Abstract
A national evaluation study was performed in 11 specialized laboratories with the objective of assessing their capacities to genotype hepatitis C virus (HCV) and define the applicability of a given genotyping method. The panel consisted of 14 samples positive for HCV RNA of different genotypes (including 3 samples with two different artificially mixed genotypes) and 1 HCV-negative sample. Seventeen sets of data were gathered from the 11 participating laboratories. The sensitivities ranged from 64.3 to 100% and from 42.7 to 85.7% for the methods that used sequencing of the NS5b region and the 5′ noncoding (5′ NC) region, respectively. When the data for the artificially mixed samples were excluded, NS5b genotyping gave correct results for 80% of the samples, 1.7% of the samples were misclassified, and 18.3% of the samples had false-negative results. By 5′ NC-region genotyping methods, 58.3% of the results were correct, 29.7% were incomplete, 8.3% were misclassifications, 1.2% were false positive, and 2.4% were false negative. Only two procedures based on NS5b sequencing correctly identified one of the three samples with mixtures of genotypes; the other methods identified the genotype corresponding to the strain with the highest viral load in the sample. Our results suggest that HCV 5′ NC-region genotyping methods give sufficient information for clinical purposes, in which the determination of the subtype is not essential, and that NS5b genotyping methods are more reliable for subtype determination, which is required in epidemiological studies.
Hepatitis C virus (HCV) is responsible for chronic liver disease, with a risk of evolution toward severe diseases such as cirrhosis and hepatocellular carcinoma (34). More efficient antiviral treatments have been developed in recent years (21), but their efficacies are largely influenced by several biological parameters, such as the virus genotype. For this reason, HCV genotyping is used to predict the response to antiviral therapy (12, 23, 30) and, in association with the determination of the viral load and related markers in different hosts, to optimize the duration of treatment (2, 31). Furthermore, HCV genotyping is an essential tool for epidemiological studies (3, 22, 29) and for tracing a source of contamination by HCV (1, 18-20, 27).
HCV isolates are characterized by a high degree of heterogeneity: six main genotypes and more than 70 subtypes have been described (35). Many genotyping methods focused on the 5′ noncoding (5′ NC) region have been developed, and some of them are commercially available. However, the ability of the sequence of this region to discriminate isolates at the subtype level is disputed (8), and alternative genomic regions have been proposed for use in genotyping (11, 26, 33).
Thus, before the initiation of large-scale epidemiological or therapeutic studies, the Action Coordonnée 11 group of the Agence Nationale de Recherches pour le SIDA initiated an evaluation of the HCV genotyping methods used in 11 specialized laboratories involved in multicenter clinical trials. The aim of this study was to assess the HCV genotyping capacities of these specialized laboratories and to define the best applicability of a given genotyping method.
MATERIALS AND METHODS
Panel.
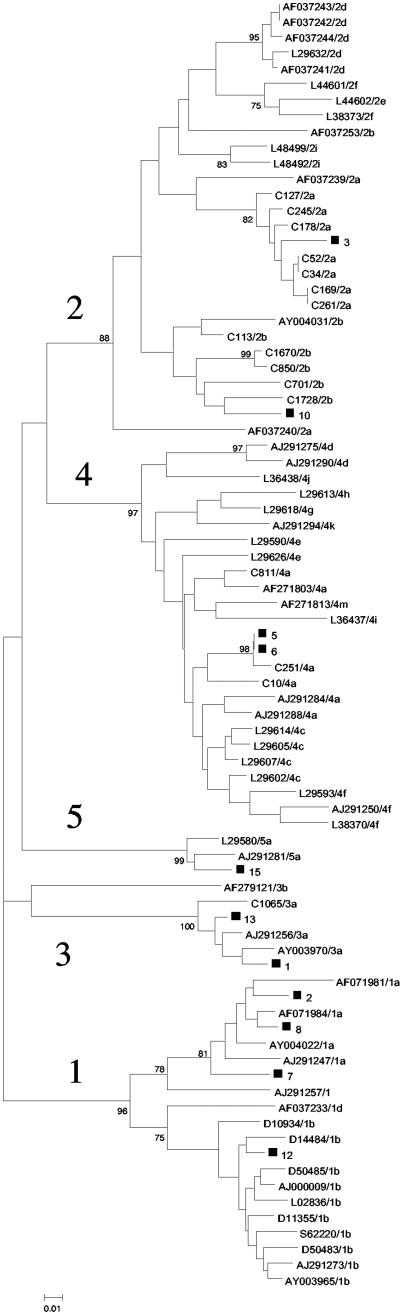
