Abstract
Small cell lung cancer (SCLC) has been associated with loss of heterozygosity at several distinct genetic loci including chromosomes 3p, 13q, and 17p. To determine whether the retinoblastoma gene (Rb) localized at 13q14, might be the target of recessive mutations in lung cancer, eight primary SCLC tumors and 50 cell lines representing all major histologic types of lung cancer were examined with the Rb complementary DNA probe. Structural abnormalities within the Rb gene were observed in 1/8 (13%) primary SCLC tumors, 4/22 (18%) SCLC lines, and 1/4 (25%) pulmonary carcinoid lines (comparable to the 20 to 40% observed in retinoblastoma), but were not detected in other major types of lung cancer. Rb messenger RNA expression was absent in 60% of the SCLC lines and 75% of pulmonary carcinoid lines, including all samples with DNA abnormalities. In contrast, Rb transcripts were found in 90% of non-SCLC lung cancer lines and in normal human lung. The finding of abnormalities of the Rb gene in SCLC and pulmonary carcinoids (both neuroendocrine tumors) suggests that this gene may be involved in the pathogenesis of a common adult malignancy.
In several childhood tumors, including retinoblastoma and Wilm’s tumor, there is growing evidence to indicate that the inactivation of both alleles of certain genes triggers tumorigenesis (1–3). The genomic locus determining susceptibility to retinoblastoma has been mapped to chromosome 13q14 (4), and several groups have obtained complementary DNA (cDNA) clones derived from this region that detect a DNA segment with properties of the putative retinoblastoma (Rb) gene (5–7). Evidence for this gene being the site of recessive mutations leading to tumor formation in retinoblastoma is based on the finding of structural changes within the gene, including internal homozygous deletions in a number of retinoblastomas, and the presence of altered or absent messenger RNA (mRNA) expression in the majority of both sporadic and familial forms of the tumor.
Specific chromosomal deletions have now been reported in various adult tumors (8), suggesting that “recessive oncogenes” may be important in the pathogenesis of these malignancies. In the case of small cell lung cancer (SCLC), a consensus deletion of DNA in the region 3p14–3p21 has been identified in virtually all cases (9–11). Several studies have also identified nonrandom changes involving other chromosomes, including chromosome 13. For example, one cytogenetic study of SCLC reported that chromosome 13 was the most frequently underrepresented chromosome, with 17 out of 21 SCLC lines having absent or hypodiploid numbers of chromosome 13 (12). In two recent studies, polymorphic probes from the long arm of chromosome 13 that spanned the region 13q12–13q33 demonstrated a reduction to homozygosity at one or more informative loci in primary tumor tissue from 18 of 23 patients with SCLC (11, 13). SCLC is an aggressive adult tumor which phenotypically resembles retinoblastoma in that both display properties of neural or neuroendocrine differentiation (14, 15) and both can have deregulated N-myc expression (16, 17). These observations, coupled with the recent report of structural abnormalities of the Rb gene in ~20% of osteosarcomas and other mesenchymal tumors (18) prompted us to examine the DNA and RNA status of the Rb gene in primary SCLC tumor tissue and in lung cancer cell lines to investigate the possible role of the Rb locus in the pathogenesis of these tumors.
Using the p0.9R and p3.8R cDNA probes (5), which represent the 4.7-kb Rb transcript that spans more than 180 kb of genomic DNA (6, 18), we analyzed DNA from eight SCLC primary tumor samples. We also studied DNA and RNA from 50 lung cancer cell lines (19), including 26 SCLC lines, 20 non-SCLC lines (eight adenocarcinomas, five large cell carcinomas, four bronchioloalveolar carcinomas, two adenosquamous carcinomas, and one squamous carcinoma), and four pulmonary carcinoid tumors (Table 1).
Table 1.
