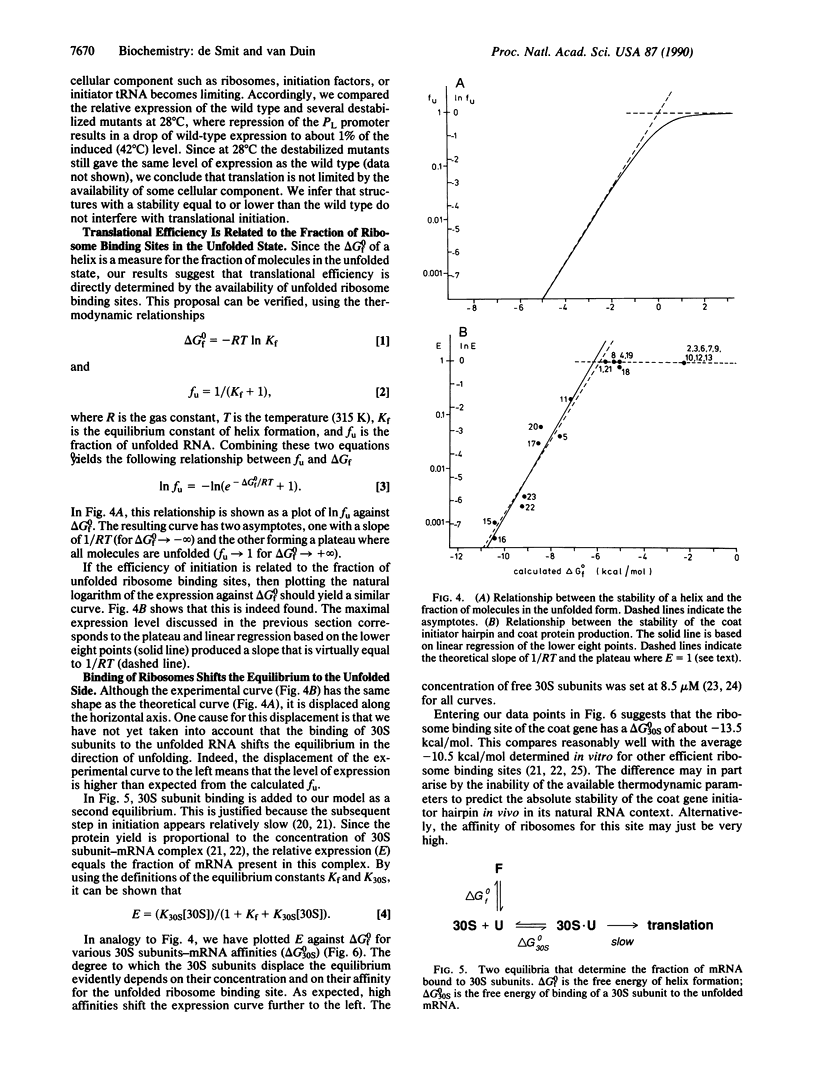
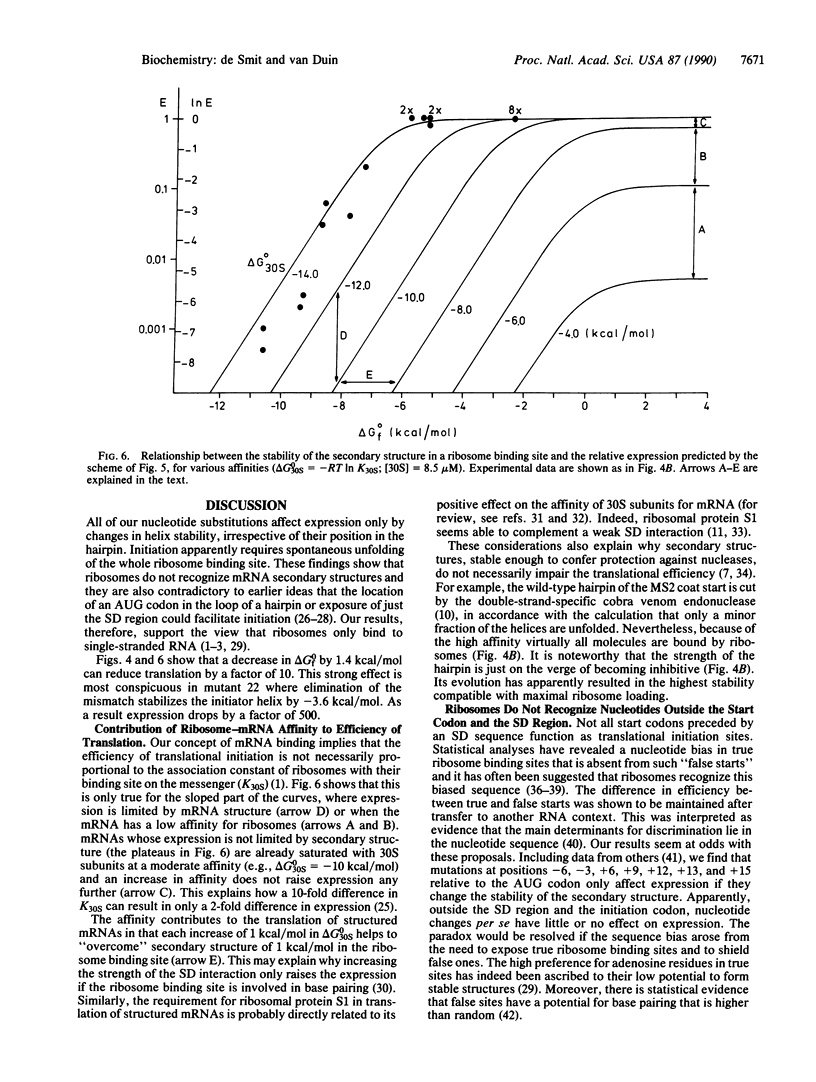
Abstract
We have quantitatively analyzed the relationship between translational efficiency and the mRNA secondary structure in the initiation region. The stability of a defined hairpin structure containing a ribosome binding site was varied over 12 kcal/mol (1 cal = 4.184 J) by site-directed mutagenesis and the effects on protein yields were analyzed in vivo. The results reveal a strict correlation between translational efficiency and the stability of the helix. An increase in its delta G0 of -1.4 kcal/mol (i.e., less than the difference between an A.U and a G.C pair) corresponds to the reduction by a factor of 10 in initiation rate. Accordingly, a single nucleotide substitution led to the decrease by a factor of 500 in expression because it turned a mismatch in the helix into a match. We find no evidence that exposure of only the Shine-Dalgarno region or the start codon preferentially favors recognition. Translational efficiency is strictly correlated with the fraction of mRNA molecules in which the ribosome binding site is unfolded, indicating that initiation is completely dependent on spontaneous unfolding of the entire initiation region. Ribosomes appear not to recognize nucleotides outside the Shine-Dalgarno region and the initiation codon.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angenent G. C., Posthumus E., Bol J. F. Biological activity of transcripts synthesized in vitro from full-length and mutated DNA copies of tobacco rattle virus RNA 2. Virology. 1989 Nov;173(1):68–76. doi: 10.1016/0042-6822(89)90222-5. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Schmidt B. F., van Strien A., van Boom J., van Westrenen J., van Duin J. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J Mol Biol. 1987 Jun 5;195(3):517–524. doi: 10.1016/0022-2836(87)90180-x. [DOI] [PubMed] [Google Scholar]
