Abstract
Tracking the behavior of leukemic samples both in vitro and in vivo plays an increasingly large role in efforts to better understand the leukemogenic processes and the effects of potential new therapies. Such work can be accelerated and made more efficient by methodologies enabling the characterization of leukemia samples in multiplex assays. We recently developed three sets of lentiviral fluorescent genetic barcoding (FGB) vectors that create 26, 14, and 6 unique immunophenotyping-compatible color codes from GFP-, yellow fluorescent protein (YFP)-, and monomeric kusabira orange 2 (mKO2)-derived fluorescent proteins. These vectors allow for labeling and tracking of individual color-coded cell populations in mixed samples by real-time flow cytometry. Using the prototypical Hoxa9/Meis1 murine model of acute myeloid leukemia, we describe the application of the 6xFGB vector system for assessing leukemic cell characteristics in multiplex assays. By transplanting color-coded cell mixes, we investigated the competitive growth behavior of individual color-coded populations, determined leukemia-initiating cell frequencies, and assessed the dose-dependent potential of cells exposed to the histone deacetylase inhibitor Entinostat for bone marrow homing. Thus, FGB provides a useful tool for the multiplex characterization of leukemia samples in a wide variety of applications with a concomitant reduction in workload, processing times, and mouse utilization.
Keywords: fluorescent genetic barcoding, lentiviral gene transfer, acute myeloid leukemia, Hoxa9, Meis1, H9M, leukemic stem cell, LSC
Introduction
Acute myeloid leukemia (AML) represents a severe acquired hematopoietic disease with low overall survival rates. Much effort is, therefore, being directed at better characterizing the AML mutational landscape, mutation-dependent transcriptional programs, and epigenetic signatures within the disease driving leukemic stem cell (LSC) population, which contains the functionally defined leukemia-initiating cells (LICs).1, 2, 3, 4, 5, 6, 7, 8, 9 In addition, major thrusts of current research are focused on establishing optimized in vitro cultivation systems for primary AML cells, as well as developing de novo AML models.10, 11, 12, 13, 14, 15, 16 With these advances, there is an increasing imperative for functional investigations. However, these often rely on labor- and cost-intensive in vitro and in vivo experiments. For example, pre-clinical in vitro drug efficacy testing requires subsequent in vivo evaluation assays to dissect the effects on blast cells and LSC properties with regards to engraftment, self-renewal, and survival, especially since these parameters may not be directly accessible by in vitro assays.17, 18
Historically, hematopoietic research has extensively benefitted from the development of innovative marking and tracking techniques. While initial studies utilized congenic or autologous markers for distinguishing donor- and host-derived cells in transplantation assays,19, 20, 21 the advent of retroviral gene marking facilitated the stable genetic modification of target cells and fate tracking by fluorescent proteins and integration site analyses.22, 23 The establishment of barcoded vector libraries further increased resolution and enabled the ability to track hundreds of cell clones with regards to size, differentiation potential, and dynamics of hematopoietic reconstitution in a single animal.24, 25 The opportunity to achieve real-time resolution and to prospectively isolate cells of interest for consecutive studies, such as gene expression analyses or secondary transplantations, originated from the transition toward advanced fluorescence-based tracking approaches. Seminal studies utilized stochastic combinatorial expression methods of various fluorescent proteins in transgenic animals or in retrovirally transduced cells for creating defined hues compatible with fluorescent microscopy-assisted clonal cell tracking.26, 27, 28 Refinement of these techniques enabled the flow cytometric characterization of multiple uniquely labeled cell populations in multiplex assays by employing pre-defined fluorescent color codes as recently demonstrated by the in vivo tracking of transgenic murine HSC and lentivirally marked glioblastoma clones.29, 30, 31, 32 In our work, we further expanded the scope of flow cytometric tracking applications by developing three lentiviral fluorescent genetic barcoding (FGB) vector systems.32 These FGB vectors encode for GFP, YFP, and mKO2 fluorescent proteins and variants (e.g., YFPe and meKO3), which facilitate the production of 26, 14, and 6 different color codes. In contrast to DNA barcoding, the 14x and 6xFGB systems require the initial labeling of separate cultures with single color codes expressed from individual vectors before mixing of samples and their flow cytometric deconvolution in multiplex studies. Additionally, the 6xFGB vector system links a defined DNA barcode (BC) to each color code (GFP-BCA, YFP-BC5, meKO3-BC6, YFPe-2A-GFP-BC8, GFP-2A-meKO3-BC28*, and YFP-2A-meKO3-BC31*). This approach thus enables immunophenotyping-compatible highly specific cell tracking by fluorescent signature, as well as by molecular techniques as demonstrated in cell lines and primary human and murine HSC in vitro and in vivo.32 Additionally, color-coded vectors equipped with microRNA (miRNA) co-expression cassettes revealed microRNA 126 (miR-126) induced in vitro expansion of a murine bone marrow (BM) cell line.32 These examples predict that multiplexing of color-coded cells will also have considerable potential for AML research. Here, the potential to directly compare properties of multiple samples within a single animal promises a reduction in host-dependent variability and thus offers the opportunity to reduce animal consumption in line with the replace, reduce, and refine (3R) guidelines without compromising on data quality. Potential applications of FGB include the investigation of LSC repopulation kinetics, LSC frequencies, and the competitive fitness of defined cell populations under normal conditions or in response to drug treatment. Additionally, applying FGB to longitudinal in vitro studies may lead to a reduction in processing times, culture-induced variability, and reagent expenses. To further explore the unique multiplexing capabilities of the 6xFGB system, we here describe its application in the Hoxa9/Meis1 (H9M) murine AML model.33 H9M leukemic cell lines support long-term in vitro cultivation and trigger aggressive leukemia with a latency of 40–60 days after transplantation.33, 34 Through transplantation of color-coded H9M cell mixes, we monitored the dynamic competition between differently color-coded populations in recipient mice, established multiplex limiting dilution assays (MLDAs) and studied drug responses in short-term assays. Together, these experiments provide proof-of-principle that FGB enables the characterization of leukemia samples in multiplex assays and provides increased accuracy of data, while concomitantly allowing for a reduction in sample numbers. Therefore, FGB provides a powerful platform for accelerating leukemia research through multiplexing applications.