Summary of DNA and RNA status of the Rb gene in 50 lung cancer lines.
| Cell line* | DNA changes | RNA expression† | Cell line (histology) | DNA changes | RNA expression | |
|---|---|---|---|---|---|---|
| Small cell lung cancer | Pulmonary carcinoids | |||||
| H187 | 5′ rearrangement | − | H679 | (Atypical) | 3′ rearrangement | − |
| H345 | 3′ rearrangement | − | H720 | (Atypical) | None | − |
| H378 | 3′ rearrangement | − | H835 | (Atypical) | None | − |
| H889 | 3′ rearrangement | − | H727 | (Typical) | None | + |
| H82 | None | − | ||||
| N417 | None | − | Non-small celllung cancer | |||
| H510 | None | − | H125 | (Adenosquamous) | None | + |
| H524 | ND | − | H226 | (Squamous) | None | + |
| H526 | None | − | H322 | (Bronchioloalveolar) | None | + |
| H735 | None | − | H358 | (Bronchioloalveolar) | None | + |
| H748 | ND | − | H460 | (Large cell) | None | + |
| H774 | None | − | H522 | (Adenocarcinoma) | None | + |
| H1284 | None | − | H661 | (Large cell) | None | + |
| H1304 | None | − | H810 | (Large cell) | None | + |
| H1450 | ND | − | H820 | (Bronchioloalveolar) | None | + |
| H69 | None | tr | H838 | (Adenocarcinoma) | None | + |
| N592 | None | tr | H1373 | (Adenocarcinoma) | None | + |
| H711 | ND | tr | H1385 | (Large cell) | None | + |
| H847 | None | tr | H1404 | (Bronchioloalveolar) | None | + |
| H1622 | None | tr | H1435 | (Adenocarcinoma) | None | + |
| H209 | None | + | H1437 | (Adenocarcinoma) | None | + |
| H841 | None | + | H23 | (Adenocarcinoma) | None | tr |
| H1092 | None | + | H1355 | (Adenocarcinoma) | None | tr |
| H1105 | None | + | H596 | (Adenosquamous) | None | − |
| H1184 | None | + | H1155 | (Large cell) | None | − |
| H1436 | None | + | H1445 | (Adenocarcinoma) | None | ND |
Full designation of the cell lines includes the prefix “NCI-”
+, easily detectable transcript (comparable to normal lung expression);tr, trace; −, absent; ND, not determined.
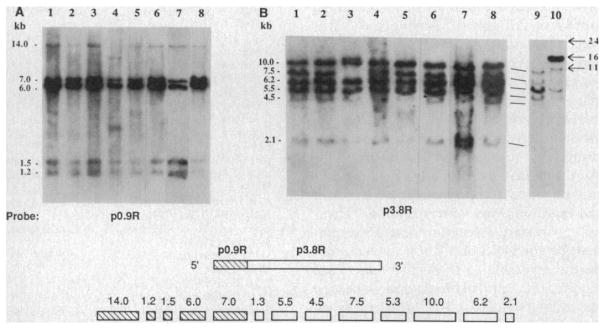
Structural rearrangements of the Rb gene were detected in DNA from one of the SCLC primary tumors (Fig. 1B, lane 10). Tumor DNA from this specimen exhibited novel Hind III fragments at 24, 16, and 11 kb. The 16-kb band was amplified several-fold relative to the other bands. In addition, the normal 10-kb band was markedly reduced in intensity, while the 7.5-, 6.2-, 5.5-, 4.5-, and 2.1-kb bands appeared normal. As the hybridization signal of the 10-kb fragment was much less than 50% of the expected intensity, this fragment may have been homozygously deleted in the tumor, with the residual band seen on DNA blot analysis representing contaminating normal cells in the biopsy sample. Similar abnormalities were also noted when the DNA was digested with Sst I (20). To expand on these observations and to examine correlations between DNA and RNA abnormalities, we studied the DNA and RNA status of Rb in lung cancer cell lines (Table 1).
Fig. 1.
Genomic restriction endonuclease analysis of the Rb in a representative panel of lung cancer lines. DNA (10 μg) extracted from seven lung cancer cell lines (H345, H510, H889, H187, H378, H209, and H679; lanes 1 to 7, respectively), normal human thymus (lane 8), and two primary SCLC tumor specimens (lanes 9 to 10, panel B) were digested to completion with Hind III and transferred to nitrocellulose after 0.8% agarose gel electrophoresis. Filters were then hybridized with either the p0.9R (A) or the p3.8R (B) 32P random-primed cDNA probes for 18 hours under standard hybridization conditions (26). For p0.9R, a 675-bp Eco RI–Hpa I fragment was used as a probe since this removed a GC-rich region that conferred a high background on hybridization. Filters were washed twice at room temperature in 2× SSC/0.1% SDS for 30 minutes and then twice in 0.1× SSC/0.1% SDS at 55°C for 1 hour. A diagram at the bottom of the figure depicts the previously described Rb cDNA clone and the Hind III genomic fragments (5, 6) detected by the p0.9R probe (hatched boxes) and the p3.8R probe (open boxes).
