Abstract
Desktop Genetics is a bioinformatics company building a gene-editing platform for personalized medicine. The company works with scientists around the world to design and execute state-of-the-art clustered regularly interspaced short palindromic repeats (CRISPR) experiments. Desktop Genetics feeds the lessons learned about experimental intent, single-guide RNA design and data from international genomics projects into a novel CRISPR artificial intelligence system. We believe that machine learning techniques can transform this information into a cognitive therapeutic development tool that will revolutionize medicine.
Keywords: : bioinformatics, biomedical research, CRISPR, design, genetics, genomics, sgRNA
Desktop Genetics was founded in 2012 by three University of Cambridge (UK) graduates with the goal of marrying modern genomics with rapid advancements in data science. CRISPR, a cheap and precise tool to manipulate the genome both in vitro and in vivo, has opened the door to new basic, preclinical and translational research studies. In 2013, with the emergence of CRISPR, the company shifted its focus to gene editing.
CRISPR works by associating a target-specific single-guide RNA (sgRNA) with an RNA-guided endonuclease (RGEN) such as Cas9. The sgRNA directs the RGEN to a specific locus in the genome where the complex induces double-stranded breaks in the DNA. The cell then endogenously repairs the genome through either nonhomologous end joining or homology-directed repair (HDR). Nonhomologous end joining happens at a higher (and inversely proportional) frequency to HDR and introduces indel mutations, while HDR uses an exogenous donor molecule to make precision edits [1].
The promise of CRISPR
Over the last decade, clinical genomics data have provided insight into the origins of human disease. Personal genome databases have revealed genetic variations, such as chromosomal inversions and SNPs, across human populations. The advent of CRISPR allows us to empirically establish relationships between mutations and disease pathogenesis. Taking this further, CRISPR can be used as a therapeutic option to correct these events in the clinic for somatic cell and gene therapy.
For example, hemophilia A is caused by chromosomal inversions, which knockout the Factor VIII clotting protein. In the hemophiliac population, inversions occur in major and minor forms. In a study by Park et al., researchers isolated endothelial cells from hemophilia patients and reprogrammed them into pluripotent stem cells. The group then corrected both inversions with CRISPR and injected them into FVIII-deficient mice. This approach successfully ameliorated disease symptoms [2].
In another landmark study, researchers used therapeutic CRISPR editing in live organisms. Yin et al. hydrodynamically injected a CRISPR plasmid into the livers of mice suffering from hereditary tyrosinemia type 1 (HT1). HT1 is caused by cytotoxic build-up of Fah proteins in liver cells due to an SNP. Owing to the regenerative nature of hepatocytes and the fact that healthy cells were selected for, a healthy phenotype was restored after 30 days [3].
Challenges of CRISPR research
While these therapeutic studies are promising, there are still barriers to CRISPR research. Many sgRNA design algorithms do not take into account experimental intent and provide broad scoring algorithms which, while generally helpful for CRISPR experiments, may not meet the specific needs of a given investigation.
For example, exploiting HDR for precise nucleotide adjustment remains a challenge. While some researchers have suggested solutions such as asymmetric DNA donors, these options are not offered by most online tools [4–6]. Such roadblocks prevent advanced gene-editing options from reaching therapeutic development.
Therapeutic delivery options are also limited. Delivering CRISPR components into specific patient tissues will likely require viruses or nanoparticles with restrictive cargo size. This is problematic when trying to fit both a CRISPR nuclease, such as the standard 4.2-kb Streptococcus pyogenes Cas9, and an sgRNA into a relatively small vector such as an adeno-associated virus [7].
Some studies suggest that noncoding DNA can regulate gene function [8]. With this in mind, researchers must work to limit CRISPR off-target events both in the coding and noncoding regions of the genome. We can further interrogate the so-called epigenome with CRISPR functional assays. This may produce potential drug targets and help us better understand the ramifications of off-target editing.
Another challenge is that most guide RNAs are designed against the reference genome of the model organism. In reality, cell-line genomes tend to differ due to perturbations such as cell-line-specific SNPs and copy number variants. Not only do these changes have an effect on sgRNA on-target activity, but they may also introduce unexpected off-target events. A lack of cell-line-specific genotypes stymies both basic and clinical CRISPR research. Understanding genetic variations in cells and human populations will help investigators design more effect guides and address the off-target effect [9,10].
Gold standard assays for investigating the intended (on-target) and unintended (off-target) effects of CRISPR guides on in vitro and in vivo models are in their infancy. This uncertainty makes it difficult to reproduce experimental outcomes and form consensus around effective guide design strategies. This also raises safety concerns about using CRISPR in humans.