The panel was made up of 15 samples, including 10 undiluted samples (collected from HCV-infected blood donors and selected as a subset of HCV subtypes mainly encountered in clinical practice in Europe), 1 diluted HCV-positive sample (sample 6, which was a 1-in-200 dilution of sample 5), 3 samples with mixtures of two different genotypes (to mimic coinfections), and 1 HCV-negative sample. The characteristics of these samples are given in Table 1. Each HCV-positive sample was characterized by its viral load (Amplicor HCV Monitor, version 2.0; Roche, Meylan, France) and by its HCV genotype, determined by a method based on sequence analysis of the NS5b region (25). Briefly, after viral RNA extraction (QI Amp Viral RNA; Qiagen, Hilden, Germany) and reverse transcription, performed by a random priming method (cDNA ramdom hexamers kit; Amersham Pharmacia Biotech, Orsay, France), cDNA was amplified by a heminested PCR based on primers PR3 and PR4 in the first round, followed by PCR with primers PR3 and PR5 in the second round (Table 2). The DNA of each strain obtained from the purified PCR products (Quick Spin; Qiagen) was sequenced by PCR with primers PR3 and PR5. Cycle sequencing was undertaken by use of the fluorescent dye terminator technology (Big Dye terminator cycle sequencing; Applied Biosystems, Courtaboeuf, France) with AmpliTaq DNA polymerase, according to the instructions of the manufacturer. Electrophoresis and data collection were performed with an ABI 3100 genetic analyzer. The genotype of each sample was determined by comparison with those of HCV prototype strains from GenBank. Figure 1 shows the phylogenetic tree in which the samples of the panel are represented.
TABLE 1.
Characteristics of the panel
| Sample no. | Genotype (by NS5b sequencing) | HCV RNA load (log IU/ml) |
|---|---|---|
| 1 | 3a | 4.01 |
| 2 | 1a | 4.93 |
| 3 | 2a | 5.39 |
| 4 | −a | 0 |
| 5 | 4a | 5.62 |
| 6 | 4a | 3.41 |
| 7 | 1a | 3.33 |
| 8 | 1a | 6.00 |
| 9 | 3a-1b | 5.86/14.31 |
| 10 | 2b | 5.44 |
| 11 | 1a-1b | 4.93/14.31 |
| 12 | 1b | 5.68 |
| 13 | 3a | 5.86 |
| 14 | 1b-3a | 5.68/4.01 |
| 15 | 5a | 6.65 |
The sample was HCV negative.
TABLE 2.
PCR primers used in NS5b-based in-house methods
| Gene region and primer name | Polarity | Sequencea | Positionb | Reference |
|---|---|---|---|---|
| NS5b | ||||
| PR1 | Sense | 5′-TGGGGATCCCGTATGATACCCGCTGCTTTGA-3′ | 8245-8275 | 14 |
| PR2 | Antisense | 5′-GGCGGAATTCCTGGTCATAGCCTCCGTGAA-3′ | 8616-8645 | 14 |
| PR3 | Sense | 5′-TATGAYACCCGCTGYTTTGACTC-3′ | 8256-8278 | 25 |
| PR4 | Antisense | 5′-GCNGARTAYCTVGTCATAGCCTC-3′ | 8622-8644 | 25 |
| PR5 | Antisense | 5′-GCTAGTCATAGCCTCCGT-3′ | 8619-8636 | 33 |
| 1203 | Sense | 5′-ATGGGGTTCTCGTATGATACCCGCTGCTTTGACTC-3′ | 8244-8278 | 24 |
| 1204 | Antisense | 5′-GGAGGGGCGGAATACCTGGTCATAGCCTCCGTCAA-3′ | 8616-8650 | 24 |
| 122 | Sense | 5′CTCAACCGTCACTGAGAGAGACAT-3′ | 7935-7958 | 24 |
| 123 | Antisense | 5′-CCTCCTGCTCGCCTTGGACTCTCG-3′ | 8614-8591 | 24 |
| 242 | Antiense | 5′-GGCGGAATTCCTGGTCATAGCCTCGCTGAA-3′ | 8275-8304 | 14 |
| 243 | Sense | 5′-TGGGGATCCCGTATGATACCCGCTGCTTTGA-3′ | 7904-7934 | 14 |
| 5′ NC | ||||
| KY 80 | Sense | 5′-GCAGAAAGCGTCTAGCCATGGCGT-3′ | −274 to −250 | 40 |
| KY 78 | Antisense | 5′-CTCGCAAGCACCCTATCAGGCAGT-3′ | −31 to −55 | 40 |
| SF1 | Sense | 5′-GCCATGGCGTTAGTATGAGT-3′ | −261 to −240 | 13 |
| DOG-1 | Antisense | 5′-CAGGCAGTACCACAAGGC-3′ | −54 to −77 | 13 |
Y = C or T; R = A or G; V = A, C, or G; N = A, T, G, or G.
