Abstract
A real-time PCR and pyrosequencing method was developed to detect Aspergillus fumigatus in whole blood by amplifying the cyp51A gene and sequencing the codon for glycine 54. Mutations in this codon can result in amino acid substitutions that confer reduced susceptibility to itraconazole and posaconazole.
Invasive aspergillosis is a major cause of morbidity and mortality in immunocompromised patients. The survival of these individuals depends on early diagnosis and prompt initiation of effective antifungal treatment (5, 8). Unfortunately, conventional laboratory tests, such as culture and galactomannan detection, lack sensitivity and are rarely conclusive, resulting in true-positive results only at advanced stages of infection or necessitating invasive procedures for formal microbiological evaluation (2, 5). Furthermore, the emergence of clinical resistance to commonly used azole drugs impedes successful treatment (3, 8, 12). Therefore, the development of techniques to facilitate the early and reliable diagnosis of invasive aspergillosis and to predict susceptibility to antifungal drug treatment is currently under active investigation.
In this study, we developed a specific and sensitive method for detecting Aspergillus fumigatus in DNA extracted from whole blood and for rapidly identifying amino acid substitutions at residue 54 of cytochrome P450 14α-demethylase, Cyp51A, by sequencing the corresponding codon in the cyp51A gene. Cyp51A is involved in the synthesis of ergosterol, a bulk sterol component of fungal cell membranes, and is the target of azole-based compounds (14), which have become the most commonly used antifungal treatment. Other studies have identified clinical isolates and in vitro laboratory-selected mutants of A. fumigatus that exhibit reduced susceptibility to both itraconazole and posaconazole (4, 7, 9). Analysis of these isolates identified mutations in the cyp51A gene generating amino acid substitutions at glycine residue 54 of Cyp51A. Expression of cyp51A alleles containing these mutations in azole-sensitive A. fumigatus strains conferred reduced susceptibility. Three-dimensional modeling of the drug-and-protein interaction suggests that residue 54 resides in a channel that accommodates the long side chains of itraconazole and posaconazole (15).
Using the Rotor-Gene 3000 platform (Corbett Research, Sydney, Australia), a 269-bp region of the A. fumigatus cyp51A gene (GenBank accession number AF338659) was amplified. A dual-labeled DNA probe was selected for real-time monitoring of PCR amplification. PCRs were carried out in a volume of 25 μl containing a 300 nM concentration of each primer (forward, 5′-TCATTGGGTCCCATTTCTGGGTAG-3′; reverse, 5′-biotin/TAGACCTCTTCCGCATTGACATCC-3′), 100 nM probe (5′-6-FAM/AAACCACAGTCTACCTGGGCGTTCA/BHQ-1-3′), and 12.5 μl of a 2× concentration of Platinum Quantitative PCR Supermix-UDG (Invitrogen, Carlsbad, Calif.). PCR parameters were as follows: an initial incubation at 50°C for 2 min for UDG activity followed by 95°C for 2 min to inactivate the UDG and activate the Taq DNA polymerase. Next, 45 cycles of denaturation (95°C, 20 s) and annealing and extension (60°C, 60 s) were performed with fluorescence acquisition (excitation, 470 nM; emission, 510 nM) immediately following each annealing-extension step. A final extension (72°C, 10 min) was performed. Fluorescence curves were analyzed with dynamic tube normalization, slope correction, and automatic threshold determination by a best-fit line of at least three standards using Rotor-Gene version 5.0 software. The specificity of the real-time PCR was assessed by carrying out the reaction with DNA from a panel of 44 different species of viral, bacterial, and fungal pathogens, including the Aspergillus species A. fumigatus (SRRC 2006), A. flavus (MC 21), A. nidulans (NRRL 187), A. niger (SN 26), A. oryzae (NRRL 1989), A. terreus (NRRL 255), and A. versicolor (NRRL 238) (all isolated cultures were purchased from American Type Culture Collection and DNA extracted by using standard methods [1]), as well as human DNA. Only DNA from A. fumigatus was amplified (data not shown).