- Buell G., Schulz M. F., Selzer G., Chollet A., Movva N. R., Semon D., Escanez S., Kawashima E. Optimizing the expression in E. coli of a synthetic gene encoding somatomedin-C (IGF-I). Nucleic Acids Res. 1985 Mar 25;13(6):1923–1938. doi: 10.1093/nar/13.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero R. A., Pon C. L., Canonaco M. A., Gualerzi C. O. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos R., van Emmelo J., Contreras R., Fiers W. Construction and characterization of a plasmid containing a nearly full-size DNA copy of bacteriophage MS2 RNA. J Mol Biol. 1979 Mar 15;128(4):595–619. doi: 10.1016/0022-2836(79)90295-x. [DOI] [PubMed] [Google Scholar]
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988 Nov 5;204(1):79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- Ellis S., Conway T. W. Initial velocity kinetic analysis of 30 S initiation complex formation in an in vitro translation system derived from Escherichia coli. J Biol Chem. 1984 Jun 25;259(12):7607–7614. [PubMed] [Google Scholar]
- Forchhammer J., Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J Mol Biol. 1971 Feb 14;55(3):563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza M. C., Kofoid E. C., Marlière P., Louis B. G. Potential secondary structure at translation-initiation sites. Nucleic Acids Res. 1987 Jan 12;15(1):345–360. doi: 10.1093/nar/15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gouy M., Grantham R. Polypeptide elongation and tRNA cycling in Escherichia coli: a dynamic approach. FEBS Lett. 1980 Jun 30;115(2):151–155. doi: 10.1016/0014-5793(80)81155-0. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Risuleo G., Pon C. L. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry. 1977 Apr 19;16(8):1684–1689. doi: 10.1021/bi00627a025. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Looman A. C., Bodlaender J., Comstock L. J., Eaton D., Jhurani P., de Boer H. A., van Knippenberg P. H. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987 Aug;6(8):2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Munson L. M., Stormo G. D., Niece R. L., Reznikoff W. S. lacZ translation initiation mutations. J Mol Biol. 1984 Aug 25;177(4):663–683. doi: 10.1016/0022-2836(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Precup J., Parker J. Missense misreading of asparagine codons as a function of codon identity and context. J Biol Chem. 1987 Aug 15;262(23):11351–11355. [PubMed] [Google Scholar]