Numbering according to Choo et al. (10).
FIG.1.
Phylogenetic analysis of the NS5b sequences of the samples containing HCV strains included in the panel (excluding those artificially coinfected) compared with NS5b reference sequences from GenBank and in-house NS5b sequences. HCV genotypes are designated by the numbers 1 to 5. Genomes from this study are indicated with black squares and with the sample number. The phylogenetic tree was constructed by using the neighbor-joining method in the PHYLIP package (15). Bootstrap values are shown at each main branch.
Among the three samples containing mixtures of genotypes, sample 11 had equivalent viral loads of two genotypes (genotypes 1a and 1b), whereas samples 9 and 14 had greater viral loads of genotypes 3a and 1b, respectively. All samples in the panel were prepared and aliquoted by an external investigator and were randomly coded and sent under appropriate transportation conditions to each participating laboratory for blind testing. Each laboratory was free to use any genotyping method of its choice.
Participating laboratories.
The 11 participating laboratories were coded as laboratories A to K. Six of them used one genotyping method, four used two genotyping methods, and one used three genotyping methods, resulting in a total of 17 results. The different genotyping methods used consisted of four in-house protocols of the NS5b sequencing assay (16, 20, 24, 33) in eight laboratories, a newly developed NS5b sequencing assay (Trugene HCV NS5b genotyping kit; Bayer Health Care Diagnostics, Puteaux, France) (28) in two laboratories, an in-house 5′NC region sequencing assay in two laboratories (13, 39), and two commercially available 5′ NC region genotyping assays (Inno-LIPA [Innogenetics, Ghent, Belgium] and Trugene HCV genotyping kit [Bayer Health Care Diagnostics]) in five laboratories (32, 37). Table 2 includes the details for the PCR primers used only with the in-house methods.
Interpretation of results.
For each HCV-positive sample, the result was considered correct when both the correct genotype and the correct subtype were identified and, for samples with mixtures of genotypes, when both the genotype and the subtype of the strain with the highest viral load were identified. An incomplete result was defined as an exact genotype result with an unidentified subtype or with the absence of discrimination between two subtypes. A correct genotype in association with the incorrect subtype defined a misclassification. The sensitivity was defined as the percentage of correct results (correct genotype and correct subtype) among the 14 HCV-positive samples. The quality score was calculated by the percentage of correct results among all samples in the panel.
RESULTS
Performance of HCV genotyping.
Overall, 17 sets of data were generated by five different technical approaches (Table 3). In order to simplify the presentation, we considered each set of data as coming from an independent laboratory.
TABLE 3.