There were structural abnormalities within the Rb gene in four SCLC cell lines and in one carcinoid line (Fig. 1). These structural changes were of two types: (i) the homozygous loss of a normal Hind III fragment, associated with the appearance of one or two novel-sized bands, and (ii) reduced intensity of one or more normal bands by at least 50%, confirmed by densitometry, suggesting the presence of hemizygous deletions. SCLC line H889 displayed a complete loss of the 7.5-kb fragment with the appearance of a new band at 10.6 kb as identified with the p3.8R probe (Fig. 1B, lane 3), while H187 had a complete loss of the 14.0-kb Hind III fragment associated with the appearance of two new bands of 12.0 and 2.8 kb as determined with the p0.9R probe (Fig. 1A, lane 4). Additionally, in the carcinoid line H679, the p3.8R probe detected a doublet at 2.25 and 1.95 kb in place of the normal 2.1-kb band (Fig. 1B, lane 7). In all three cases, these changes were present in DNA obtained from the cell lines at different time points up to 28–50 passages in culture, suggesting that they were stable abnormalities. To exclude the possibility that the different patterns seen in H187, H679, and H889 represented Hind III restriction fragment length polymorphisms (RFLP), we digested DNA from these cell lines with Eco RI and Sst I, and by comparing to normal thymus DNA we observed rearrangements and homozygous loss of bands in these digests as well (20). In addition, 18 samples of normal human DNA probed with both p0.9R and p3.8R demonstrated a uniform pattern of restriction fragments with no Hind III polymorphisms (5), and no polymorphism has been reported for these restriction fragments among over 80 retinoblastomas and other tumors now examined. The homozygous internal deletion involving the 7.5-kb Hind III fragment, seen in line H889, has been observed in a large number of retinoblastomas and has been implicated as the possible site of a “mutational hotspot” (7).
SCLC lines H345 and H378 (Fig. 1, lanes 1 and 5, respectively) contained all expected Hind III bands detected by p3.8R, but on visual inspection both lines appeared to contain several bands of decreased intensity. To evaluate the possibility of partial Rb deletions on one allele in H345 and H378, densitometric analysis was performed to quantitate the DNA band intensities in both lines. Reproducible densitometric tracings were obtained and quantitated on autoradiograph films of DNA blots (hybridized either with p0.9R or p3.8R) from H345 and H378, 13 other lung cancer cell lines with normal-appearing DNA patterns, a normal B-lymphoblastoid line, and a normal human thymus specimen (fresh uncultured tissue). For DNA blots hybridized with p0.9R, all lung cancer lines, including H345 and H378, displayed densitometric tracings very similar to those for the normal human DNA samples. For blots hybridized with p3.8R, the tracings for H345 and H378 were markedly abnormal (Fig. 2), while the 13 other lung cancer lines showed tracings similar to those for normal human DNA. To quantitate these abnormalities, the areas under the peaks corresponding to each of the Hind III bands detected by p3.8R were calculated and normalized to the 10.0-kb peak, which served as an internal control. Each of the 13 lung cancer lines with normal densitometric tracings had quantitative estimates of band intensity ratios similar to those for the normal DNA samples, with minimal variability. In H345, however, there appeared to be a decrease in intensity by approximately 50% of the 6.2-kb band, suggesting that an internal hemizygous deletion of this genomic fragment may have occurred. In H378, the abnormalities were more complex but included a decrease in intensity of about 50% for the 6.2-kb and 2.1-kb bands, consistent with a hemizygous deletion of the 3′-most end of the gene (see Fig. 1 for genomic structure). The homozygous and hemizygous DNA structural abnormalities described above are similar to those recently published for retinoblastoma and associated mesenchymal tumors, and the presence of such changes has been interpreted as evidence to implicate the Rb locus as the target of recessive mutations in these tumors (5–7).
Fig. 2.
