Abstract
Rotaviruses of species A (RVA) are a common cause of diarrhoea in children and the young of various other mammals and birds worldwide. To investigate possible interspecies transmission of RVAs, whole genomes of 18 human and 6 domestic animal RVA strains identified in Uganda between 2012 and 2014 were sequenced using the Illumina HiSeq platform. The backbone of the human RVA strains had either a Wa- or a DS-1-like genetic constellation. One human strain was a Wa-like mono-reassortant containing a DS-1-like VP2 gene of possible animal origin. All eleven genes of one bovine RVA strain were closely related to those of human RVAs. One caprine strain had a mixed genotype backbone, suggesting that it emerged from multiple reassortment events involving different host species. The porcine RVA strains had mixed genotype backbones with possible multiple reassortant events with strains of human and bovine origin.Overall, whole genome characterisation of rotaviruses found in domestic animals in Uganda strongly suggested the presence of human-to animal RVA transmission, with concomitant circulation of multi-reassortant strains potentially derived from complex interspecies transmission events. However, whole genome data from the human RVA strains causing moderate and severe diarrhoea in under-fives in Uganda indicated that they were primarily transmitted from person-to-person.
Introduction
Rotaviruses belong to the genus Rotavirus of the family Reoviridae, comprising nine species (groups) designated as A, B, C, D, E, F, G, H and I [1, 2] and possibly a tenth species J [3]. Group A rotaviruses (RVA) are a leading cause of diarrhoea in children and young animals worldwide [4, 5]. In children, the infection may lead to severe dehydration and may cause death if the condition is not well managed [6]. Rotavirus infections in animals may affect productivity and have important economic consequences [7].
The rotavirus genome consists of eleven segments of double-stranded RNA (dsRNA). All RNA segments, with the exception of segment 11, are monocistronic, encoding either structural viral proteins (VP1 to VP4, VP6 and VP7) or non-structural proteins (NSP1 to NSP4). Genome segment 11 codes for two proteins: NSP5 and NSP6 [8]. Rotaviruses can be differentiated by a dual classification system, based on the two outer capsid proteins, VP7 and VP4, that determine the G (VP7, glycoprotein) and P (VP4, protease sensitive) genotypes, respectively [9]. At least 35 G types and 50 P types have so far been identified in humans and animals (rega.kuleuven.be/cev…/virus classification/newgenotype) [10, 11].
Globally, genotypes G1, G2, G3, G4, G9 and G12 in combination with P[4], P[6] or P[8] constitute more than 90% of the circulating human RVA strains [12]. The most common combinations of the G and P genotypes are G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8] [12, 13]. However, regional variability has been observed. In Africa, RVA genotypes such as G8P[6] and G8P[8] are highly prevalent but uncommon elsewhere [14]. These uncommon rotavirus strains are thought to have arisen from host interspecies transmission [15, 16].
More recently, RVAs have been classified based on the sequence diversity of all 11 segments, assigning specific genotypes according to established nucleotide homology cut-off values [10, 17, 18]. This classification system, combined with whole genome sequencing and phylogenetic analysis, has been used to trace interspecies transmission events and potential origins of new and emerging strains [10, 19, 20].
Interspecies transmission of RVAs is thought to be an important contributor to rotavirus evolution, contributing to the diversity of viruses in both humans and animals [5, 8]. Other mechanisms for rotavirus evolution include accumulation of point mutations, gene reasortment or rearrangement, and gene recombination [5]. Genome evolution may occur either intragenically or intergenogroup [21, 22]. The combination of interspecies transmission and reassortment between RVAs of different species can lead to the emergence and spread of novel rotavirus strains [23].
Globally, few RVA co-surveillance studies in animals and humans in the same geographical region have been carried out. A study in the Netherlands found no evidence of interspecies transmission, the animal and human rotaviruses appeared to evolve separately [24]. By contrast, a study in Slovenia found interspecies transmission, with evidence of transmission from pigs to humans [25]. A study in Southern India found evidence of human-to-animal transmission of a G2 RVA strain [26]. Another study in Northern India found possible reassortment between genes of animals and human RVAs resulting in circulation of unusual rotavirus genotypes [27].
None of the above studies characterised whole genomes of RVAs from different species, hence provided only a partial picture of a potentially more widespread phenomenon. In the present study, whole genomes of RVAs identified from faecal samples of humans and animals living in the same region in Uganda during 2012–2014 were sequenced and analysed in order to investigate possible interspecies transmission events of RVAs in this setting.
Materials and methods
Ethical approval
This study was approved by the Research and Ethics Committees of the School of Medicine, College of Health Sciences, Makerere University (REF 2011–061); Uganda Virus Research Institute (GC/127/319); Mulago National Referral Hospital; St. Francis Hospital Nsambya; and Uganda National Council for Science and Technology (HS 1186). The caretakers/guardians of the children gave written consent for the children to participate in the study. In addition, the animal owners gave written consent for their animals to be included in the study.
Human and animal recruitment, and sample collection
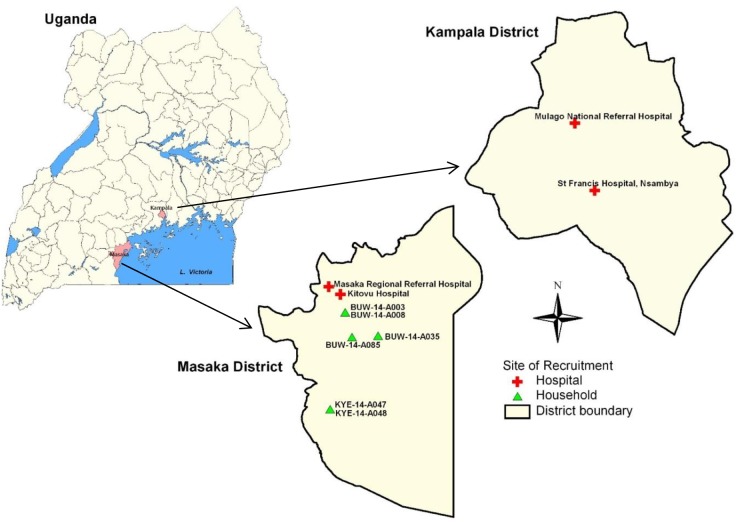
Human stool samples were collected from children under-five years old hospitalised with acute diarrhoea in four hospitals located in Kampala and Masaka districts in central Uganda (Fig 1). The study was carried out from September 2012 through September 2013. The stools were investigated as previously described [28]. Eighteen out of 208 human RVA-positive samples were selected for whole genome sequencing. Selection was based on the availability of sufficient material and adequate viral load for unbiased sequencing directly from the stool sample, presence of G and P types found in animal RVAs, or the presence of unusual G and/or P types.
Fig 1. The maps of Uganda, Kampala and Masaka districts showing the hospitals and households at which the study children and animals (rotavirus positives that were sequenced) were recruited respectively.
Animal stool samples were collected from 116 symptomatic (with history of diarrhoea in previous two weeks) and 984 asymptomatic (without history of diarrhoea in previous two weeks) domestic animals (cattle, goats and pigs) in homes located in Bukoto county, Masaka district from December 2013 through January 2014 (Fig 1, S1 Table). Out of the 41 RVA-positive animal samples whole genome analysis was possible on six samples (one bovine, one caprine and four porcine). Among these, only the bovine RVA was associated with a history of diarrhoea lasting four days in the two weeks prior to sample collection.
Rotavirus dsRNA extraction, cDNA synthesis and amplification from human and animal samples
Rotavirus dsRNA was extracted from human stool suspension (approximately 100 mg of stool were suspended in 200 μl of PBS or 200 μl of semi-formed stool was mixed with 150 μl of PBS) using TRIZOL LS Reagent (Invitrogen, Carlsbad, CA, USA). The procedure of extraction was as previously described [29]. The guanidinium isothiocyanate silica method was used to extract rotavirus dsRNA from 10% faecal suspensions in PBS for all animal samples [30].