Performance of the laboratories in HCV genotype determination
| Gene region and genotyping technique | Labora- tory | Primers used for PCRa or reference | Primers used for sequencing or reference | Results (no. of samples) for HCV RNA-positive samples (n = 14)
|
No. of samples with false-positive results (n = 1) | Quality scoree (n = 15) (no. [%] of samples) | |||
|---|---|---|---|---|---|---|---|---|---|
| Correct resultsb | Incomplete resultsc | Misclassified samplesd | False-negative results | ||||||
| NS5b | |||||||||
| In-house | A | PR3-PR4/PR3-PR5 | PR3-PR5 | 10 (71.4) | 0 | 0 | 4 | 0 | 11 (73.3) |
| In-house | B | PR1-PR2/PR3-PR5 | PR3-PR5 | 12 (85.7) | 0 | 1 | 1 | 0 | 13 (86.7) |
| In-house | C1 | PR3-PR4/PR3-PR5 | PR3-PR5 | 12 (85.7) | 0 | 0 | 2 | 0 | 13 (86.7) |
| Trugene | C2 | 28 | 13 (92.9) | 0 | 0 | 1 | 0 | 14 (93.3) | |
| In-house | D | 1203-1204 | 123 | 9 (64.3) | 0 | 0 | 5 | 0 | 10 (66.7) |
| In-house | E1 | 1203-1204 | 1203-1204 (−20 ntf at 3′ end) | 11 (78.6) | 1 | 0 | 2 | 0 | 12 (80) |
| In-house | F1 | 1203-1204/1203-123 | 1203-1204 | 11 (78.6) | 1 | 0 | 2 | 0 | 12 (80) |
| Trugene | G1 | 28 | 12 (85.7) | 0 | 1 | 1 | 0 | 13 (86.7) | |
| In-house | H1 | 243-242/122-123 | 122-123 | 9 (64.3) | 0 | 0 | 5 | 0 | 10 (66.7) |
| In-house | I | PR3-PR4 | PR3-PR4 | 14 (100) | 0 | 0 | 0 | 0 | 15 (100) |
| 5′ NC | |||||||||
| In-house | E2 | K80-K78 | K80-K78 | 9 (64.3) | 3 | 0 | 1 | 0 | 10 (66.7) |
| In-house | F2 | K80-K78 | DOG1-SF1 | 9 (64.3) | 3 | 0 | 1 | 1 | 10 (60) |
| Trugene | G2 | 32 | 32 | 12 (85.7) | 1 | 1 | 0 | 0 | 13 (86.7) |
| InnoLipa | G3 | 37 | 9 (64.3) | 2 | 2 | 1 | 0 | 10 (66.7) | |
| InnoLipa | H2 | 37 | 6 (42.7) | 7 | 1 | 0 | 0 | 7 (46.7) | |
| Trugene | J | 32 | 32 | 6 (42.7) | 7 | 1 | 0 | 0 | 7 (46.7) |
| InnoLipa | K | 37 | 6 (42.7) | 7 | 1 | 0 | 0 | 7 (46.7) | |
Heminested PCR or nested PCR was used when two pairs of primers are mentioned.
Percentage of correct results among the HCV RNA-positive samples. Percent sensitivity is presented in parentheses.
Correct genotype with a subtype not identified.
Correct genotype but incorrect subtype.
Correct results among the 15 samples in the panel.
nt, nucleotides.
The sensitivities ranged from 64.3 to 100% and from 42.7 to 85.7% for laboratories using the NS5b and 5′ NC-region genotyping methods, respectively. Among the 10 NS5b region-based methods, incomplete results were observed for two (14.3%) samples, whereas the seven 5′NC-region-based methods provided incomplete results for one to seven (0.7 to 50%) samples. Two of the NS5b-based analyses misclassified the genotype in one sample, while five of the 5′NC-region-based analyses misclassified the genotype in one or two samples. False-negative results were more commonly observed by NS5b-based analyses (9 of 10; 90%), which failed to identify the genotype in one to five samples. The virus in only one sample could not be genotyped by three of seven 5′NC-region-based methods. A false-positive result was observed for only one sample (by an in-house 5′ NC-region-based assay). The quality scores ranged from 66.7 to 100% for the NS5b-based analyses and from 46.7 to 86.7% for the 5′NC-region-based analyses.
Comparison of NS5b- and 5′NC-region-based genotyping results by sample (excluding coinfected samples).
A further analysis that excluded the samples with artificially mixed genotypes was selectively performed (Table 4). By the 10 approaches based on NS5b analysis, 120 results were expected. Among these, 96 (80%) results were correct. For each sample, the proportion of correct results ranged from 10 to 100%. Of the 24 incorrect results, 2 could be attributed to misclassifications (in two samples) and 22 could be attributed to false-negative results (seven samples from which sequences could not be amplified by one to nine genotyping assays).
TABLE 4.