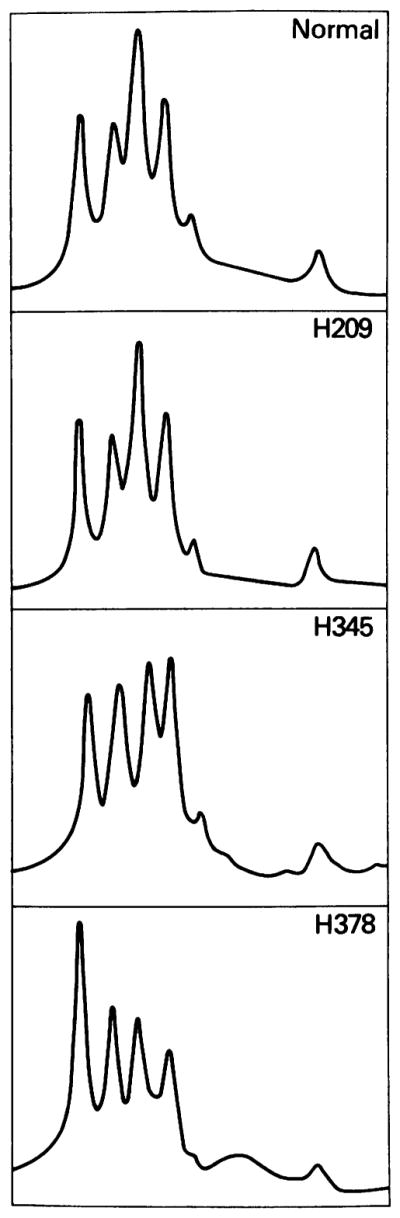
Densitometric analysis. The top panel shows the densitometric tracing observed with normal human DNA (from fresh thymus tissue) digested to completion with Hind III and hybridized with the p3.8R probe. The five peaks from left to right represent, respectively, the Hind III fragments at 10, 7.5, 6.2, 5.5, 4.5, and 2.1 kb (see Fig. 1B). The tracings were obtained on a Hoefer scanning densitometer (model GS300). Thirteen tumor lines, including H209 shown above, demonstrated densitometric patterns similar to that obtained for normal human DNA. In contrast, cell lines H345 and H378, shown in the lower two panels, had anomalous patterns.
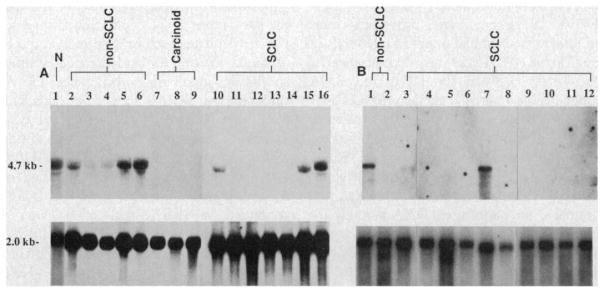
Total and poly(A)-selected RNA were prepared from the lung cancer lines and RNA analysis was performed with the p3.8R probe (Table 1 and Fig. 3). Of 26 SCLC lines, 15 expressed no detectable Rb mRNA, five expressed trace levels, and six had easily detectable levels of the Rb transcript. In addition, three of four pulmonary carcinoids expressed no detectable Rb transcript. Pulmonary carcinoid tumors are similar to SCLC in that both contain neurosecretory granules and exhibit neuroendocrine differentiation. Thus, these two tumors are distinctive from other types of lung cancer and may represent variants of malignant transformation in the same neuroendocrine precursor cell (21). All three tumors lacking the transcript were characterized as “atypical” carcinoids, with clinical and histologic features intermediate between SCLC and classic carcinoid (14). In contrast to SCLC and carcinoid, 15 of 19 non-SCLC lines expressed abundant steady-state levels of the 4.7-kb transcript, two expressed trace amounts, and two had undetectable levels of the Rb mRNA. These data demonstrated a correlation between the DNA and RNA status (Table 1) in that the four SCLC lines and one carcinoid line with gross structural changes within the Rb locus expressed no detectable Rb transcript, while the SCLC lines which did express abundant levels of mRNA, and all of the non-SCLC lines, had normal DNA patterns on DNA blots. Eight SCLC lines, however, expressed no detectable mRNA but had normal DNA patterns with the probes used in this study. As postulated for retinoblastoma, some or all of these cases may have contained small mutations in the Rb gene which accounted for the absent mRNA but which were undetectable by our DNA analysis.