Oligonucleotide ligation to enable cDNA synthesis, and unbiased PCR amplification and purification of the entire rotavirus genome were carried out on the human samples that yielded more than 2 ng/μl dsRNA as described previously [29, 31]. The purified rotavirus cDNA PCR amplicons were subjected to standard bar-coding and library construction for Illumina sequencing using the Nextera XT DNA Library Preparation Kit following the manufacturer’s recommendations (Illumina Inc., CA, USA). For all animal samples and human samples that yielded less than 2 ng/μl dsRNA, the ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre, Chicago, IL, USA) was used, following the manufacturer’s instructions with the slight modification of an initial denaturation step (95°C for 5 min). Each library was indexed with Illumina compatible barcodes to allow multiplexing (http://dx.doi.org/10.17504/protocols.io.h4vb8w6). The quality of the libraries was assessed using the VP6-specific qPCR [32] and the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were quantified with the Qubit dsDNA High Sensitive assay (Life Technologies, Carlsbad, CA, USA), and sequenced using the HiSeq 2500 Illumina platform at the Centre for Genomic Research, University of Liverpool, UK.
Nucleotide sequence assembly, genotype assignment and phylogenetic analyses
Illumina adapter sequences were trimmed from the raw Fastq sequence data using Cutadapt version 1.2.1 and Sickle version 1.2 software [33]. Both de novo and mapping assembly tools embedded in Geneious software [34] were employed to generate consensus sequences for all analysed strains. To ensure that the multiple sequences detected in some of the samples were not due to assembly artifacts, sequence reads that had more than one contig were mapped separately to both Wa and DS-1 rotavirus prototype strains using both medium and high custom sensitivity parameters where only sequence reads with more than 80% overlap identity were used to build the consensus. Mixed populations were only accepted as true populations when the two consensus sequences generated through mapping and de novo assemblers were identical, could be translated to a functional protein without need for editing and had coverage of at least 200. The presence of multiple sequences in a single specimen was confirmed at the J. Craig Venter Institute by the Virology Project Team who blindly and independently assembled the sequence reads on CLC command-line assembly module (CLC Bio’s clc_novo assemble and CLC Bio’s clc_ref_assemble_long_program) [35].
RotaC version 2 (http://rotac.regatools.be/) [36], a classification tool for RVAs, was used to assign genotypes to all eleven genome segments. The nucleotide sequences generated in this study were deposited into the NCBI GenBank under the accession numbers KX632243-632352, KX655437-KX655538, KX988264-KX988283, KY055416-KY055437, KY077640-KY077650 (S2 Table).
Phylogenetic analysis was conducted using MEGA version 6.06 [37]. Multiple alignments of sequences from the study strains and reference strains from GenBank were carried out using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) software [38]. The phylogenetic trees were constructed using the Maximum-Likelihood method with the best-fit substitution models. The substitution models that best fitted the sequence data were determined using the corrected Akaike Information Criterion (AICc). The models used in this study were: GTR+G+I for VP1, VP2 and VP3; T92+G for VP4, VP6, VP7, NSP1, NSP3 and NSP5; TN93+G+I for NSP2; and HKY +G for NSP4. The bootstrap (1000 replicates) values were used to determine the reliability of each node in the tree. The lineages for VP4 P[6] lineage I, P[4] lineage II and IV, P[8] lineage III, were assigned as previously suggested [16, 19, 39–42]. No literature was found with regard to the classification of P[1], P[7] and P[13] genes into lineages. The lineages for VP7: G1, G3, G6, G8, G9, G12 were assigned as previously suggested [39, 42–45]. Nucleotide distance matrices for each of the characterized RVA genomes were determined using BioEdit program [46].
Results
The Illumina Hiseq sequencing yielded mean read lengths of 72.3 (SD 35) -119.1(SD 16.0) bp for the human and 34.0 (SD 20.5) -79.4 (SD 32.4) bp for animal RVA strains. The maximum expected read length was 125 bp. Complete nucleotide sequences were obtained for all the 11 segments of the 18 human strains and one bovine strain. Partial sequences were obtained for some genome segments of the porcine and caprine rotavirus strains (S3 Table). The fragments of the partial sequences ranged from 33.1% to 99.8% of the expected gene lengths (S3 Table). Nonetheless, these sequence lengths were adequate for assigning genotypes (Table 1).
Table 1. Whole genome constellation of characterised human and animal rotavirus strains circulating in Uganda, 2012–2014.
| Species | Strain Nomenclature | VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | RVA/Human-wt/UGA/KTV-13-023/2012/G12P[6] | G12 | P[6] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-13-183/2013/G12P[6] | G12 | P[6] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/NSA-13-043/2013/G9P[8] | G9 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-13-163/2013/G9P[8] | G9 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-12-147/2012/G9P[8] | G9 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-12-093/2012/G9P[8] | G9 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-13-285/2013/G9P[8] | G9 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-13-157/2013/G1P[8] | G1 | P[8] | I1 | R1 | C2 | M1 | A1 | N1 | T1 | E1 | H1 |
| Human | RVA/Human-wt/UGA/MUL-12-104/2012/G3P[6] | G3 | P[6] | 12 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-308/2013/G8P[6] | G8 | P[6] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-166/2013/G3P[6] | G3 | P[6] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-171/2013/G3P[6]** | G3/G3 | P[6]/P[6] | I2 | R2/R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-204/2013/G8P[6] | G8 | P[6] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-496/2013/G8P[4] | G8 | P[4] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-12-117/2012/G3P[6] | G3 | P[6] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-160/2013/G8P[4] | G8 | P[4] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MSK-13-048/2013/G9P[8] | G9 | P[8] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Human | RVA/Human-wt/UGA/MUL-13-427/2013/G8P[4] | G8 | P[4] | 12 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| Goat | RVA/Goat-wt/UGA/BUW-14-A085/2014/G6P[1] | G6 | P[1] | I2 | R2 | C2 | M2 | A11 | N2 | T6 | E2 | H3 |
| Cattle | RVA/Cow-wt/UGA/BUW-14-A035/2014/G12P[8] | G12 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Pig | RVA/Pig-wt/UGA/BUW-14-A008/2014/G12P[8] | G12 | P[8] | I1 | R1 | C1 | M1 | A8 | N1 | T1 | E1 | H1 |
| Pig | RVA/Pig-wt/UGA/BUW-14-A003/2014/G3P[13] | G3 | P[13] | I1 | R1 | C1 | M1 | A8 | N1 | T7 | E1 | H1 |
| Pig | RVA/Pig-wt/UGA/KYE-14-A047/2014/G3P[13] | G3 | P[13] | I1 | R1 | C1 | M1 | A8 | N1 | T1 | E1 | H1 |
| Pig | RVA/Pig-wt/UGA/KYE-14-A048/2014/G3P[13] | G3 | P[13] | I1 | R1 | C1 | M1 | A8 | N1 | T1 | E1 | H1 |
** Sample contained mixed sequences of the same genotype in VP1, VP4 and VP7 genes
Colours represent the genome constellation: Green(Wa-like), Red(DS-1 like), White(non 1, non 2 genotype)
VP: viral structural protein, NSP: Viral non structural protein
Whole genome classification of the analysed rotavirus strains
Complete genotype constellation of human strains
All but the genome segments encoding VP7 and VP4 for seven of human rotavirus strains (KTV-13-023, MUL-13-183, NSA-13-043, MUL-13-163, MUL-12-147, MUL-12-093 and MUL-13-285) had a Wa-like genotype constellation (-I1-R1-C1-M1-A1-N1-T1-E1-H1). One of the human Wa-like RVAs strain, MUL-13-157, contained a DS-1-like VP2 gene and was therefore classified as Wa-DS-1-like mono-reassortant (Table 1). The non-G and non-P genes of the other 10 human rotaviruses (MUL-12-104, MUL-13-308, MUL-13-166, MUL-13-171, MUL-13-204, MUL-13-496, MUL-12-117, MUL-13-160, MSK-13-048 and MUL-13-427) were assigned a DS-1-like genotype constellation (-I2-R2-C2-M2-A2-N2-T2-E2-H2), and hence were classified as DS-1-like human strains (Table 1).