Results obtained for the 12 samples in the panel, infected only with single genotypes
| Sample no. | HCV RNA load (log IU/ml) | Genotype by NS5b-based genotyping | No. of samples
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS5b-based genotyping (n = 10)
|
5′ NC-region-based genotyping (n = 7)
|
|||||||||||
| Correct resultsa | Incomplete results | Misclassified samples | False-positive results | False-negative results | Correct resultsa | Incomplete results | Misclassified samples | False-positive results | False-negative results | |||
| 1 | 4.01 | 3a | 7 (70) | 3 | 6 (86) | 1 | ||||||
| 2 | 4.93 | 1a | 6 (60) | 4 | 6 (86) | 1 | ||||||
| 3 | 5.39 | 2a | 9 (90) | 1 | 2 (29) | 5 | ||||||
| 4 | 0 | Negative | 10 (100) | 6 (86) | 1 (1b)b | |||||||
| 5 | 5.62 | 4a | 7 (70) | 1 (4c)b | 2 | 2 (29) | 4 | 1 (4c)b | ||||
| 6 | 3.41 | 4a | 1 (10) | 9 | 5 | 1 (4c)b | 1 | |||||
| 7 | 3.33 | 1a | 7 (70) | 1 (1b)b | 2 | 1 | 5 (1b)b | 1 | ||||
| 8 | 6.00 | 1a | 10 (100) | 5 (71) | 2 | |||||||
| 10 | 5.44 | 2b | 10 (100) | 5 (71) | 2 | |||||||
| 12 | 5.68 | 1b | 9 (90) | 1 | 6 (86) | 1 | ||||||
| 13 | 5.86 | 3a | 10 (100) | 5 (71) | 2 | |||||||
| 15 | 6.65 | 5a | 10 (100) | 6 (86) | 1 | |||||||
| Totalc | 96/120 (80) | 2/120 (1.7) | 22/120 (18.3) | 49/84 (58.3) | 25/84 (29.7) | 7/84 (8.3) | 1/84 (1.2) | 2/84 (2.4) | ||||
Values in parentheses in this column are in percent.
Designations in parentheses are the genotype identified.
Data represent the total number of samples with the indicated result/total number of results (percent).
By the seven approaches based on 5′ NC-region analysis, 84 results were expected. Among these, 49 (58.3%) results were correct. For each sample, the proportion of correct results ranged from 0 to 86%. Most (71%) of the incorrect results were due to incomplete genotype identification, while the genotypes in seven samples were misclassified. Two false-negative results and one false-positive result were also noted.
Comparison of NS5b and 5′NC-region genotyping results for coinfected samples.
The results of the comparison of the NS5b and 5′NC-region genotyping results for the coinfected samples are depicted in Table 5. Sample 9 (which contained genotypes 3a and 1b) was never identified as containing a mixed infection, and the genotype present in the sample was classified as 3a in 14 cases (all 10 NS5b-based analyses and 4 of the 7 5′NC-region-based analyses). One laboratory, using an NS5b-based method, identified the genotype 3a strain and mentioned the presence of a double population without giving the other subtype. All three incorrect results were obtained by 5′ NC-region-based methods: two because incomplete data were provided and one due to a lack of amplification.
TABLE 5.
Results obtained with the three samples containing mixtures of genotypes
| Sample no. | Genotypes by NS5b-based genotyping | Viral load (log IU/ml) | Genotype(s) identified by method based on the following region and procedurea:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS5b
|
5′ NC
|
||||||||||||||||||
| IH | IH | IH | Tru | IH | IH | IH | Tru | IH | IH | IH | IH | Tru | Inno | Inno | Tru | Inno | |||
| 9 | 3a + 1b | 5.86 + 4.31 | 3a | 3a | 3a | 3a | 3a | 3a | 3ab | 3a | 3a | 3a | 3 | 0 | 3a | 3a | 3a | 3a | 3 |
| 11 | 1a + 1b | 4.93 + 4.31 | 0 | 1a | 1ab | 1a + 1b | 1a + 1b | 1b | 1b | 1a | 1a | 1a | 1a | 1 | 1ab | 1 | 1 | 1a | 1 |
| 14 | 1b + 3a | 5.68 + 4.01 | 1b | 1b | 1b | 1b | 1b | 1bb | 1b | 1b | 1b | 1b | 1b | 1b | 1b | 1b | 1b | 1 | 1b |
IH, in-house genotyping method; Tru, Trugene procedure; Inno, InnoLipa line probe assay procedure.
The presence of a double population was mentioned.