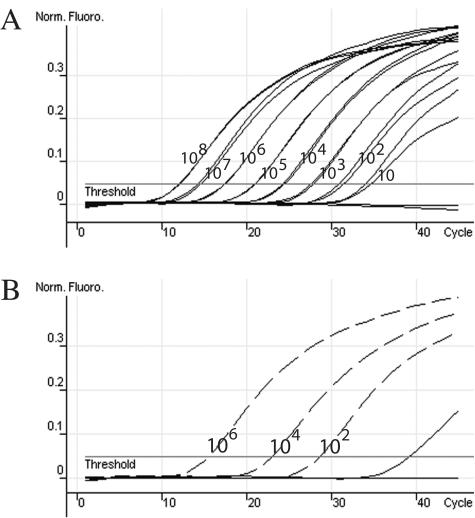
A positive control plasmid was constructed by subcloning the PCR amplicon generated with genomic DNA extracted from isolated A. fumigatus as a template into the pCR II-TOPO vector (Invitrogen). The linear detection range was from 108 to 10 copies of positive control plasmid per reaction in the presence of 500 ng of DNA extracted from Aspergillus negative whole-blood samples (Fig. 1A). All extractions of whole blood were performed as described previously (13). The real-time PCR was also able to detect 50 μg of A. fumigatus (mycelia from freeze-dried culture; American Type Culture Collection) spiked into 1-ml whole blood samples from two healthy donors with no clinical symptoms of aspergillosis, using the donors' nonspiked whole blood as negative controls (data not shown).
FIG. 1.
Positive control plasmid dilution detection range and analysis of clinical samples by real-time PCR. (A) Normalized fluorescence curves (Norm. Fluoro.) of the positive control plasmid containing the 269-bp fragment of A. fumigatus cyp51A, which was quantified by use of PicoGreen and diluted 10-fold from 108 to 10 copies per reaction in the presence of 500 ng of human DNA (respective CT curves for each 10-fold dilution, in duplicate, from left to right; r2 = 0.997). (B) Normalized fluorescence curves of three concentrations of positive control plasmid (dashed curves; r2 = 0.999), a negative control, and 32 DNA extracts from whole-blood samples. One positive specimen (solid curve) was detected with a calculated concentration of nine copies per reaction.
To assess the functionality of the real-time PCR with actual clinical samples, DNA extracts from 56 whole-blood samples obtained from different patients (sent to our lab for routine A. fumigatus detection) and three dilutions of the positive control plasmid were used as templates (Fig. 1B). A. fumigatus DNA was detected in 2 of the 56 samples, which correlates to the results obtained from an independent A. fumigatus conventional PCR assay targeting the 26S/internal transcribed spacer rRNA gene region (11). Neither PCR assay generated a positive result for the other 54 samples. The quality of clinical sample DNA and the absence of PCR inhibitors were validated by amplification of the human glyceraldehyde-3-phosphate dehydrogenase gene by real-time PCR (Biosearch Technologies, Novato, Calif.) (data not shown). These data support the use of this quantitative real-time PCR method for specific and sensitive detection of A. fumigatus in whole-blood DNA extracts. It is currently being determined if the methods are amenable to use with bronchoalveolar lavage samples to extend the clinical applicability.
Our approach to acquiring sequence data from real-time PCRs involves a bioluminometric, nonelectrophoretic technique called pyrosequencing, which employs a cascade of coupled enzymatic reactions to monitor DNA synthesis (10). The technique has the advantages of speed, accuracy, and parallel processing. The 5′ end of the reverse PCR primer was biotinylated to facilitate amplicon capture and preparation for pyrosequencing directly from the PCR using streptavidin Sephadex (Amersham Biosciences, Uppsala, Sweden) and the Pyrosequencing Vacuum Prep tool (Biotage, Uppsala, Sweden).