Two distinct complete gene sequences of the same genotype were generated for genome segments encoding VP7, VP4 and VP1 for human rotavirus strain MUL-13-171, compatible with a mixed infection with two variant strains, by contrast, single sequences were generated for the remaining eight genome segments (Table 1).
Complete genotype constellation of animal strains
The characterised bovine strain had a Wa-like gene constellation. The G3P[13] porcine strains had predominantly Wa-like gene constellation, with the exception of the NSP1 gene (A8), and also the NSP3 gene in one of the strains (T7). The G12P[8] porcine strain also had a predominant Wa-like gene constellation with the exception of the NSP1 gene (A8). The G6P[1] caprine strain had a predominantly DS-1-like gene constellation with the exception of the genes encoding NSP1, NSP3 and NSP5 (Table 1).
Phylogenetic analysis
In order to identify the relationships among the RVA strains detected from human and animal species in Uganda and investigate potential origin and evidence of interspecies transmission, phylogenetic analyses were conducted for each gene of the investigated RVA strains and compared with cogent RVA sequences available in the GenBank database.
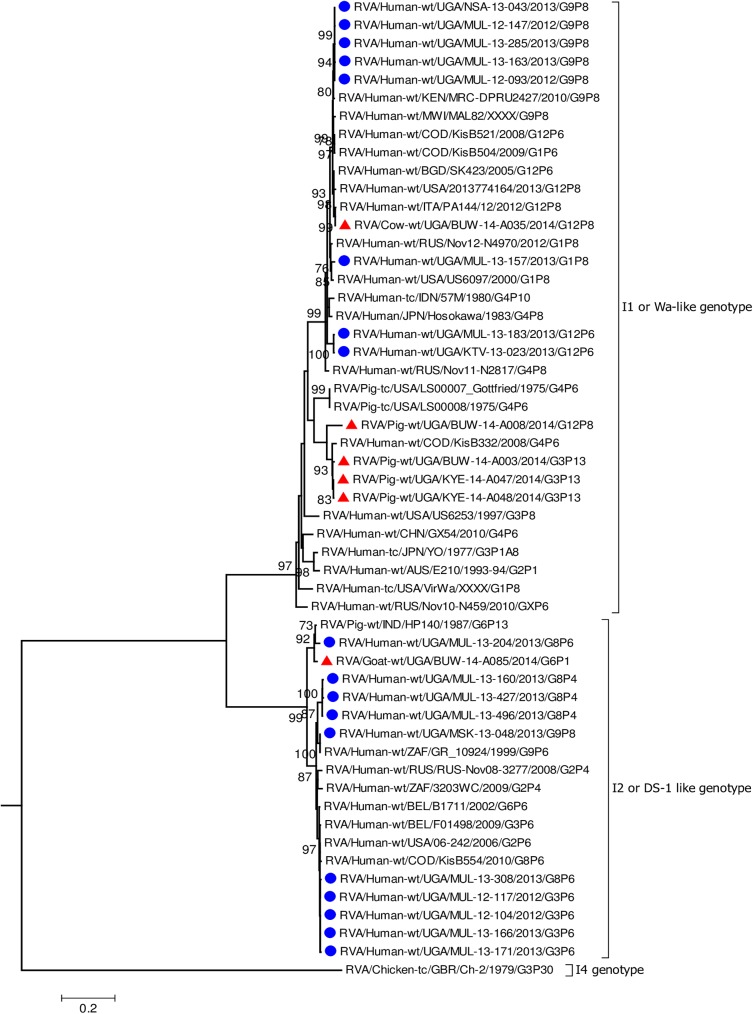
In all the genes, except VP4 and VP7, the human strains clustered with Wa-like and DS-1 like human strains found in Africa including Democratic Republic of Congo (DRC), Tanzania, and Kenya which are neighbouring countries to Uganda (Fig 2, Fig 3, Figs 4–6, S1–S4 Figs) [16, 19, 29, 31, 39, 47–51].
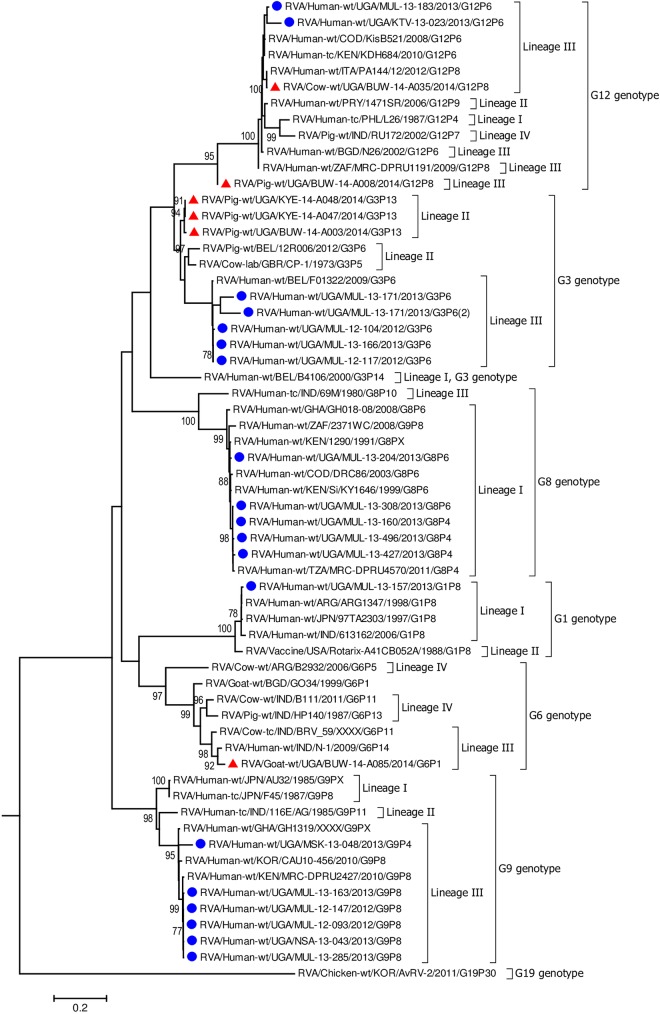
Fig 2. VP1 gene (segment 1).
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 1 of humans and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. The Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
Fig 3. VP6 gene (segment 6).
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 6 of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Chicken strain RVA/Chicken-tc/GBR/Ch-2/1979/G3P[30] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
Fig 4. NSP2 gene (segment 8).
Maximum Likelihood phylogenetic trees of nucleotide sequences of genome segment 8 of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
Fig 6. NSP5 gene (segment 11).
Maximum Likelihood phylogenetic trees of nucleotide sequences of genome segment 11 of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
The nucleotide sequences of the genes derived from human RVAs were 82.2–100% identical to each other (S4 Table, S2 Table). Some genes of a few human strains were closely related to animal strains identified in this study or elsewhere. The VP1 gene sequences of MUL-13-204 clustered with the cogent genes of the goat strain GO34 with a nucleotide identity of 97% (Fig 2) [52]. The VP6 of human strain MUL-13-204 clustered with cogent genes of the porcine RVA strain HP140 which may be a bovine-human reassortant and caprine strain BUW-14-A085 and had 98% and 88.9% nucleotide identity, respectively (Fig 3) [53]. The nucleotide sequences of the NSP4 gene of MUL-13-204 clustered with the cogent gene of a caprine rotavirus strain GO34 and had 97% nucleotide identity (Fig 5) [52].
Fig 5. NSP4 gene (segment 10).