Testing CRISPR dogma with DESKGEN
DESKGEN is our regularly curated cloud platform. It incorporates the latest thinking in sgRNA design algorithms and parameters. DESKGEN also serves as a proof-of-concept testing ground for an ever changing CRISPR dogma.
The Knockout and Knockin tools accommodate a range of genomes including both eukaryotes and prokaryotes, as well as alternative CRISPR nucleases such as Cas9 orthologs and Cpf1 [11]. Offering RGEN options in Knockin and Knockout mode gives investigators options for therapeutic delivery. Further, adjusting homology arm length and symmetry in Knockin mode can lead to more efficient HDR editing experiments. In both cases, these are parameters that can all be found in a unified suite of cloud tools.
Guide Picker, our third DESKGEN tool, can directly compare literature-based sgRNA design rules. For example, the Doench 2016 Full function incorporates a percent peptide score, which represents the target locus in the coding DNA sequence [12]. When we plotted Doench 2016 Full against percent peptide score, we saw a distinct position-based effect on Doench scoring; toward the 3′ end of the gene, Doench scores tend to drop off. Monitoring the correlation of design parameters can illuminate trends in our prediction algorithms.
Once designed using either Knockout, Knockin or Guide Picker, investigators can use these sequences to order sgRNA oligos and perform CRISPR experiments at the bench. We gather feedback from scientists who use DESKGEN to continuously improve our design principles.
High-throughput CRISPR screens
CRISPR screens are an excellent way to target large panels of genes or genomic regions with a pool of sgRNAs to test for function or essentiality. Researchers use these screens to understand which genes play key roles in phenomena like tumorigenesis (e.g., constitutive mutants of KRAS) and cytotoxic build-up (e.g., Fah in HT1). This approach equips scientist with tools to rapidly elucidate novel disease pathways, identify new drug targets and investigate the causality of genomic variants in human disease pathogenesis.
The effectiveness of a screen is reliant on sgRNA design rules. No individual scoring function can completely predict the behavior of a guide, but by combining different parameters, we can create a library that is well suited to a researcher's experiment. Our ongoing conversations with the CRISPR community influence how we adapt scoring functions from the literature. Not only do we use scoring functions found on DESKGEN, but we also incorporate and modify other sgRNA design parameters into our design process.
We understand that as versatile as DESKGEN is, researchers often benefit from support in designing high-throughput experiments. Accordingly, our bioinformatics team is free to design libraries to meet experimental intent by including score thresholds and parameter adjustments not inherently built into our online software. Table 1 includes a snapshot of just a few of these parameters.
Table 1. . Desktop Genetics CRISPR library design parameters.
| Score | Purpose |
|---|---|
| Doench (2014)/(2016) |
Predicted on-target score |
| Chari (2015) |
Predicted on-target score |
| Xu (2015) |
Predicted on-target score |
| Hsu (2013) |
Predicted on-target score |
| Percent peptide score |
Target location in coding DNA sequence |
| RGEN selection | Cas9 orthologs, Cpf1 with varying PAM sequences |
A snapshot of only a few of the scores and other thresholds used by Desktop Genetics to design CRISPR guide RNA libraries. These literature-based parameters address some of the fundamental concerns facing the CRISPR field.
PAM: Protospacer adjacent motif; RGEN: RNA-guided endonuclease.
For example, our cloud tools do not use on-target activity scores developed by Chari et al. [13] or Xu et al. [14]. These scores differ from the Doench 2016 score used on DESKGEN; they do not factor in percent peptide score and were trained on log2 fold change rather than a ranking system. Depending on the experiment, one score may be more appropriate to address a given intent.
We also work internally with a more precise off-target scoring system based on Hsu et al. [15]. The Hsu score evenly considers off-target hits across the genome. Our libraries are designed to evaluate off-target effects on a broad, weighted scale; this means that we look at separate Hsu scores for coding and noncoding regions and weight them according to factors such as noncanonical PAM sequences.
As an example, we know that NAG PAM sequences tend to be far less active CRISPR editing sites for Streptococcus pyogenes Cas9 [15]. Therefore, we give NAG off-targets less weight than NGG. This allows more accurate evaluation of guide specificity and, as a result, produces more reliable screen data. Knowing that the sgRNAs designed to target a given gene are not giving false-positive results by affecting other parts of the genome and epigenome is essential for building a case for causality and generating reproducible results. More robust conclusions about causality will help move the field toward safe and effective CRISPR therapeutics in years to come.
Next-generation sequencing data drive better guide design
Standardization of editing experiments is important for ensuring that laboratory data are reproducible across cell lines. CRISPR studies also need to be robust enough to meet stringent clinical regulations. However, current methods for validating experimental outcomes post-CRISPR can compromise accuracy and experimental throughput [16].
