Abstract
The LREX’ prostate cancer model is resistant to the antiandrogen enzalutamide via activation of an alternative nuclear hormone receptor (NHR), glucocorticoid receptor (GR), which has similar DNA binding specificity to the androgen receptor (AR). Small molecules that target DNA to interfere with protein-DNA interactions may retain activity against enzalutamide-resistant prostate cancers where ligand binding domain antagonists are ineffective. We reported previously that a pyrrole-imidazole (Py-Im) polyamide designed to bind the consensus androgen response element half-site has antitumor activity against hormone-sensitive prostate cancer. In enzalutamide-resistant LREX’ cells, Py-Im polyamide interfered with both androgen receptor- and glucocorticoid receptor-driven gene expression, while enzalutamide interfered with only that of androgen receptor. Genomic analyses indicated immediate interference with the androgen receptor transcriptional pathway. Long-term treatment with Py-Im polyamide demonstrated a global decrease in RNA levels consistent with inhibition of transcription. The polyamide was active against two enzalutamide-resistant xenografts with minimal toxicity. Overall, our results identify Py-Im polyamide as a promising therapeutic strategy in enzalutamide-resistant prostate cancer.
Keywords: Genitourinary cancers: prostate, minor-groove binder, androgen-receptor, CRPC
Introduction
Prostate cancer (CaP) is the second leading cause of cancer death in American men with 26,000 deaths annually (1), the majority from metastatic, castrate resistant CaP (mCRPC), in which androgen deprivation therapy (ADT), which suppresses AR signaling, is ineffective. Enzalutamide, a potent AR-ligand binding domain (LBD) antagonist, is effective against mCRPC and is a current standard of care (2). Unfortunately, de novo or acquired resistance to enzalutamide is common (3); overcoming this is an unmet need.
Mechanisms of enzalutamide resistance include restoration of AR signaling through LBD mutations or expression of transcriptionally active splice variants lacking the LBD (4), bypass of AR signaling through alternative NHRs (5), or development of complete independence from AR signaling (6). GR is a NHR with a sequence preference similar to AR (7). After enzalutamide treatment, the LREX’ cell line highly expresses GR, which drives enzalutamide resistance by regulating gene expression significantly overlapping that of AR, suggesting CaPs co-opt GR to progress through AR antagonism (5). Furthermore, GR expression in mCRPC associates with poor response to enzalutamide (5). Therefore, interference with the NHR-DNA interface may overcome enzalutamide resistance.
A Py-Im polyamide (ARE-1) is effective against hormone sensitive LNCaP xenografts with minimal host toxicity (8). Py-Im polyamides are minor groove DNA binding small molecules with modular sequence specificity and high affinity (9). Polyamide-DNA binding induces widening of the minor groove and compression of the opposing major groove (10), interfering with transcription factor-DNA interactions and the transcriptional machinery (11,12). A polyamide targeted to the ARE might prevent AR and GR signaling, and transcription.
We hypothesized that ARE-1 may be effective against enzalutamide-resistant CaP. We report ARE-1 efficacy against enzalutamide resistant VCaP and LREX’ CaP models in cell culture and xenografts. Mechanistic studies reveal immediate interference with androgen induced gene expression and reduced transcription after long-term treatment.
Materials and Methods
Cell culture conditions and cytotoxicity assays
The LREX’ and LNCaP/AR cell lines were gifts from Charles Sawyers (Memorial Sloan Kettering) and received 2014 and 2007, respectively. The VCaP cell line was a gift from Kenneth Pienta (University of Michigan Medical School) and received 2012. Cells were maintained as previously described (5,8,11–13), and were used within 10 passages from thawing. Cells were validated to parental cell lines by STR profile at IDEXXX Bioresearch following experimentation and confirmed to be Mycoplasma free. WST-1 assay (Roche) was used to measure cytotoxicity. Long-term toxicity in VCaP cells was assayed by cell counting.
Confocal imaging
Imaging was as described (8). Briefly, 2μM of ARE-1-FITC was added for 16hours, washed with PBS, and imaged on a Zeiss LSM 5 Exciter.