Maximum Likelihood phylogenetic trees of nucleotide sequences of genome segment 10 gene of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
Some genes of the human strains were closely related to strains reported to have zoonotic origin. The VP2 gene sequences of MUL-13-204 and MUL-13-157 were closely related (99.2% and 91% nucleotide similarity, respectively), to that of human strain 1473 from Malawi, which is artiodactyl-like; a human-bovine reassortant strain (S1 Fig) [29].
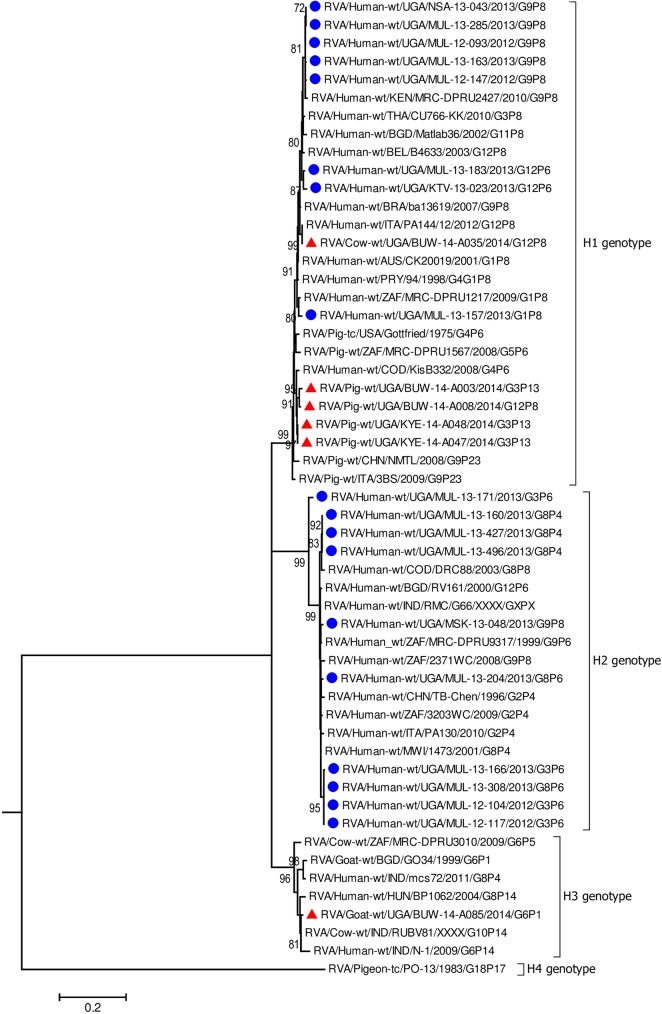
The NSP4 gene sequences of MUL-13-183 and KTV-13-023 clustered with porcine-like human RVA strains KDH684 and KisB332 from Kenya and DRC, respectively, and had 99% and 98% nucleotide identity, respectively (Fig 5) [16, 50].
Among the animal strains, the bovine strain, BUW -14- A035 showed high identity with human RVA strains in all genes with nucleotide identities of 96.9% to 98.5% (Figs 2–8, S1–S4 Figs).
Fig 8. VP7 gene (segment 9).
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 9 of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Chicken strain RVA/Chicken-wt/KOR/ArRv-2/2011/G19P[30] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
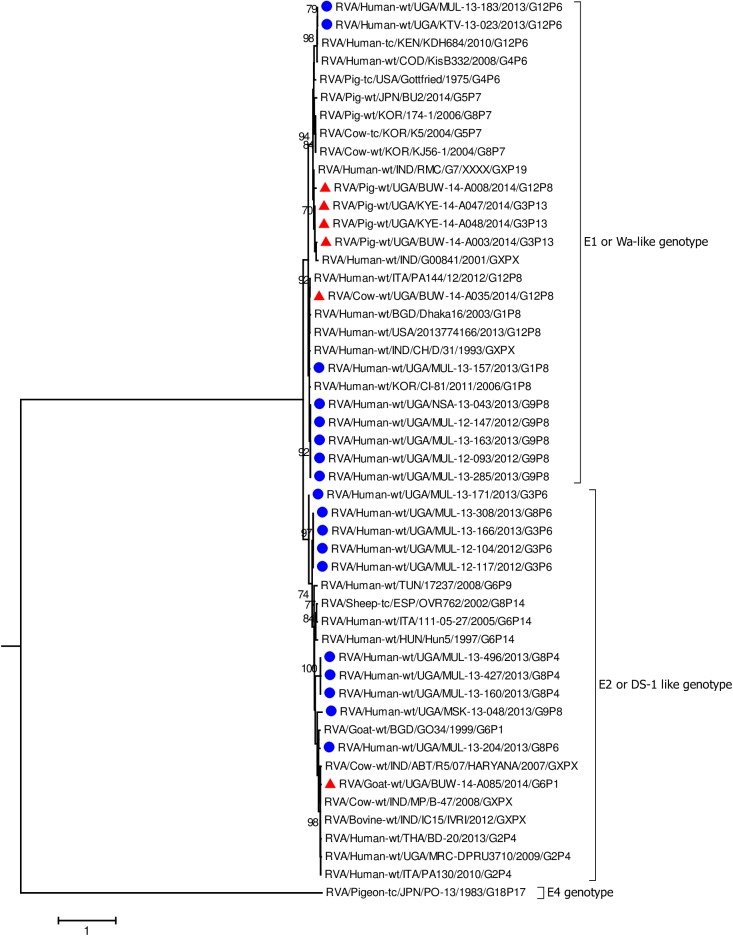
Porcine strains KYE-14-A047 and KYE-14-A048 collected from the same homestead clustered together with nucleotide identity of 95.5–99.6% across all genes. Porcine strains BUW-14-A003 and BUW-14-A008 were also collected from the same household, and the nucleotide sequences of the genes: VP2, VP3, NSP2, NSP3, NSP4 and NSP5 clustered together (S1 Fig, S2 Fig, S4 Fig, Figs 4–6).
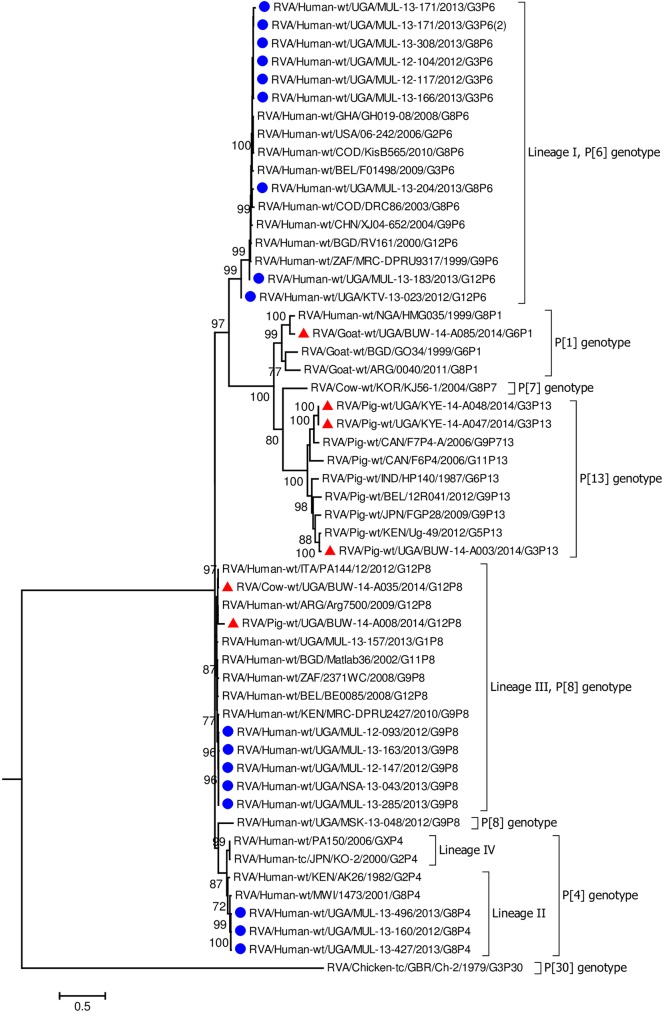
Each individual gene across the porcine strains clustered with porcine strains of this study and from elsewhere, and with strains that had been identified in humans but had evidence of zoonotic transmission (Figs 2–4, Fig 6, S1–S3 Figs) [16, 54–56]. The only exceptions were the genes encoding the VP7 and VP4 of strain BUW-14-008, which were more closely related to those from human strains (Fig 7, Fig 8) [57].