Fig. 3.
Rb gene expression in a representative panel of lung cancer lines. (A) Total RNA (15 μg) was extracted (27) from normal lung tissue obtained from a surgical specimen (lane 1), five non-SCLC lines (H820, H1355, H1373, H1404, and H1437; lanes 2 to 6), three carcinoid lines (H720, H835, and H679; lanes 7 to 9), and seven SCLC lines (H1092, H1622, H748, H1304, H774, H1436, and H1105; lanes 10 to 16). (B) Two micrograms of poly(A)-selected RNA (28) was extracted from two additional non-SCLC lines (H125 and H23; lanes 1 and 2) and ten additional SCLC lines (N592, H345, H378, H510, H209, H187, H526, H524, H82, and N417; lanes 3 to 12). The above RNA samples were transferred to nitrocellulose after formaldehyde-denaturing gel electrophoresis (29) and filters were hybridized sequentially with the 32P random-primed p3.8R probe which detects the 4.7-kb Rb transcript (5–7) or with a β-actin cDNA probe which detects a 2-kb transcript (30). Autoradiographs were developed on XAR-5 film after 5 days at −70°C for p3.8R and after overnight exposure for β-actin. N, normal lung.
Expression of Rb mRNA was detected in total RNA from normal adult human lung (Fig. 3A, lane 1) and poly(A)-selected RNA from bovine lung (20). Although the cell of origin of SCLC remains controversial, the finding of Rb gene expression in lung tissue, as well as in a few SCLC lines and in all other normal tissues examined thus far (6), suggests that Rb may normally be expressed in most cells, including the SCLC precursor cell. Therefore, the lack of expression in the SCLC and carcinoid lines supports the idea that the Rb gene has been inactivated in these lines.
In light of our finding of DNA and RNA abnormalities in these cell lines, we analyzed the karyotypes of 11 of the SCLC lines for which adequate metaphase spreads could be obtained, looking for evidence of alterations specifically involving chromosome 13. All three of the SCLC lines with detectable rearrangements of the Rb gene that were examined cytogenetically contained abnormalities involving the region 13q14. H889 displayed a del(13)(q14>ter) and had no normal copy of chromosome 13. H187 contained a t(8;13)(q24;q14) and a t(9;13)(q13;q14), and H378 contained a del(13)(q14>ter) and a t(3;13)(p21;q14). In H510, a cell line with no Rb RNA expression and a normal DNA blot pattern, a portion of chromosome 13 was attached at band q14 to an unidentified chromosomal fragment, and there was no normal copy of 13. Two other cell lines with trace or absent Rb expression (H69 and H526) had lost one copy of chromosome 13 on the basis of expected modal number. In two SCLC lines with apparently intact structure and expression of Rb (H209 and H841) and in three lines with absent or trace expression (H82, N417, and H847) no abnormalities of number or structure of chromosome 13 were detected.
The presence of deletions within the Rb locus and lack of expression of the gene in SCLC is analogous to observations recently made in retinoblastoma and associated mesenchymal tumors where the gene is believed to contribute to tumorigenesis by a recessive “two-hit” mechanism (1). However, in contrast to retinoblastoma, which occurs in a familial form in 40% of cases (22), SCLC is not known to have a clear hereditary pattern. Although it is possible that an inherited abnormality of the Rb gene might predispose to SCLC (23), there is no established clinical association between retinoblastoma and SCLC, either in the same patient or within families. This would suggest that mutations at both alleles of the Rb gene in SCLC occur as somatic events. SCLC is closely associated with heavy cigarette smoking (24), and carcinogen exposure from tobacco may generate an increased mutation rate in the bronchial epithelium. Extremely large genes, such as Rb, may be especially susceptible to mutation and, thus, may commonly become inactivated in the bronchial epithelium of smokers.