Gene expression analysis
LNCaP/AR and LREX’ cells were cultured for 72hours after plating in phenol-red free RPMI1640 (10% CT-FBS) in six well plates at 40,000 and 50,000cells/mL, respectively. LNCaP/AR cells were treated with 10μM ARE-1, bicalutamide (bic), or enzalutamide (enz, Aurum Pharmatech) for an additional 48, 2, and 2hours, respectively, prior to treatment with 1nM DHT or ethanol for 16hours. LREX’ cells were treated with 10μM ARE-1 for 16hours prior to induction with 1nM DHT or 100nM dexamethasone (dex) for 8hours. RNA extraction (RNEasy columns, Qiagen), cDNA generation (Transcriptor First Strand cDNA kit, Roche), and qRT-PCR (SYBR Green Master Mix, Applied Biosystems, ABI7300 instrument) were as described (8,11,12). Expression was normalized to β-glucuronidase.
RNAseq analysis
LREX’ cells were plated at 50,000cells/mL in 10cm2 dishes, treated with or without 10μM of ARE-1 in fresh media, incubated 16hours, and induced with 1nM DHT for 8hours. Tumor samples were homogenized mechanically. Total RNA was triazol extracted, sequenced (Illumina HiSeq2000), mapped against the human genome (hg19) with Tophat2 using Ensembl GRCh37 gene annotations. Human and mouse reads from tumor samples were parsed with BBSplit and unique reads were mapped. Htseq-count was used for exon alignment and DESeq2 for differential expression. Gene set enrichment analysis (GSEA) was performed on genes with padj<0.05 and fold change≥1.6 for cell samples and padj<0.05 for tumor samples.
Nascent RNA measurement
LREX’ cells were plated at 100,000cells/mL in 96 well plates in RPMI1640 (20% FBS and 1μM enz), adhered for 24hours, dosed with ARE-1, incubated for 48hours. The Click-iT® RNA Alexa Fluor® 488 HCS kit was used for dye conjugation and incorporation of 5-ethynyl uridine (5-EU) was measured on a Flexstation 3 plate reader.
Flow cytometry
LREX’ cells were plated at 100,000cells/mL in 175cm2 flasks, adhered 24hours, incubated with 10μM ARE-1 24, 48, and 72hours, then with 300μM 5-EU in fresh media. Cells were detached by Accumax or Accutase, and Alexa Fluor® 488 azide dye was conjugated. Cells were passed through 35μm mesh prior to flow, sorted on a FACSCalibur instrument (Beckman-Dickinson), analyzed using FlowJo.
Animal experiments
Animal experiments were performed at Caltech under IACUC approval. VCaP and LREX’ cells were engrafted as 1:1 mixtures of 3×106 cells in Matrigel (BD Biosciences) into flanks of intact and castrated male SCID mice (Charles River), respectively. LREX’ engrafted mice received 10mg/kg enz (oral gavage) daily. Once tumors were 100mm3 (0.5*l*w*w), ARE-1 was administered subcutaneously to opposing flanks in 20% DMSO:saline. For circulation studies, four C57BL6/J animals were injected subcutaneously with ARE-1 at 30mg/kg and blood collected retroorbitally. Plasma concentrations of ARE-1 were analyzed by HPLC, area under the curve (AUC) approximated by the linear trapezoidal method, as described (8).
Immunohistochemistry
Tumors were fixed in neutral-buffered formalin, paraffinized, sectioned, stained as described (12). Quantification of five random fields per slice was performed by ImmunoRatio.
Statistical analysis
Cell culture experiments represent ≥3 independent biological replicates. Sequencing analyses were duplicates for cell culture and quadruplicates for tumor samples. For xenografts, animals were randomly assigned to groups. For circulation experiments, concentrations of ARE-1 were duplicate measurements. Measurements in cell culture, animal, and immunohistochemistry experiments were assessed by Student’s t-test.
Results
ARE-1 is more potent than enzalutamide against CaP cell growth and is not rescued by GR activation
ARE-1 (Fig. 1A) targets the sequence 5̀-WGWWCW-3̀ (W=A or T), similar to the consensus half site recognized by either AR or GR. Nuclear uptake in LNCaP/AR, LREX’, and VCAP cells was evaluated using fluorescent analog ARE-1-FITC (Supplementary Fig. S1). The LNCaP/AR cell line overexpresses full length AR, modeling castration resistance (14). ARE-1 reduced proliferation of LNCaP/AR cells more than bicalutamide (Fig. 1B). The VCaP cell line overexpresses AR with modest GR expression, the activation of which reduces the antiproliferative effects of enzalutamide (5). ARE-1 reduced proliferation of both VCaP and LREX’ cells regardless of induction of AR signaling by 1nM DHT, induction of GR signaling by 100nM dex, or both (Fig. 1C–D). Long-term cell viability studies in VCaP cells show ARE-1 is more potent than enzalutamide and insensitive to GR activation (Fig. 1D).