Sample 11 (which contained genotypes 1a and 1b) was correctly identified as containing a mixture of genotypes by two NS5b-based analyses; it was subtyped as genotype 1a in eight cases (5 of the 10 NS5b-based analyses and 3 of the 7 5′NC-region-based analyses; a double population was mentioned in two cases. Among the seven remaining analyses, one provided a result of subtype 1b, five analyses could not completely characterize the genotype, and one gave a negative result.
Sample 14 (which contained genotypes 1b and 3a) was never identified as containing a mixture of genotypes by any of the assays; genotype 1b was detected in 16 analyses, and 1 analysis gave an incomplete result.
DISCUSSION
The data obtained in this multicenter study have shown a wide heterogeneity of genotyping results, depending on the laboratory and the genotyping method used. Indeed, as the objective of the study was to compare the ability of expert laboratories to provide correct HCV genotyping results, no method was imposed. All laboratories used commercial RNA purification methods, and some of them used the same sets of primers; but all procedures were different. However, the performance variations observed allowed us to suggest two different strategies according to the HCV genotype determined.
The discrimination between major HCV genotypes, which is the strategy commonly adopted in clinical practice, was successfully performed independently of the method used by all laboratories for all samples except those containing artificially mixed genotypes. However, 5′NC-region genotyping-based methods showed higher sensitivities. Indeed, in our study, while the assays based on analysis of the NS5b region missed 23 (16.4%) of the 140 expected positive results, 5′NC-region-based assays missed only 3 (3.1%) of the 98 expected positive results. Such false-negative results are probably due to the low levels if viremia in the samples tested, as described elsewhere (17, 26), as well as to the difficulty of amplifying products from samples infected with genotype 4 by NS5b-based methods (38). Therefore, unless the sensitivities of the present NS5b-based genotyping methods are improved (especially in the choice of the primers used) or unless genotype C and E1 genotyping methods are developed, as described elsewhere (4, 5, 11, 36), procedures based on the 5′ NC-region gene (and, essentially, those that use commercial standardized assays) can be considered the most adequate for genotyping in clinical practice, at least in countries where genotypes 1 to 5 are mainly encountered. Indeed, it was shown that 5′ NC-region-based genotyping methods cannot distinguish certain isolates of genotype 6 from isolates of genotype 1 (9). This point must be emphasized, especially for countries, such as Southeastern Asian countries, where genotype 6 is frequently found.
In the case of epidemiological studies requiring the precise determination of the HCV subtype, our results confirm that NS5b-based genotyping procedures are preferable to 5′ NC-region-based ones (6, 26, 33). Indeed, in our series, NS5b-based procedures were more accurate than 5′ NC-region-based methods, with the genotypes in only 2 samples misclassified by NS5b-based procedures, whereas 27 incomplete results or misclassifications were obtained by 5′ NC-region-based genotyping. One of these two misclassifications was for a sample infected with genotype 4a, which was misclassified as genotype 4c. The laboratory implicated in this misclassification observed after the study that this error was due to confusion over the nomenclature for genotype 4 in the sequence database. The database was subsequently modified to take this misclassification into account. Interestingly, we observed some previously described failures of 5′ NC-region-based genotyping, such as misclassification of genotype 1a as genotype 1b (7, 38), the absence of discrimination between subtypes 2a and 2c, and the lack of typing or subtyping of the strains in samples containing genotype 4 (38). For this reason, the use of NS5b-based genotyping methods is preferable to the use of 5′ NC-region-based genotyping methods for epidemiological investigations of HCV.
Although the results obtained with mixtures must be interpreted cautiously due to their artificial constitution, we have observed that when mixtures of strains of two genotypes were present in the same sample, the identification of both genotypes may be compromised if one of the two viral strains is present at a lower load. Although some participating laboratories indicated the existence of a double population in such samples, none of them identified the genotype 1b strain in sample 9 or the genotype 3a virus in sample 14. Techniques based on direct sequencing, as well as commercial line probe assays, are not appropriate in all cases for the detection of mixtures of genotypes in the samples studied (only two NS5b-based techniques were able to discriminate genotypes 1a and 1b in sample 11).
A consensus HCV sequencing method would be useful. The divergences observed in our study could be linked to differences in procedures (such as the RNA extraction methods, the primers, the types of enzymes, and the components in the mixture preparation used) and to the HCV sequence databases. However, the influence of each of these parameters is difficult to define in practice from the results of a multicenter study.