Mismatch cleavage assays such as Surveyor, while simple and cost effective, are unable to identify the sequence changes at the target site and are insensitive to low (<5%) allelic frequencies [17]. Comparatively, Sanger sequencing is highly accurate but not amenable to high-throughput applications in mixed cell populations [18]. Researchers are adopting next-generation sequencing as the new standard quality threshold.
Options such as amplicon deep sequencing are efficient, high-throughput approaches for validating gene-editing outcomes. Deep sequencing assays are sensitive enough to detect the identity and distribution of CRISPR edits within heterogeneous cell populations even at low frequencies. This provides comprehensive data on unintended edits across the coding and noncoding genome of the model cell line or organism.
Next-generation sequencing is becoming increasingly important in improving and demonstrating the accuracy of improved guide design techniques. Data suggest that genetic variation influences guide activity. Whole-genome sequencing of the experimental cell line prior to gene editing can provide model-specific data, improving predictions of guide efficiency and specificity. Data provided from personal genomes can improve clinical therapeutic design by addressing safety concerns.
This approach not only standardizes the characterization of guide activity, but also acts as a validation of guide design. Sequencing datasets that pass through our algorithms improve our cognitive machine learning tool. This creates a positive feedback loop that enhances our predictive capabilities. This is an essential step as we move toward a clinically relevant artificial intelligence CRISPR design system.
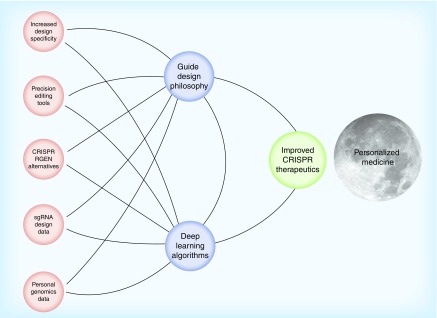
We imagine our gene-editing artificial intelligence system as a rocket (Figure 1). The only way to propel it forward is to give it fuel; the more we can provide, the better our rocket will perform. The data we gather through library screen analyses and sequencing data power our machine learning cognitive tool. As Desktop Genetics begins to cement itself as the go-to CRISPR bioinformatics resource in the field, our prediction software will only improve.
Figure 1. . Machine learning fuels Desktop Genetics.
Desktop Genetics uses data from CRISPR experiments and literature to fuel our cognitive machine learning algorithms. In concert with the moon shot goals of personal genomics initiatives, this artificial intelligence system will efficiently design CRISPR therapeutics tailored to the needs of individual patients.
The more scientists we collaborate with, the further we can move toward a new paradigm for genomics. Personal genome initiatives generate mountains of data that need to be analyzed and validated. We believe that combining CRISPR and deep learning approaches will allow us to meet the goals of personalized medicine.
Executive summary.
CRISPR and personal genomics promise to revolutionize medicine, but several roadblocks must be addressed beforehand.
The DESKGEN platform features multiple experiment-focused design settings while most online single guide RNA design tools do not.
The DESKGEN platform incorporates modern strategies to improve precision editing rates via homology-directed repair.
Vector/cargo barriers to therapeutic delivery may resolve with the use of alternative CRISPR endonucleases.
Desktop Genetics designs CRISPR libraries with weighted off-target scoring to more precisely edit the genome.
Next-generation sequencing (NGS) provides personal genomics that can be interrogated with CRISPR libraries.
NGS sequencing can be used to characterize model (or patient) genomes leading to better single-guide RNA design.
CRISPR libraries can be designed to target coding and noncoding (regulatory) regions of the genome.
Validating CRISPR experiments with NGS deep sequencing can ensure causality and identify or confirm off-target effects.
The more data that flow through Desktop Genetics, the better the CRISPR cognitive tool gets due to machine learning.
Footnotes
Financial & competing interests disclosure
All authors are employed by Desktop Genetics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park CY, Kim DH, Son JS, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17(2):213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratz SJ, Ukken FP, Rubinstein CD, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33(5):543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 6.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34(3):339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 7.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2016;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Sanjana NE, Wright J, et al. High-resolution interrogation of functional elements in the noncoding genome. bioRxiv. 2016 doi: 10.1126/science.aaf7613. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015;12(9):823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Xiao T, Chen CH, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25(8):1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vouillot L, Thélie A, Pollet N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 2015;5(3):407–415. doi: 10.1534/g3.114.015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell CC, Magor GW, Gillinder KR, Perkins AC. A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC. Genomics. 2014;15(1):1–7. doi: 10.1186/1471-2164-15-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]