Figure 1.
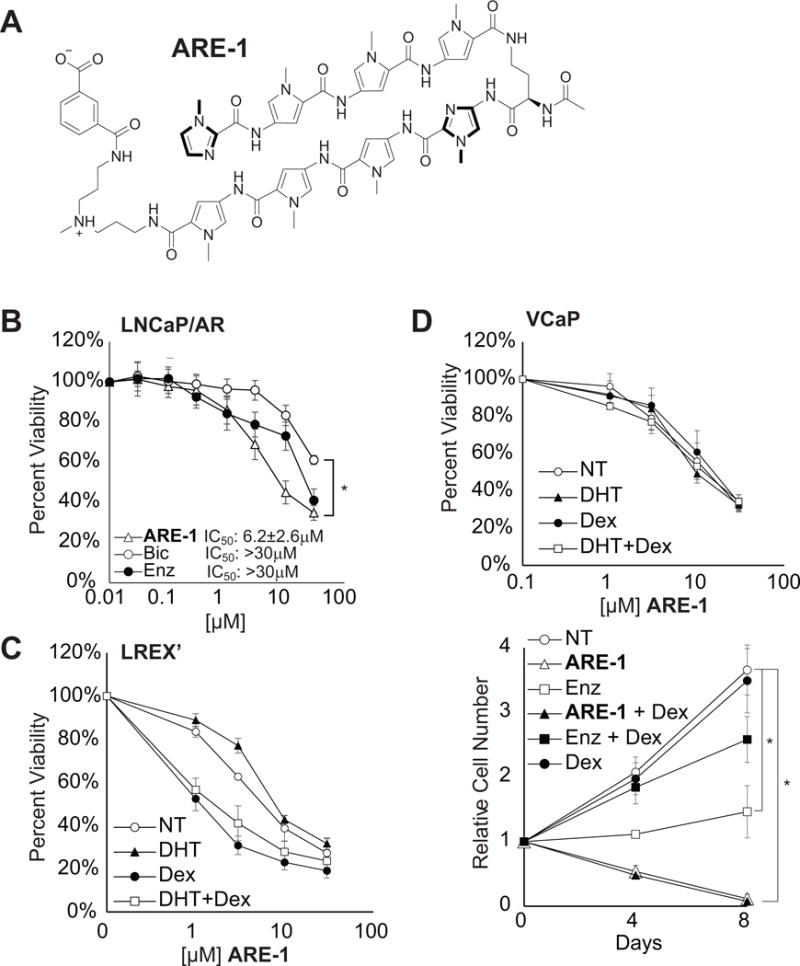
A, structure of ARE-1. B, cytotoxicity of ARE-1, Bic, and Enz in LNCaP/AR cells over 72hours. C, cytotoxicity of ARE-1 in LREX’ cells is not attenuated by AR or GR activation. D, top, cytotoxicity of ARE-1 in VCaP cells is not affected by Dex, bottom, long-term incubation of ARE-1 or Enz in VCaP cells. Error bars are SEM. * p<0.05.
Py-Im polyamide attenuates androgen and glucocorticoid driven gene expression
In androgen-depleted conditions, bicalutamide activates AR in the LNCaP/AR cell line (14). Enzalutamide and ARE-1 demonstrate no agonist activity; ARE-1 reduced baseline expression of KLK3 (Fig. 2A). In LREX’ cells, ARE-1 represses KLK3 and HOMER2 expression, which are co-regulated by AR and GR (Fig. 2B). While enzalutamide was more potent than ARE-1 in reducing DHT induced transcription, the opposite was observed with dex induction. Furthermore, co-administration of enzalutamide and ARE-1 was additive, suggesting ARE-1 may potentiate enzalutamide’s activity.
Figure 2.