Fig 7. VP4 gene (segment 4).
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 4 of human and animal RVA strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains with red triangles. Chicken strain, RVA/Chicken-tc/GBR/Ch-2/1979/G3P[30] served as the outgroup. The scale bar at the bottom of the tree calibrates the genetic distance expressed as nucleotide substitution per site.
In addition, the NSP4 genes of the studied porcine strains clustered with the Ugandan human RVA strains: MUL-13-183 and KTV-13-023 (Fig 5).
None of the nucleotide sequences of the caprine strain BUW-14-A085 clustered with those of other animal and human sequences in this study, except the nucleotides sequences of the gene encoding VP6 which clustered with the human strain MUL-13-204 (Fig 3).
All the remaining genes of the caprine strain BUW-14-A085 clustered with those identified in other caprine strains or primarily shared among bovine and caprine strains. The NSP2 cluster, however, included cogent genes of RVAs from diverse animal species: caprine, bovine, pigs, dogs, sheep and antelope RVA strains (Fig 4). Sequences of VP1, VP2, VP3 and NSP5 genes of BUW-14 A085 clustered with human strains of zoonotic origin (Fig 2, Fig 6, S1 Fig, S2 Fig) [18, 53, 58, 59].
Discussion
Interspecies transmission of rotaviruses is thought to occur frequently due to the close proximity or sharing of animal and human dwellings in some communities, particularly in low income countries [16]. In view of this, we sought to investigate whether interspecies transmission of RVAs was occurring and possibly contributing to the genetic diversity of RVA strains in Uganda.
The human rotaviruses analysed in the present study were closely related to RVAs from other parts of Africa including neighbouring countries: Kenya, Democratic Republic of the Congo (DRC) and Tanzania. Direct reassortment with porcine or porcine-like human RVAs such as those found in Kenya and DRC may potentially have led to the emerging of NSP4 genes in the human strains MUL-13-183 and KTV-13-023 [16, 50]. By contrast, the VP2 genes of MUL-13-204 and MUL-13-157 strains, because of their close relationship to human RVA strain 1473 from Malawi [29], may have a different origin. The VP6 genes of MUL-13-204 may have been a result of interspecies transmission and reassortment events. Although the NSP4 gene of the human strain MUL-13-204 was closely related to those of other human strains characterised in the present study, it clustered with the NSP4 gene of strain GO34, a caprine rotavirus strain from Bangladesh that is thought to be of bovine origin [52]. These findings highlight the challenges in identifying the geographical and temporal origin of such interspecies transmission through sequence analysis of data obtained in discrete cross-sectional studies.
In some of the RVA genes there was close similarity among the Ugandan human and animal strains. However, phylogenetic analysis of sporadic strains is not enough to elucidate where and when interspecies transmission and reassortment events took place. Nonetheless, in this study, the identification of one bovine strain (BUW-14-A035) in which all gene segments were highly related to human strains circulating in Uganda, was highly suggestive of a direct anthroponotic transmission event.
In order to establish the origin and, or timing of such RVA transmission events, future studies analysing large numbers of RVA strains collected from humans and other mammalian species over a longer period are warranted. The relative small number of samples is one limitation of this study. In addition, the human strains were all from moderate to severe cases of gastroenteritis. Therefore, zoonotic RVAs may have been missed if they circulate at low frequency or were associated with mild disease or asymptomatic infections. However, this study provides confirmation that RVAs causing moderate to severe diarrhoea in humans in Uganda are of common genotypes that have been detected globally and are primarily transmitted from person-to-person. Therefore rotavirus control measures targeting humans should be expected to significantly reduce RVA transmission and burden of moderate or severe rotavirus diarrhoea in Uganda.
Failure to detect two distinct variants for all 11 genes of human strain MUL-13-171 may be due to the variant strains sharing the same sequence in the remaining genes. Also the variants may represent drift and the accumulation of point mutations during the infection and shedding period. Although the presence of quasispecies is expected among RNA viruses, in the acute phase of infections minority species are present at very low levels [60]. It is potentially possible that we were only able to detect variants among those genes that through immune pressure may be driven to hyper variability, such as VP7 and VP4. The heterozygosity detected among the three genes may therefore be associated with prolonged shedding and selective pressure, or as discussed above, with mixed infection [35].
In this study, G12P[8] strains, commonly associated with infection in humans [13, 61, 62] were detected in a pig and a cow. Furthermore, the cow had recent history of diarrhoea. All 11 genes of the bovine RVA strain were of likely human origin due to their high degree of nucleotide identity to those of human RVA in this study and human RVAs strains collected elsewhere [41]. Whereas most interspecies transmission reports observed bovine to human transmission [58, 59], the present study found evidence of human to bovine RVA transmission in Uganda. Since the bovine RVA infection may have been associated with disease, this warrants further research on animal husbandry and feeding practices that may promote inter-species transmission of RVA in this region and drive the emergence of reassortant strains.
G12P[8] strains have been occasionally characterised in pigs, especially those living in close proximity with humans and cows [62, 63]. In this study, we identified one porcine G12P[8] RVA strain (BUW-14-A008) from a household that also housed cattle. Although cattle and human samples collected from this household were rotavirus negative at the time of sampling, it is conceivable that this strain, which had 5 genes (VP1, VP4, VP7, NSP1 and NSP4) closely related to those from human RVAs, may have been derived through human-to-animal transmission and reassortment, as G12P[8] RVAs have the potential to infect all three hosts in the household. In addition, a second porcine strain from the same household (BUW-14-A003: G3P[13]) showed that despite the differences in their VP4 and VP7 genotypes, some genes (VP2,VP3, NSP2, NSP3, NSP4 and NSP5) clustered closely suggesting possible complex exchanges of RVA genes between pigs and humans. A study on RVAs in pigs in East Africa showed possible human to pig transmission of RVAs, where P[8] strains were closely related genetically to human RVAs [64]. These finding are in agreement with the hypothesis on the common origin of porcine and Wa-like human RVAs by Matthijnssens J et al [18].
The high nucleotide identity between all gene segments of porcine strains KYE-14-A047 and KYE -14-A048 suggests that the two pigs were infected by the same strain and provides evidence of ongoing transmission within a household. Similar observations have been reported in South Africa, where five sequences of rotavirus isolates from two calves on the same farm had high nucleotide identity [65].
There are few reports on whole genome sequencing of caprine RVA strains [52, 66]. Our study found that a potentially complex series of reassortment events may have led to the origin of the caprine RVA strain BUW-14-A085 with RVA genes of potential human, bovine and porcine origin. This was similar to what was reported for a caprine RVA strain, GO34, in Bangladesh [52]. In Uganda, most of the animals live in close proximity with each other. This could explain the observed RVA reassortment events. The caprine strain, BUW-14-A085 had an overall genotype constellation of G6-P[1]-I2-R2-C2-M2-A11-N2-T6-E2-H3, which has been found in cattle [58, 67] and thus, may have originated through direct bovine-to-goat transmission.