In conclusion, our results suggest, in agreement with previous studies (6, 17, 33, 38), that HCV 5′ NC-region-based genotyping methods give sufficient information for clinical purposes and that NS5b-based genotyping methods are more reliable for the subtype determination required in epidemiological studies.
Acknowledgments
This work was supported by a grant from the Agence Nationale de Recherches sur le SIDA et les Virus des Hépatites.
We thank Françoise Bouchardeau, Sandrine Castelain, Sébastien Hantz, Nadine Le Marrec, Joëlle Lerable, Virginie Morel, Christopher Payan, Annie Razer, and Henia Saoudin for technical assistance and Camille Sureau for helpful reading of the manuscript.
REFERENCES
- 1.Ackerman, Z., E. Ackerman, and O. Paltiel. 2000. Intrafamilial transmission of hepatitis C virus: a systematic review. J. Viral Hepat. 7:93-103. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Consensus conference. Treatment of hepatitis C virus. Gastroenterol. Clin. Biol. 26:B312-B320. [PubMed] [Google Scholar]
- 3.Bourliere, M., J. M. Barberin, M. Rotily, V. Guagliardo, I. Portal, L. Lecomte, S. Benali, C. Boustiere, H. Perrier, M. Jullien, G. Lambot, R. Loyer, O. LeBars, R. Daniel, H. Khiri, and P. Halfon. 2002. Epidemiological changes in hepatitis C virus genotypes in France: evidence in intravenous drug users. J. Viral Hepat. 9:62-70. [DOI] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Purcell, and R. H. Miller. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 90:8234-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantaloube, J., H. Venault, J. Zappitelli, P. Gallian, M. Touinssi, H. Attoui, P. Biagini, X. de Lamballerie, and P. de Micco. 2000. Molecular analysis of HCV type 1 to 5 envelope gene: application to investigations of posttransfusion transmission of HCV. Transfusion 40:712-717. [DOI] [PubMed] [Google Scholar]
- 7.Cantaloube, J. F., P. Gallian, H. Attoui, P. Biagini, P. De Micco, and X. De Lamballerie. 2001. Erroneous HCV genotype assignment by a hybridization typing assay in a case of posttransfusion HCV infection. Transfusion 41:429-430. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinchai, T., J. Labout, S. Noppornpanth, A. Theamboonlers, B. L. Haagmans, A. D. Osterhaus, and Y. Poovorawan. 2003. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J. Virol. Methods 109:195-201. [DOI] [PubMed] [Google Scholar]
- 10.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26:122S-127S. [DOI] [PubMed] [Google Scholar]
- 13.Doglio, A., C. Laffont, S. Thyss, and J. C. Lefebvre. 1998. Rapid genotyping of hepatitis C virus by direct cycle sequencing of PCR-amplified cDNAs and capillary electrophoresis analysis. Res. Virol. 149:219-227. [DOI] [PubMed] [Google Scholar]
- 14.Enomoto, N., A. Takada, T. Nakao, and T. Date. 1990. There are two major types of hepatitis C virus in Japan. Biochem. Biophys. Res. Commun. 170:1021-1025. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1989. PHYLIP—phylogenic interference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 16.Gault, E., P. Soussan, Y. Morice, L. Sanders, A. Berrada, B. Rogers, and P. Deny. 2003. Evaluation of a new serotyping assay for detection of anti-hepatitis C virus type-specific antibodies in serum samples. J. Clin. Microbiol. 41:2084-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halfon, P., P. Trimoulet, M. Bourliere, H. Khiri, V. de Ledinghen, P. Couzigou, J. M. Feryn, P. Alcaraz, C. Renou, H. J. Fleury, and D. Ouzan. 2001. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J. Clin. Microbiol. 39:1771-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izopet, J., C. Pasquier, K. Sandres, J. Puel, and L. Rostaing. 1999. Molecular evidence for nosocomial transmission of hepatitis C virus in a French hemodialysis unit. J. Med. Virol. 58:139-144. [DOI] [PubMed] [Google Scholar]