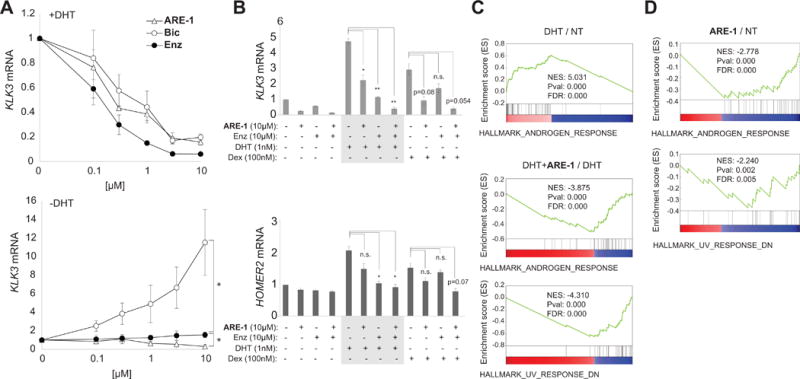
A, effects of ARE-1, Bic, and Enz on DHT induced KLK3 mRNA in LNCaP/AR cells (top), and baseline (charcoal stripped media) (bottom). B, effects of ARE-1 and Enz against select genes co-regulated by AR and GR in LREX’ cells. C, GSEA analysis in LREX’ cells. Top, DHT enriches for the Androgen Response. Bottom, ARE-1 with DHT negatively enriches for Androgen Response and the UV Response Down. D, ARE-1 treatment of LNCaP cells negatively enriches for Androgen Response and the UV Response Down. * p<0.05.
Global transcriptomic effects of Py-Im polyamides
We performed RNA-seq analysis on three treatment conditions in LREX’ cells: vehicle, DHT treatment, and co-treatment with ARE-1 and DHT, and two conditions in parental LNCaP cells: vehicle, and ARE-1 treatment. GSEA of affected genes in LREX’ cells using the hallmark pathways in the Molecular Signatures Database revealed DHT treatment enriched for the AR signaling pathway as expected (Fig. 2C, Supplementary Fig. S2 and Table S1). DHT-induced LREX’ cells treated with ARE-1 negatively enriched for the AR signaling pathway (NES −3.875) (Fig. 2C, Supplementary Table S1), consistent with interference in AR-driven gene expression by ARE-1. Additionally, ARE-1 treatment negatively enriched for the UV DNA damage response pathway down (NES −4.310) (Fig. 2C). Similarly, ARE-1 treatment in LNCaP cells negatively enriched for the AR signaling pathway (NES −2.778) and the UV DNA damage response pathway down (NES −2.240) (Fig. 2D, Supplementary Table S1). UV radiation induces DNA helical distortions through formation of pyrimidine dimers and 6–4 photoproducts, which arrest RNA Polymerase II (RNAP2) during elongation, triggering degradation of RPB1. ARE-1 reduced nascent RNA in LREX’ cells as measured by 5-EU incorporation (Fig. 3), and we have previously observed RPB1 degradation after long-term treatment with ARE-1 and related polyamides (8,12). This suggests long-term treatment with ARE-1 reduces global transcription in LREX’ cells.
Figure 3.
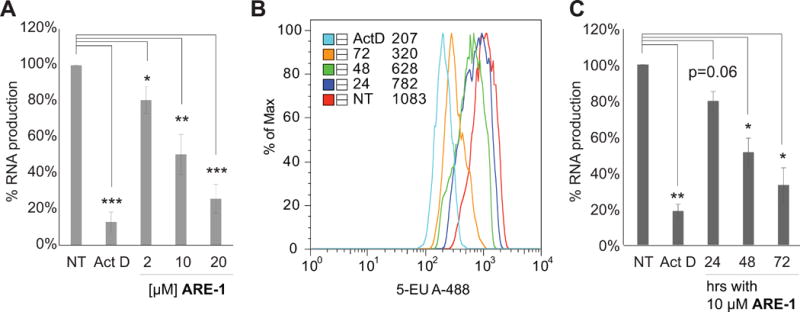
A, nascent RNA in LREX’ cells treated with ARE-1 for 48hours. Actinomycin D (Act D):positive control. B, nascent RNA in LREX’ cells by flow cytometry after treatment with 10μM ARE-1 for 24, 48, 72hours. C, composite of flow cytometry results. Error bars are SEM. *p<0.05; **p<0.005; ***p<0.0005.
