. Laribacter hongkongensis: a potential cause of infectious diarrhea. Diagn. Microbiol. Infect. Dis. 47:551-556. [DOI] [PubMed] [Google Scholar]
- 4.Woo, P. C. Y., S. K. P. Lau, J. L. L. Teng, T. L. Que, R. W. H. Yung, W. K. Luk, R. W. M. Lai, W. T. Hui, S. S. Y. Wong, H. H. Yau, and K. Y. Yuen. 2004. Association of Laribacter hongkongensis in community-acquired gastroenteritis with travel and eating fish: a multicentre case-control study. Lancet 363:1941-1947. [DOI] [PubMed] [Google Scholar]
- 5.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]