Conclusions
The present study shows occurrence of interspecies transmission of RVAs of human and animal origins in Uganda with possible reassortment among rotaviruses from different host species. Whereas previous reports on RVA evolution have been mainly on RVA transmission from animals to humans, this study suggests that domestic animals may also become infected by RVAs from humans. The complex reassortment events of rotaviruses from different host species may lead to the emergence of novel rotavirus strains with the potential to influence the epidemiology of rotaviruses in this setting. Therefore, continued surveillance of rotavirus strains from both animals and humans is necessary to monitor changes in the rotavirus epidemiology over time. Whole genome sequencing of rotaviruses from domestic animals and humans living in close proximity can increase our understanding of the molecular epidemiology and evolution of RVAs in Uganda and other countries. Such studies, if conducted in a systematic way, will help to elucidate the complex interspecies transmission patterns that lead to the diversity of rotavirus strains seen among the different species. Ultimately, this should lead to a better understanding of the genes or gene combinations that govern successful transmission between hosts or that are likely to result in host restriction.
Supporting information
(DOC)
(XLS)
(DOCX)
(XLSX)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 2 encoding VP2 gene of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 3 encoding VP3 gene of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 5 encoding NSP1 of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 7 encoding NSP3 of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Acknowledgments
We thank the caretakers who allowed their children to participate in this study. We also thank the animal owners for allowing us sample from their animals. We thank the research assistants at Mulago National Referral Hospital, St. Francis Hospital Nsambya, Masaka Regional Referral Hospital and Kitovu Hospital, who assisted with participant recruitment and data collection. We thank the veterinary assistants from Masaka district, who assisted with the sample and data collection from animals. We thank Phionah Tushabe and Joseph Gaizi, who assisted with sample processing and laboratory testing at the Uganda Virus Research Institute. We are grateful to Rebecca A. Halpin, Nadia B. Fedorova,Karla M. Sucker and Tim Stockwell from J Craig Venter Institute, Adriana Luchs from Adolfo Lutz Institute, Jesus F. Salazar-Gonsalez from MRC/UVRI Uganda Research Unit on AIDS and Nasan Natseri from WHO, Uganda for their support.
Data Availability
The nucleotide sequences generated in this study are available in the NCBI GenBank under the accession numbers KX632243-632352, KX655437-KX655538, KX988264-KX988283, KY055416-KY055437, KY077640-KY077650.
Funding Statement
This research was funded by THRiVE, Wellcome Trust grant no. 087540 and Cambridge Alborado Research funds to JB, WHO funds for laboratory twinning initiative between Health Protection Agency (now Public Health England), Uganda Virus Research Institute and Laboratory for Viral infections, Almaty, Kazakhstan (Project no 105508) and WHO funds for rotavirus surveillance in Uganda. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Archives of virology. 2012;157(6):1177–82. Epub 2012/03/21. doi: 10.1007/s00705-012-1273-3 . [DOI] [PubMed] [Google Scholar]
- 2.Mihalov-Kovacs E, Gellert A, Marton S, Farkas SL, Feher E, Oldal M, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerging infectious diseases. 2015;21(4):660–3. Epub 2015/03/27. doi: 10.3201/eid2104.141370 ; PubMed Central PMCID: PMCPMC4378476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banyai K, Kemenesi G, Budinski I, Foldes F, Zana B, Marton S, et al. Candidate new rotavirus species in Schreiber's bats, Serbia. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;48:19–26. Epub 2016/12/10. doi: 10.1016/j.meegid.2016.12.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136(6):1939–51. Epub 2009/05/22. doi: 10.1053/j.gastro.2009.02.076 ; PubMed Central PMCID: PMCPMC3690811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desselberger U. Rotaviruses. Virus research. 2014;190:75–96. Epub 2014/07/13. doi: 10.1016/j.virusres.2014.06.016 . [DOI] [PubMed] [Google Scholar]
- 6.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2016;62 Suppl 2:S96–s105. Epub 2016/04/10. doi: 10.1093/cid/civ1013 . [DOI] [PubMed] [Google Scholar]
- 7.Midgley SE, Banyai K, Buesa J, Halaihel N, Hjulsager CK, Jakab F, et al. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Veterinary microbiology. 2012;156(3–4):238–45. Epub 2011/11/15. doi: 10.1016/j.vetmic.2011.10.027 . [DOI] [PubMed] [Google Scholar]
- 8.Estes MK, Kang G, Zeng CQ, Crawford SE, Ciarlet M. Pathogenesis of rotavirus gastroenteritis. Novartis Foundation symposium. 2001;238:82–96; discussion -100. Epub 2001/07/11. . [DOI] [PubMed] [Google Scholar]
- 9.Kirkwood CD. Genetic and antigenic diversity of human rotaviruses: potential impact on vaccination programs. The Journal of infectious diseases. 2010;202 Suppl:S43–8. Epub 2010/08/13. doi: 10.1086/653548 . [DOI] [PubMed] [Google Scholar]
- 10.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Archives of virology. 2011;156(8):1397–413. Epub 2011/05/21. doi: 10.1007/s00705-011-1006-z ; PubMed Central PMCID: PMCPmc3398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trojnar E, Sachsenroder J, Twardziok S, Reetz J, Otto PH, Johne R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. The Journal of general virology. 2013;94(Pt 1):136–42. Epub 2012/10/12. doi: 10.1099/vir.0.047381-0 . [DOI] [PubMed] [Google Scholar]
- 12.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Reviews in medical virology. 2005;15(1):29–56. Epub 2004/10/16. doi: 10.1002/rmv.448 . [DOI] [PubMed] [Google Scholar]
- 13.Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30 Suppl 1:A122–30. Epub 2012/05/02. doi: 10.1016/j.vaccine.2011.09.111 . [DOI] [PubMed] [Google Scholar]
- 14.Mwenda JM, Ntoto KM, Abebe A, Enweronu-Laryea C, Amina I, McHomvu J, et al. Burden and epidemiology of rotavirus diarrhea in selected African countries: preliminary results from the African Rotavirus Surveillance Network. The Journal of infectious diseases. 2010;202 Suppl:S5–s11. Epub 2010/08/13. doi: 10.1086/653557 . [DOI] [PubMed] [Google Scholar]
- 15.Esona MD, Geyer A, Page N, Trabelsi A, Fodha I, Aminu M, et al. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: relationship to animal rotaviruses. Journal of medical virology. 2009;81(5):937–51. Epub 2009/03/26. doi: 10.1002/jmv.21468 . [DOI] [PubMed] [Google Scholar]
- 16.Heylen E, Batoko Likele B, Zeller M, Stevens S, De Coster S, Conceicao-Neto N, et al. Rotavirus surveillance in Kisangani, the Democratic Republic of the Congo, reveals a high number of unusual genotypes and gene segments of animal origin in non-vaccinated symptomatic children. PLoS One. 2014;9(6):e100953 Epub 2014/06/27. doi: 10.1371/journal.pone.0100953 ; PubMed Central PMCID: PMCPmc4072759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Archives of virology. 2008;153(8):1621–9. Epub 2008/07/08. doi: 10.1007/s00705-008-0155-1 ; PubMed Central PMCID: PMCPmc2556306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. Journal of virology. 2008;82(7):3204–19. Epub 2008/01/25. doi: 10.1128/JVI.02257-07 ; PubMed Central PMCID: PMCPmc2268446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis FE, Fujii Y, Haga K, Damanka S, Lartey B, Agbemabiese CA, et al. Identification of novel Ghanaian G8P[6] human-bovine reassortant rotavirus strain by next generation sequencing. PLoS One. 2014;9(6):e100699 Epub 2014/06/28. doi: 10.1371/journal.pone.0100699 ; PubMed Central PMCID: PMCPMC4074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delogu R, Ianiro G, Morea A, Chironna M, Fiore L, Ruggeri FM. Molecular characterization of two rare human G8P[14] rotavirus strains, detected in Italy in 2012. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;44:303–12. Epub 2016/07/28. doi: 10.1016/j.meegid.2016.07.018 . [DOI] [PubMed] [Google Scholar]