Suppression of enzalutamide resistant, castrate resistant CaP in vivo
We further tested the efficacy of ARE-1 in VCaP xenografts, which exhibit modest response to 10mg/kg enzalutamide treatment, and in mice engrafted with enzalutamide resistant LREX’ cells (5,13). In VCaP xenografts, ARE-1 dose-dependently reduced tumor growth by 70% at 5mg/kg compared to vehicle (Fig. 4A) without significant toxicity (Supplementary Fig. S3A). In castrated mice bearing LREX’ tumors, ARE-1 and enzalutamide cotreatment reduced growth by 80% compared to enzalutamide alone (Fig. 4B) without significant toxicity (Supplementary Fig. S3B). Enzalutamide was administered daily post engraftment at 10mg/kg to maintain GR expression, which was confirmed by immunohistochemistry. LNCaP tumors, which do not express GR, were used as controls (Fig. 4C). Furthermore, LREX’ tumors treated with ARE-1 and enzalutamide showed reduced KLK3 expression (Supplementary Fig. S3C), elevated TUNEL and reduced Ki67 staining compared to enzalutamide alone (Supplementary Fig. S3D). GSEA of tumor expression profiles show ARE-1 treatment elicits similar UV response signatures as seen in cell culture, and represses ontologies associated with DNA binding dependent transcription (Supplementary Tables S2 and S3). Plasma concentration of ARE-1 from terminal blood samples from LREX’ engrafted animals was compared to the plasma concentration in C57BL6/J animals treated with 30mg/kg; AUC was 25.9 and 189.9μg*hr/mL, respectively (Supplementary Fig. S4). At 30mg/kg mice experienced a 6% weight loss but recovered within 5 days without visible signs of distress (not shown).
Figure 4.
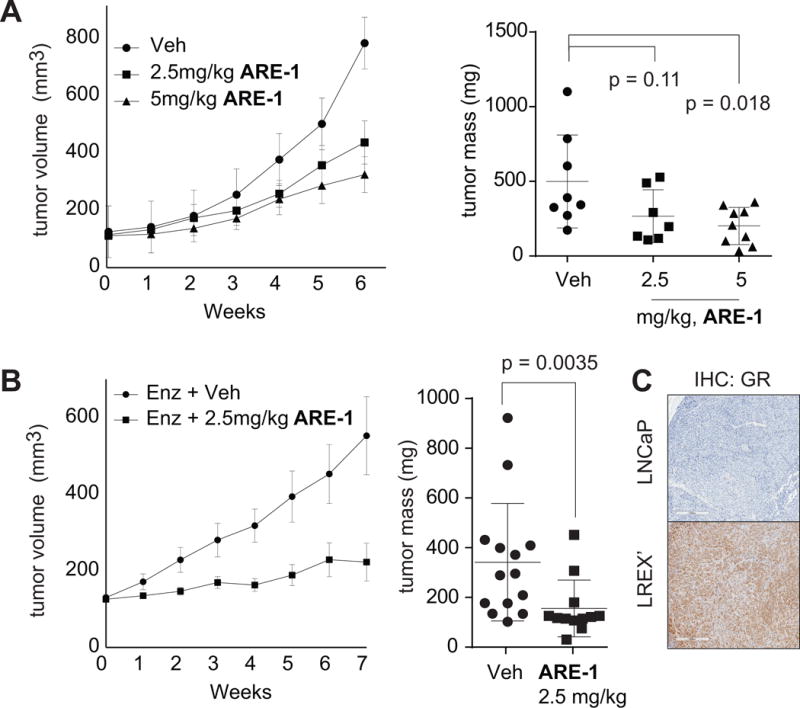
Mice were treated three times per week with ARE-1 subcutaneously to flanks opposite engrafted tumor. A, left, tumor volumes and (right) final tumor masses of VCaP xenografts treated with vehicle (Veh)(n=8), 2.5mg/kg(n=7), and 5mg/kg(n=8) ARE-1. B, left, tumor volumes and (right) final tumor masses of LREX’ xenografts in castrated animals treated daily with 10mg/kg Enz and Veh(n=14) or Enz and 2.5mg/kg ARE-1(n=12). C, GR staining of LREX’ and LNCaP tumors. All LREX’ tumors stained for GR. Error bars for tumor volumes are SEM. Whisker plots represent means, standard deviations.
Discussion
AR LBD mutations, expression of transcriptionally active splice variants lacking the LBD, co-option of NHRs with similar DNA binding specificities, or loss of reliance on AR, may drive enzalutamide resistance (3). Furthermore, different metastatic foci within a patient may resist enzalutamide through different mechanisms (15), suggesting a successful treatment strategy might use multiple therapeutics that overcome different resistance mechanisms, or alternatively, a single therapeutic capable of overcoming multiple mechanisms. Therapeutic targeting of the NHR-DNA interface may overcome most known enzalutamide resistance mechanisms.