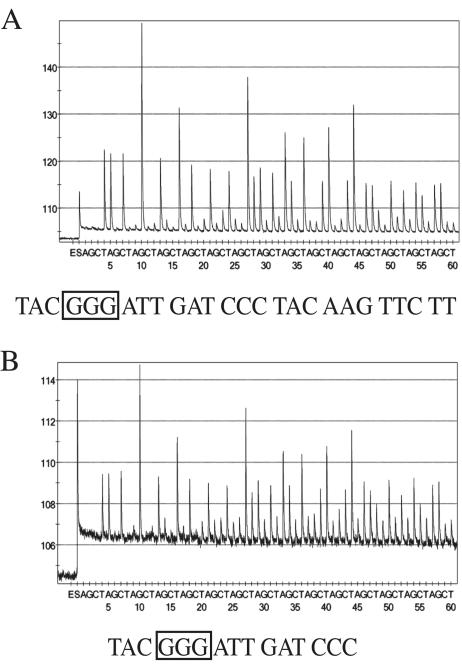
For the pyrosequencing reaction, a 0.5 μM concentration of the sequencing primer (5′-TCTGGGTAGTACCATCAGT-3′), a unique sequence within the PCR amplicon 5′ to the codon for the amino acid 54, was used. A Pyrosequencing 96MA System (Biotage) was programmed with 10 cycles of an AGCT dispensation order. The resulting pyrosequencing data, termed pyrograms, were analyzed with the PSQ 96MA version 2.1 software. The best-quality DNA sequences resolved were used in subsequent analyses. Typically, 19 to 24 bases of sequence were interpretable when genomic DNA extracted from isolated A. fumigatus was used (Fig. 2A). The lengths of the best-quality sequences were significantly shorter from clinical samples but provided enough sequence to identify the target and determine the codon for amino acid 54 (Fig. 2B). All sequences obtained by pyrosequencing were identical to the expected sequence of A. fumigatus cyp51A. These data support the use of this pyrosequencing method for identifying the codon for the amino acid at position 54 of Cyp51A.
FIG. 2.
Representative pyrosequencing data from positive controls and clinical samples. (A) Pyrosequencing analysis of the PCR amplicon from a wild-type A. fumigatus isolate purchased from American Type Culture Collection. (B) Pyrosequencing analysis of the positive clinical sample from Fig. 1B. The best-quality sequence is presented under each pyrogram, and the codon for amino acid 54 is boxed.
The continued elucidation of three-dimensional models of drug-and-substrate interaction may identify more sites of mutation that confer reduced susceptibility or resistance. The technique used here is amenable to alterations or additions that will amplify and sequence multiple regions involved in susceptibility. The data presented herein also serve as justification for expansion of the pyrosequencing analysis of cyp51A to determine other sites of mutation, including but not limited to the codons corresponding to the glycine residues near the heme cofactor at positions 138 and 448. Amino acid substitutions at either G138 or G448 confer reduced susceptibility to fluconazole and voriconazole (6, 15). Continued analysis of the correlation between the mutations and effectiveness of azole drugs with A. fumigatus isolates containing the mutations will support the usefulness of rapid identification of molecular determinants of drug susceptibility when selecting treatment.
REFERENCES
- 1.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 3.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manavathu, E. K., J. L. Cutright, D. Loebenberg, and P. H. Chandrasekar. 2000. A comparative study of the in vitro susceptibilities of clinical and laboratory-selected resistant isolates of Aspergillus spp. to amphotericin B, itraconazole, voriconazole and posaconazole (SCH 56592). J. Antimicrob. Chemother. 46:229-234. [DOI] [PubMed] [Google Scholar]
- 7.Mann, P. A., R. M. Parmegiani, S. Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marr, K. A., T. Patterson, and D. Denning. 2002. Aspergillosis. Pathogenesis, clinical manifestations, and therapy. Infect. Dis. Clin. N. Am. 16:875-894. [DOI] [PubMed] [Google Scholar]
- 9.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363, 365. [DOI] [PubMed] [Google Scholar]
- 11.Spreadbury, C., D. Holden, A. Aufauvre-Brown, B. Bainbridge, and J. Cohen. 1993. Detection of Aspergillus fumigatus by polymerase chain reaction. J. Clin. Microbiol. 31:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinbach, W. J., and D. A. Stevens. 2003. Review of newer antifungal and immunomodulatory strategies for invasive aspergillosis. Clin. Infect. Dis. 37(Suppl. 3):S157-S187. [DOI] [PubMed] [Google Scholar]
- 13.Van Burik, J. A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanden Bossche, H. 1985. Biochemical targets for antifungal azole derivatives: hypothesis on the mode of action. Curr. Top. Med. Mycol. 1:313-351. [DOI] [PubMed] [Google Scholar]
- 15.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]