From this study, it appears that abnormalities in structure and RNA expression of the Rb gene occur commonly in SCLC and pulmonary carcinoid but infrequently in other types of lung cancer. Further, the presence of DNA abnormalities of Rb in primary SCLC tumor tissue argues that these changes occur in vivo and are not an artifact of cell culture. Among the SCLC specimens examined, one primary tumor and 18% of cell lines demonstrated evidence for gross structural abnormalities of the Rb gene, and 60% expressed no detectable mRNA. Another 20% of SCLC lines expressed only trace amounts of the mRNA, suggesting that alterations in Rb gene expression may have occurred in these lines as well. Similar abnormalities of structure and expression were observed in our “atypical” pulmonary carcinoid lines. These percentages for DNA and RNA abnormalities were comparable to those that have been reported for retinoblastoma and associated mesenchymal tumors (5–7, 18). In comparison, no alterations of the Rb gene were detected in any of our non-SCLC lines, and 90% of these lines expressed Rb mRNA. Recent reports of chromosomal deletions at 3p in many non-SCLC tumors, as well as in SCLC (13, 25), suggest that the chromosome 3 deletion may be a common pathogenetic step to most or all major types of lung cancer. In contrast, a loss of heterozygosity of chromosome 13q markers appeared to occur mostly in SCLC when primary tumor tissue was examined (11, 13), and our data support this observation in that abnormalities of the Rb gene were more specific to SCLC and “atypical” pulmonary carcinoid. The finding of abnormalities of the Rb gene in a large percentage of both of these types of pulmonary neuroendocrine tumors suggests that inactivation of this gene may be an important event in the pathogenesis of lung tumors of neuroendocrine differentiation. These findings indicate that the Rb gene may be involved in the pathogenesis of a common non-ocular adult malignancy, suggesting that it may play an important physiological role and may contribute to malignant transformation in various tissues besides the retina. Ultimately, functional studies will be required to prove a definitive role for this gene in the genesis of human tumors.
Acknowledgments
We thank S. H. Friend, R. A. Weinberg, and T. P. Dryja for the generous gift of the p0.9R and p3.8R probes; B. E. Johnson for the primary tumor material; W. Nash for some of the cytogenetic data; and J. F. Battey, E. A. Sausville, I. R. Kirsch, M. J. Birrer, M. Zajac-Kaye, and M. M. Nau for critical review of the manuscript. J.W.H. is a Howard Hughes Medical Institute-NIH Research Scholar.
Contributor Information
J. William Harbour, Howard Hughes Medical Institute and NCI-Navy Medical Oncology Branch, National Cancer Institute, NIH, Bethesda, MD 20814.
Shinn-Liang Lai, NCI–Navy Medical Oncology Branch, National Cancer Institute, NIH, Bethesda, MD 20814.
Jacqueline Whang-Peng, Medical Oncology Branch, National Cancer Institute, NIH, Bethesda, MD 20814.
Adi F. Gazdar, NCI–Navy Medical Oncology Branch, National Cancer Institute, NIH, Bethesda, MD 20814
John D. Minna, NCI–Navy Medical Oncology Branch, National Cancer Institute, NIH, and Uniformed Services University of the Health Sciences, Bethesda, MD 20814
Frederic J. Kaye, NCI-Navy Medical Oncology Branch, National Cancer Institute, NIH, and Uniformed Services University of the Health Sciences, Bethesda, MD 20814
REFERENCES AND NOTES
- 1.Knudson AG., Jr Proc Natl Acad Sci USA. 1971;68:820. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedict WF, et al. Science. 1983;219:973. doi: 10.1126/science.6336308. [DOI] [PubMed] [Google Scholar]; Cavenee WK, et al. Nature. 1983;305:779. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]; Hansen MF, et al. Proc Natl Acad Sci USA. 1985;82:6216. doi: 10.1073/pnas.82.18.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friend SH, Dryja TP, Weinberg RA. N Engl J Med. 1988;318:618. doi: 10.1056/NEJM198803103181007. [DOI] [PubMed] [Google Scholar]