The GR antagonist mifepristone added to ADT was previously tested in mCRPC patients and was not effective(16). Trials for mCRPC patients combining enzalutamide with mifepristone are underway. Other NHRs may also be active in refractory CaP (3). Notably, progesterone receptor inhibitors have entered clinical trials for mCRPC. Therapeutics targeting the N-terminal domain (NTD) of AR, or that mediate degradation of AR, may overcome treatment resistance due to AR splice variants. The NTD inhibitor EPI-506 has entered clinical trials (17). However, this approach may not overcome resistance due to co-option of alternate NHRs. Others have reported small molecules that interfere with the AR DNA-binding domain (18). The clinical utility of this approach is unknown.
We report a Py-Im polyamide with activity against enzalutamide-resistant CaP in cell and animal models. Polyamide ARE-1, targeted to the sequence 5′-WGWWCW-3′, which is similar to the ARE and GRE half site, attenuates ligand induced AR and GR transcriptional activity, is more potent than enzalutamide and bicalutamide in cell culture, and is active against enzalutamide resistant xenografts. Long term treatment of LREX’ cells with ARE-1 also decreases nascent RNA synthesis. In biophysical experiments, polyamides can halt RNAP2 elongation directly upstream of a polyamide binding site (19). We hypothesize this stalling of RNAP2 promotes ubiquitination and degradation of RPB1, ultimately interfering with RNA synthesis, which may contribute to efficacy against treatment refractory CaP. Other molecules that interfere with RNA synthesis are proposed as potential drug candidates for CaP (13,20).
Supplementary Material
Acknowledgments
The authors thank Rochelle Diamond at the Caltech flow cytometry facility for assistance.
Support: NIH grant GM27681 to P.B. Dervan. NIH grant T32GM007616 to A. A. Kurmis.
Footnotes
Conflict of Interest: P.B.D., F.Y., N.G.N. founded Gene Sciences, Inc. which is developing therapeutics for enzalutamide-resistant prostate cancer.
References
- 1.American Cancer Society Cancer. Facts & Figures 2016. Atlanta: American Cancer Society; 2016. [Google Scholar]
- 2.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in Metastatic Prostate Cancer before Chemotherapy. N Engl J Med. 2014;371:424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ware KE, Garcia-Blanco MA, Armstrong AJ, Dehm SM. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer. 2014;21:T87–103. doi: 10.1530/ERC-13-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid Receptor Confers Resistance to Antiandrogens by Bypassing Androgen Receptor Blockade. Cell. 2013;155:1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–10. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang F, Nickols NG, Li BC, Szablowski JO, Hamilton SR, Meier JL, et al. Animal toxicity of hairpin pyrrole-imidazole polyamides varies with the turn unit. J Med Chem. 2013;56:7449–57. doi: 10.1021/jm401100s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–99. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 10.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc Natl Acad Sci U S A. 2009;106:13175–9. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc Natl Acad Sci U S A. 2007;104:10418–23. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Nickols NG, Li BC, Marinov GK, Said JW, Dervan PB. Antitumor activity of a pyrrole-imidazole polyamide. Proc Natl Acad Sci U S A. 2013;110:1863–8. doi: 10.1073/pnas.1222035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 15.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taplin M-E, Manola J, Oh WK, Kantoff PW, Bubley GJ, Smith M, et al. A phase II study of mifepristone (RU-486) in castration-resistant prostate cancer, with a correlative assessment of androgen-related hormones. BJU Int. 2008;101:1084–9. doi: 10.1111/j.1464-410X.2008.07509.x. [DOI] [PubMed] [Google Scholar]
- 17.Myung J-K, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–60. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem. 2014;289:26417–29. doi: 10.1074/jbc.M114.553818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L, Wang W, Gotte D, Yang F, Hare AA, Welch TR, et al. RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif. Proc Natl Acad Sci. 2016;113:12426–31. doi: 10.1073/pnas.1612745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peltonen K, Colis L, Liu H, Jäämaa S, Zhang Z, Af Hällström T, et al. Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol Cancer Ther. 2014;13:2537–46. doi: 10.1158/1535-7163.MCT-14-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