- 21.Phan TG, Okitsu S, Maneekarn N, Ushijima H. Evidence of intragenic recombination in G1 rotavirus VP7 genes. Journal of virology. 2007;81(18):10188–94. Epub 2007/07/05. doi: 10.1128/JVI.00337-07 ; PubMed Central PMCID: PMCPMC2045391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donker NC, Boniface K, Kirkwood CD. Phylogenetic analysis of rotavirus A NSP2 gene sequences and evidence of intragenic recombination. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11(7):1602–7. Epub 2011/06/22. doi: 10.1016/j.meegid.2011.05.024 . [DOI] [PubMed] [Google Scholar]
- 23.Doro R, Farkas SL, Martella V, Banyai K. Zoonotic transmission of rotavirus: surveillance and control. Expert review of anti-infective therapy. 2015;13(11):1337–50. Epub 2015/10/03. doi: 10.1586/14787210.2015.1089171 . [DOI] [PubMed] [Google Scholar]
- 24.van der Heide R, Koopmans MP, Shekary N, Houwers DJ, van Duynhoven YT, van der Poel WH. Molecular characterizations of human and animal group a rotaviruses in the Netherlands. Journal of clinical microbiology. 2005;43(2):669–75. Epub 2005/02/08. doi: 10.1128/JCM.43.2.669-675.2005 ; PubMed Central PMCID: PMCPmc548030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steyer A, Poljsak-Prijatelj M, Barlic-Maganja D, Marin J. Human, porcine and bovine rotaviruses in Slovenia: evidence of interspecies transmission and genome reassortment. The Journal of general virology. 2008;89(Pt 7):1690–8. Epub 2008/06/19. doi: 10.1099/vir.0.2008/001206-0 . [DOI] [PubMed] [Google Scholar]
- 26.Rajendran P, Kang G. Molecular epidemiology of rotavirus in children and animals and characterization of an unusual G10P[15] strain associated with bovine diarrhea in south India. Vaccine. 2014;32 Suppl 1:A89–94. Epub 2014/08/06. doi: 10.1016/j.vaccine.2014.03.026 . [DOI] [PubMed] [Google Scholar]
- 27.Chakraborty P, Bhattacharjee MJ, Sharma I, Pandey P, Barman NN. Unusual rotavirus genotypes in humans and animals with acute diarrhoea in Northeast India. Epidemiology and infection. 2016:1–10. Epub 2016/04/27. doi: 10.1017/s0950268816000807 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bwogi J, Malamba S, Kigozi B, Namuwulya P, Tushabe P, Kiguli S, et al. The epidemiology of rotavirus disease in under-five-year-old children hospitalized with acute diarrhea in central Uganda, 2012–2013. Archives of virology. 2016. Epub 2016/01/05. doi: 10.1007/s00705-015-2742-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jere KC, Mlera L, O'Neill HG, Potgieter AC, Page NA, Seheri ML, et al. Whole genome analyses of African G2, G8, G9, and G12 rotavirus strains using sequence-independent amplification and 454(R) pyrosequencing. Journal of medical virology. 2011;83(11):2018–42. Epub 2011/09/15. doi: 10.1002/jmv.22207 . [DOI] [PubMed] [Google Scholar]
- 30.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. Journal of clinical microbiology. 1990;28(3):495–503. Epub 1990/03/01. ; PubMed Central PMCID: PMCPmc269651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. The Journal of general virology. 2009;90(Pt 6):1423–32. Epub 2009/03/07. doi: 10.1099/vir.0.009381-0 . [DOI] [PubMed] [Google Scholar]
- 32.Iturriza-Gomara M, Elliot AJ, Dockery C, Fleming DM, Gray JJ. Structured surveillance of infectious intestinal disease in pre-school children in the community: 'The Nappy Study'. Epidemiology and infection. 2009;137(7):922–31. Epub 2008/11/20. doi: 10.1017/S0950268808001556 . [DOI] [PubMed] [Google Scholar]
- 33.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads EMBnet 2011;17(1):10–2. [Google Scholar]
- 34.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. Epub 2012/05/01. doi: 10.1093/bioinformatics/bts199 ; PubMed Central PMCID: PMC3371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, et al. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2015;31:321–34. Epub 2015/02/24. doi: 10.1016/j.meegid.2015.02.011 ; PubMed Central PMCID: PMCPMC4361293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC microbiology. 2009;9:238 Epub 2009/11/26. doi: 10.1186/1471-2180-9-238 ; PubMed Central PMCID: PMCPMC2785824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology Evolution. 2013;30:2725–9. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–7. Epub 2004/03/23. doi: 10.1093/nar/gkh340 ; PubMed Central PMCID: PMCPMC390337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, et al. Evolutionary history and global spread of the emerging g12 human rotaviruses. Journal of virology. 2007;81(5):2382–90. Epub 2006/12/15. doi: 10.1128/JVI.01622-06 ; PubMed Central PMCID: PMCPMC1865926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giammanco GM, Bonura F, Zeller M, Heylen E, Van Ranst M, Martella V, et al. Evolution of DS-1-like human G2P[4] rotaviruses assessed by complete genome analyses. The Journal of general virology. 2014;95(Pt 1):91–109. Epub 2013/10/01. doi: 10.1099/vir.0.056788-0 . [DOI] [PubMed] [Google Scholar]
- 41.De Grazia S, Doro R, Bonura F, Marton S, Cascio A, Martella V, et al. Complete genome analysis of contemporary G12P[8] rotaviruses reveals heterogeneity within Wa-like genomic constellation. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;44:85–93. Epub 2016/06/30. doi: 10.1016/j.meegid.2016.06.039 . [DOI] [PubMed] [Google Scholar]
- 42.Dulgheroff AC, Silva GA, Naveca FG, Oliveira AG, Domingues AL. Diversity of group A rotavirus genes detected in the Triangulo Mineiro region, Minas Gerais, Brazil. Brazilian journal of microbiology: [publication of the Brazilian Society for Microbiology]. 2016;47(3):731–40. Epub 2016/06/09. doi: 10.1016/j.bjm.2016.04.012 ; PubMed Central PMCID: PMCPMC4927641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ianiro G, Delogu R, Bonomo P, Castiglia P, Ruggeri FM, Fiore L. Molecular characterization of human G8P[4] rotavirus strains in Italy: proposal of a more complete subclassification of the G8 genotype in three major lineages. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;21:129–33. Epub 2013/11/21. doi: 10.1016/j.meegid.2013.10.029 . [DOI] [PubMed] [Google Scholar]
- 44.Ndze VN, Esona MD, Achidi EA, Gonsu KH, Doro R, Marton S, et al. Full genome characterization of human Rotavirus A strains isolated in Cameroon, 2010–2011: diverse combinations of the G and P genes and lack of reassortment of the backbone genes. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;28:537–60. Epub 2014/12/03. doi: 10.1016/j.meegid.2014.10.009 . [DOI] [PubMed] [Google Scholar]
- 45.Wangchuk S, Mitui MT, Tshering K, Yahiro T, Bandhari P, Zangmo S, et al. Dominance of emerging G9 and G12 genotypes and polymorphism of VP7 and VP4 of rotaviruses from Bhutanese children with severe diarrhea prior to the introduction of vaccine. PLoS One. 2014;9(10):e110795 Epub 2014/10/21. doi: 10.1371/journal.pone.0110795 ; PubMed Central PMCID: PMCPMC4203849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall Thomas A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series 1999;41(95–98). [Google Scholar]
- 47.Matthijnssens J, Rahman M, Ciarlet M, Zeller M, Heylen E, Nakagomi T, et al. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerging infectious diseases. 2010;16(4):625–30. Epub 2010/03/31. doi: 10.3201/eid1604.091591 ; PubMed Central PMCID: PMCPMC3321964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das SR, Halpin RA, Stucker KM, Akopov A, Fedorova N, Puri V, et al. GenBank Acc no KP7525711Feb 2015.