- 4.Knudson AG, Jr, Meadows AT, Nichols WW, Hill R. 1976;295:1120. doi: 10.1056/NEJM197611112952007. ibid. [DOI] [PubMed] [Google Scholar]; Yunis JJ, Ramsay N. Am J Dis Child. 1978;132:161. doi: 10.1001/archpedi.1978.02120270059012. [DOI] [PubMed] [Google Scholar]; Sparkes RS, et al. Science. 1983;219:971. doi: 10.1126/science.6823558. [DOI] [PubMed] [Google Scholar]
- 5.Friend SH, et al. Nature. 1986;323:643. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 6.Lee WH, et al. Science. 1987;235:1394. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 7.Fung YKT, et al. 1987;236:1657. ibid. [Google Scholar]
- 8.Klein G. 1987;238:1539. ibid. [Google Scholar]; Seizinger BR, et al. 1987;236:317. doi: 10.1126/science.3105060. ibid. [DOI] [PubMed] [Google Scholar]; Ali IU, Lidereau R, Theillet C, Callahan R. 1987;238:185. doi: 10.1126/science.3659909. ibid. [DOI] [PubMed] [Google Scholar]; Solomon E, et al. Nature. 1987;328:616. doi: 10.1038/328616a0. [DOI] [PubMed] [Google Scholar]
- 9.Whang-Peng J, et al. Cancer Genet Cytogenet. 1982;6:119. doi: 10.1016/0165-4608(82)90077-2. [DOI] [PubMed] [Google Scholar]; Whang-Peng J, et al. Science. 1982;215:181. doi: 10.1126/science.6274023. [DOI] [PubMed] [Google Scholar]
- 10.Naylor SL, Johnson BE, Minna JD, Sakaguchi AY. Nature. 1987;329:451. doi: 10.1038/329451a0. [DOI] [PubMed] [Google Scholar]; Brauch H, et al. N Engl J Med. 1987;317:1109. doi: 10.1056/NEJM198710293171803. [DOI] [PubMed] [Google Scholar]
- 11.Johnson BE, et al. J Clin Invest. in press. [Google Scholar]
- 12.Wurster-Hill DH, et al. Cancer Genet Cytogenet. 1984;13:303. doi: 10.1016/0165-4608(84)90075-x. [DOI] [PubMed] [Google Scholar]
- 13.Yokota J, Wada M, Shimosato Y, Terada M, Sugimura T. Proc Natl Acad Sci USA. 1987;84:9252. doi: 10.1073/pnas.84.24.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker KL, Gazdar AF, editors. The Endocrine Lung in Health and Disease. Saunders; Philadelphia: 1984. [Google Scholar]
- 15.Kyritsis AP, Tsokos M, Triche TJ, Chader GJ. Nature. 1984;307:471. doi: 10.1038/307471a0. [DOI] [PubMed] [Google Scholar]
- 16.Lee WH, Murphree AL, Benedict WF. 1984;309:458. doi: 10.1038/309458a0. ibid. [DOI] [PubMed] [Google Scholar]
- 17.Nau MM, et al. Proc Natl Acad Sci USA. 1986;83:1092. doi: 10.1073/pnas.83.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friend SH, et al. 1987;84:9059. doi: 10.1073/pnas.84.24.9059. ibid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carney DN, et al. Cancer Res. 1985;45:2913. [PubMed] [Google Scholar]; Gazdar AF, et al. :2924. ibid. [Google Scholar]; Gazdar AF. unpublished data. [Google Scholar]
- 20.Harbour JW, et al. data not shown. [Google Scholar]
- 21.Bensch KG, Corrin B, Pariente R, Spencer H. Cancer. 1968;22:1163. doi: 10.1002/1097-0142(196811)22:6<1163::aid-cncr2820220612>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 22.Vogel F. Hum Genet. 1979;52:1. doi: 10.1007/BF00284597. [DOI] [PubMed] [Google Scholar]
- 23.Messmer EP, Richter HJ, Hopping W, Havers W, Alberti W. Klin Mbl Augenheik. 1987;191:299. doi: 10.1055/s-2008-1050514. [DOI] [PubMed] [Google Scholar]
- 24.Weiss W, et al. J Am Med Assoc. 1972;222:799. [Google Scholar]
- 25.Kok K, et al. Nature. 1987;330:578. doi: 10.1038/330578a0. [DOI] [PubMed] [Google Scholar]
- 26.Davis LG, Dibner MD, Battey JF. Basic Methods in Molecular Biology. Elsevier/North-Holland; Amsterdam: 1986. [Google Scholar]
- 27.Chirgwin J, Przybyla A, Macdonald R, Rutter W. Biochemistry. 1979;18:5294. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 28.Aviv H, Leder P. Proc Natl Acad Sci USA. 1972;69:1408. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehrach H, Diamond D, Wozney JM, Boedtker H. Biochemistry. 1977;16:4743. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 30.Ponte P, Ng SY, Engel J, Gunning P, Kedes L. Nucleic Acids Res. 1984;12:1687. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]