- Queen C., Rosenberg M. Differential translation efficiency explains discoordinate expression of the galactose operon. Cell. 1981 Jul;25(1):241–249. doi: 10.1016/0092-8674(81)90249-x. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Roberts M. W., Rabinowitz J. C. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J Biol Chem. 1989 Feb 5;264(4):2228–2235. [PubMed] [Google Scholar]
- Rosa M. D. Structure analysis of three T7 late mRNA ribosome binding sites. J Mol Biol. 1981 Mar 25;147(1):55–71. doi: 10.1016/0022-2836(81)90079-6. [DOI] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. F., Berkhout B., Overbeek G. P., van Strien A., van Duin J. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J Mol Biol. 1987 Jun 5;195(3):505–516. doi: 10.1016/0022-2836(87)90179-3. [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Selker E., Yanofsky C. Nucleotide sequence of the trpC-trpB intercistronic region from Salmonella typhimurium. J Mol Biol. 1979 May 15;130(2):135–143. doi: 10.1016/0022-2836(79)90422-4. [DOI] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Spanjaard R. A., van Dijk M. C., Turion A. J., van Duin J. Expression of the rat interferon-alpha 1 gene in Escherichia coli controlled by the secondary structure of the translation-initiation region. Gene. 1989 Aug 15;80(2):345–351. doi: 10.1016/0378-1119(89)90298-9. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. R. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Tessier L. H., Sondermeyer P., Faure T., Dreyer D., Benavente A., Villeval D., Courtney M., Lecocq J. P. The influence of mRNA primary and secondary structure on human IFN-gamma gene expression in E. coli. Nucleic Acids Res. 1984 Oct 25;12(20):7663–7675. doi: 10.1093/nar/12.20.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Szer W. RNA-helix-destabilizing proteins. Prog Nucleic Acid Res Mol Biol. 1982;27:157–187. doi: 10.1016/s0079-6603(08)60600-5. [DOI] [PubMed] [Google Scholar]
- Tomich C. S., Olson E. R., Olsen M. K., Kaytes P. S., Rockenbach S. K., Hatzenbuhler N. T. Effect of nucleotide sequences directly downstream from the AUG on the expression of bovine somatotropin in E. coli. Nucleic Acids Res. 1989 Apr 25;17(8):3179–3197. doi: 10.1093/nar/17.8.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]