- 49.Matthijnssens J, Rahman M, Yang X, Delbeke T, Arijs I, Kabue JP, et al. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. Journal of clinical microbiology. 2006;44(5):1801–9. Epub 2006/05/05. doi: 10.1128/JCM.44.5.1801-1809.2006 ; PubMed Central PMCID: PMCPMC1479174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komoto S, Wandera Apondi E, Shah M, Odoyo E, Nyangao J, Tomita M, et al. Whole genomic analysis of human G12P[6] and G12P[8] rotavirus strains that have emerged in Kenya: identification of porcine-like NSP4 genes. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;27:277–93. Epub 2014/08/12. doi: 10.1016/j.meegid.2014.08.002 . [DOI] [PubMed] [Google Scholar]
- 51.Nakagomi T, Nakagomi O, Dove W, Doan YH, Witte D, Ngwira B, et al. Molecular characterization of rotavirus strains detected during a clinical trial of a human rotavirus vaccine in Blantyre, Malawi. GenBank Acc no JN5914111. August 2011. [DOI] [PMC free article] [PubMed]
- 52.Ghosh S, Alam MM, Ahmed MU, Talukdar RI, Paul SK, Kobayashi N. Complete genome constellation of a caprine group A rotavirus strain reveals common evolution with ruminant and human rotavirus strains. The Journal of general virology. 2010;91(Pt 9):2367–73. Epub 2010/05/28. doi: 10.1099/vir.0.022244-0 . [DOI] [PubMed] [Google Scholar]
- 53.Ghosh S, Varghese V, Samajdar S, Bhattacharya SK, Kobayashi N, Naik TN. Evidence for independent segregation of the VP6- and NSP4- encoding genes in porcine group A rotavirus G6P[13] strains. Archives of virology. 2007;152(2):423–9. Epub 2006/09/29. doi: 10.1007/s00705-006-0848-2 . [DOI] [PubMed] [Google Scholar]
- 54.Papp H, Borzak R, Farkas S, Kisfali P, Lengyel G, Molnar P, et al. Zoonotic transmission of reassortant porcine G4P[6] rotaviruses in Hungarian pediatric patients identified sporadically over a 15 year period. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;19:71–80. Epub 2013/06/25. doi: 10.1016/j.meegid.2013.06.013 . [DOI] [PubMed] [Google Scholar]
- 55.Ghosh S, Urushibara N, Taniguchi K, Kobayashi N. Whole genomic analysis reveals the porcine origin of human G9P[19] rotavirus strains Mc323 and Mc345. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12(2):471–7. Epub 2012/01/10. doi: 10.1016/j.meegid.2011.12.012 . [DOI] [PubMed] [Google Scholar]
- 56.Dong HJ, Qian Y, Huang T, Zhu RN, Zhao LQ, Zhang Y, et al. Identification of circulating porcine-human reassortant G4P[6] rotavirus from children with acute diarrhea in China by whole genome analyses. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;20:155–62. Epub 2013/09/10. doi: 10.1016/j.meegid.2013.08.024 . [DOI] [PubMed] [Google Scholar]
- 57.Stupka JA, Degiuseppe JI, Parra GI. Increased frequency of rotavirus G3P[8] and G12P[8] in Argentina during 2008–2009: whole-genome characterization of emerging G12P[8] strains. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2012;54(2):162–7. Epub 2012/03/14. doi: 10.1016/j.jcv.2012.02.011 . [DOI] [PubMed] [Google Scholar]
- 58.Tacharoenmuang R, Komoto S, Guntapong R, Ide T, Haga K, Katayama K, et al. Whole Genomic Analysis of an Unusual Human G6P[14] Rotavirus Strain Isolated from a Child with Diarrhea in Thailand: Evidence for Bovine-To-Human Interspecies Transmission and Reassortment Events. PLoS One. 2015;10(9):e0139381 Epub 2015/10/01. doi: 10.1371/journal.pone.0139381 ; PubMed Central PMCID: PMCPMC4589232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez M, Phan TG, Galeano ME, Russomando G, Parreno V, Delwart E, et al. Genomic characterization of a rotavirus G8P[1] detected in a child with diarrhea reveal direct animal-to-human transmission. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;27:402–7. Epub 2014/08/30. doi: 10.1016/j.meegid.2014.08.015 . [DOI] [PubMed] [Google Scholar]
- 60.Duarte EA, Novella IS, Weaver SC, Domingo E, Wain-Hobson S, Clarke DK, et al. RNA virus quasispecies: significance for viral disease and epidemiology. Infectious agents and disease. 1994;3(4):201–14. Epub 1994/08/01. . [PubMed] [Google Scholar]
- 61.Martella V, Banyai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Veterinary microbiology. 2010;140(3–4):246–55. Epub 2009/09/29. doi: 10.1016/j.vetmic.2009.08.028 . [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi N, Ishino M, Wang Y, Chawla-sarkar M, Krishnan T. Diversity of G-type and P-type of human and animal rotaviruses and its genetic background. Communicating Current Research and Educational Topics and Trends in Applied Microbiology. 2007:847–58. [Google Scholar]
- 63.Malik Y, Kumar N, Sharma K, Sircar S, Bora D. Rotavirus diarrhea in piglets: A review on epidemiology, genetic diversity and zoonotic risks. Indian Journal of Animal Sciences. 2014;84(10):1035–942. [Google Scholar]
- 64.Amimo JO, Junga JO, Ogara WO, Vlasova AN, Njahira MN, Maina S, et al. Detection and genetic characterization of porcine group A rotaviruses in asymptomatic pigs in smallholder farms in East Africa: predominance of P[8] genotype resembling human strains. Veterinary microbiology. 2015;175(2–4):195–210. Epub 2014/12/30. doi: 10.1016/j.vetmic.2014.11.027 . [DOI] [PubMed] [Google Scholar]
- 65.Jere KC, Mlera L, O'Neill HG, Peenze I, van Dijk AA. Whole genome sequence analyses of three African bovine rotaviruses reveal that they emerged through multiple reassortment events between rotaviruses from different mammalian species. Veterinary microbiology. 2012;159(1–2):245–50. Epub 2012/05/01. doi: 10.1016/j.vetmic.2012.03.040 . [DOI] [PubMed] [Google Scholar]
- 66.Louge Uriarte EL, Badaracco A, Matthijnssens J, Zeller M, Heylen E, Manazza J, et al. The first caprine rotavirus detected in Argentina displays genomic features resembling virus strains infecting members of the Bovidae and Camelidae. Veterinary microbiology. 2014;171(1–2):189–97. Epub 2014/04/20. doi: 10.1016/j.vetmic.2014.03.013 . [DOI] [PubMed] [Google Scholar]
- 67.Doan YH, Nakagomi T, Aboudy Y, Silberstein I, Behar-Novat E, Nakagomi O, et al. Identification by full-genome analysis of a bovine rotavirus transmitted directly to and causing diarrhea in a human child. Journal of clinical microbiology. 2013;51(1):182–9. Epub 2012/11/02. doi: 10.1128/JCM.02062-12 ; PubMed Central PMCID: PMCPMC3536204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLS)
(DOCX)
(XLSX)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 2 encoding VP2 gene of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 3 encoding VP3 gene of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 5 encoding NSP1 of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Maximum Likelihood phylogenetic trees of nucleotide sequences of rotavirus genome segment 7 encoding NSP3 of human and animal strains circulating in Uganda, 2012–2014. Bootstrap values above 70 are shown for 1000 replicates. The Ugandan human strains are labelled with blue circles and the Ugandan animal strains are labelled with red triangles. Pigeon strain RVA/Pigeon-tc/PN/PO-13/1983/G18P[17] served as the outgroup. The scale bars at the bottom of the trees calibrate the genetic distance expressed as nucleotide substitution per site.
(TIF)
Data Availability Statement
The nucleotide sequences generated in this study are available in the NCBI GenBank under the accession numbers KX632243-632352, KX655437-KX655538, KX988264-KX988283, KY055416-KY055437, KY077640-KY077650.