- 20.Le Pogam, S., D. Le Chapois, R. Christen, F. Dubois, F. Barin, and A. Goudeau. 1998. Hepatitis C in a hemodialysis unit: molecular evidence for nosocomial transmission. J. Clin. Microbiol. 36:3040-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerebours, E., P. Marcellin, and D. Dhumeaux. 2002. Treatment of hepatitis C: advances and consensus. Gastroenterol. Clin. Biol. 26(Spec. No. 2):B5-B6. [PubMed] [Google Scholar]
- 22.Martial, J., Y. Morice, S. Abel, A. Cabie, C. Rat, F. Lombard, A. Edouard, S. Pierre-Louis, P. Garsaud, O. Bera, R. Chout, E. Gordien, P. Deny, and R. Cesaire. 2004. Hepatitis C virus (HCV) genotypes in the Caribbean island of Martinique: evidence for a large radiation of HCV-2 and for a recent introduction from Europe of HCV-4. J. Clin. Microbiol. 42:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 24.Mellor, J., E. C. Holmes, L. M. Jarvis, P. L. Yap, P. Simmonds, et al. 1995. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J. Gen. Virol. 76:2493-2507. [DOI] [PubMed] [Google Scholar]
- 25.Morice, Y., D. Roulot, V. Grando, J. Stirnemann, E. Gault, V. Jeantils, M. Bentata, B. Jarrousse, O. Lortholary, C. Pallier, and P. Deny. 2001. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J. Gen. Virol. 82:1001-1012. [DOI] [PubMed] [Google Scholar]
- 26.Nolte, F. S., A. M. Green, K. R. Fiebelkorn, A. M. Caliendo, C. Sturchio, A. Grunwald, and M. Healy. 2003. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5′ noncoding region. J. Clin. Microbiol. 41:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norder, H., A. Bergstrom, I. Uhnoo, J. Alden, L. Weiss, J. Czajkowski, and L. Magnius. 1998. Confirmation of nosocomial transmission of hepatitis C virus by phylogenetic analysis of the NS5-B region. J. Clin. Microbiol. 36:3066-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Othman, S. B., A. Trabelsi, A. Monnet, N. Bouzgarrou, F. Grattard, A. Beyou, T. Bourlet, and B. Pozzetto. 2004. Evaluation of a prototype HCV NS5b assay for typing strains of hepatitis C virus isolated from Tunisian haemodialysis patients. J. Virol. Methods 119:177-181. [DOI] [PubMed] [Google Scholar]
- 29.Pawlotsky, J. M., L. Tsakiris, F. Roudot-Thoraval, C. Pellet, L. Stuyver, J. Duval, and D. Dhumeaux. 1995. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J. Infect. Dis. 171:1607-1610. [DOI] [PubMed] [Google Scholar]
- 30.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 31.Poynard, T., J. McHutchison, Z. Goodman, M. H. Ling, J. Albrecht, et al. 2000. Is an “a la carte” combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? Hepatology 31:211-218. [DOI] [PubMed] [Google Scholar]
- 32.Ross, R. S., S. O. Viazov, C. D. Holtzer, A. Beyou, A. Monnet, C. Mazure, and M. Roggendorf. 2000. Genotyping of hepatitis C virus isolates using CLIP sequencing. J. Clin. Microbiol. 38:3581-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandres-Saune, K., P. Deny, C. Pasquier, V. Thibaut, G. Duverlie, and J. Izopet. 2003. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J. Virol. Methods 109:187-193. [DOI] [PubMed] [Google Scholar]
- 34.Seeff, L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35-S46. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 36.Simmonds, P., J. Mellor, T. Sakuldamrongpanich, C. Nuchaprayoon, S. Tanprasert, E. C. Holmes, and D. B. Smith. 1996. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J. Gen. Virol. 77:3013-3024. [DOI] [PubMed] [Google Scholar]
- 37.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamalet, C., P. Colson, H. Tissot-Dupont, M. Henry, C. Tourres, N. Tivoli, D. Botta, I. Ravaux, I. Poizot-Martin, and N. Yahi. 2003. Genomic and phylogenetic analysis of hepatitis C virus isolates: a survey of 535 strains circulating in southern France. J. Med. Virol. 71:391-398. [DOI] [PubMed] [Google Scholar]
- 39.Young, K. K., J. J. Archer, O. Yokosuka, M. Omata, and R. M. Resnick. 1995. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J. Clin. Microbiol. 33:654-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young, K. K., R. M. Resnick, and T. W. Myers. 1993. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol. 31:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]