Results
Lentiviral FGB Allows for Stable Color Coding of H9M AML Cells
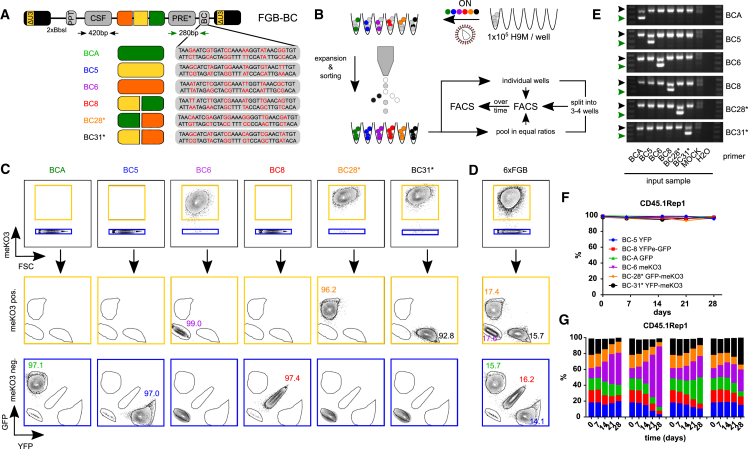
To further evaluate the utility of the 6xFGB vector system for functional in vitro and in vivo multiplex assays (Figure 1A), we applied FGB to the prototypical H9M AML model by transducing four independently generated H9M lines with all six vectors in separate wells (Figure 1B). Four H9M replicate (Rep) lines, two derived from Pep3b (CD45.1Rep1 and CD45.1Rep2) and two derived from C57BL/6 (CD45.2Rep1 and CD45.2Rep2) mice, were employed to minimize potential cell line-associated artifacts and enable the separation of the donor- and host-derived cell content in congenic transplantation experiments. After expansion, each of the 24 transduced samples was subjected to fluorescence-activated cell sorting (FACS) and sorting purities reached ≥95% in aliquots of single color-coded cells (Figures 1C and S1A). Also, flow cytometric analysis allowed for deconvolution of all six FGB input populations in mixed samples when first gating on meKO3 positive and negative cells before assessing the GFP and YFP expression in these gates (Figure 1D). Furthermore, single color-coded cells could be accurately detected by their BC in PCRs (Figure 1E). Longitudinal tracking of multiple aliquots per single color-coded population in isolation showed stable vector expression indicative of robust vector performance in H9M cells (Figures 1F and S1B). Additionally, the preparation of cell mixes containing all six FGB color-coded cell aliquots per parental H9M line in equal ratios offered the opportunity to track competitive growth behavior in culture. Therefore, assessment of the color code distribution on day 0 (d0) before splitting cells into four wells for subsequent cultivation allowed for investigating heterogeneous growth potential in aliquots from the same parental cell mixes. Flow cytometric analyses showed the stable maintenance of color codes at highly reproducible ratios in all replicates for the first 7 days of observation (Figures 1G and S1C). Interestingly, over time, some aliquots manifested the expansion of a single color-coded population, which was nevertheless stably maintained in other aliquots of the same parental cell mix. Statistical testing did not detect a significant growth advantage of any of the color codes, indicating that the assay reliably detects dynamic changes in the intrinsic competitiveness of color-coded cell populations rather than transgene-induced effects (Figure S1D).
Figure 1.
Lentiviral FGB Vectors Stably Mark H9M Cells In Vitro
(A) Schematic design of six lentiviral FGB vectors. Each vector contains a chimeric CSF promoter consisting of a CBX3-derived anti-silencing element and a spleen focus forming virus promoter for expression of GFP (BCA), YFP (BC5), meKO3 (BC6), YFPe-2A-GFP (BC8), GFP-2A-meKO3 (BC28*), or YFP-2A-meKO3 (BC31*) color codes. YFPe and meKO3 constitute a codon-diversified YFP and a modified mKO2, respectively. The unique DNA barcode (BC) linked to each fluorescent color code facilitates its molecular detection. (B) Experimental design. After separate transduction of H9M cells with single FGB vectors, cultures were expanded and color-coded cells were purified by fluorescent-activated cell sorting. Purified populations were than either mixed or directly tracked over time by flow cytometry. (C) Exemplified flow cytometry profiles of FGB vector transduced and sorted H9M populations. Cells were first gated on cells positive or negative for meKO3 expression prior to assessing GFP and YFP signals. The numbers in the plots indicate the size of each population (in %) within the total viable cell fraction. (D) Exemplified cell mix consisting of all six FGB vector transduced populations from (C). (E) Molecular detection of purified color-coded cells by BC-directed PCR. The black and green arrows indicate the 420 bp loading control and the 280 bp BC-specific amplicon, respectively, with primer positions schematically indicated as black and green arrows in (A). (F) Longitudinal tracking of single color-coded populations. On day 0, cells were analyzed by flow cytometry and subsequently split into four wells for tracking of at least three wells until the end of the observation period. The data are shown as mean ± SD. (G) Tracking of color-coded cell mixes over time. Equal ratios of color-coded cells were combined, analyzed by flow cytometry, and split into four wells for longitudinal analysis. The color codes are as in (F).
In summary, single color-coded H9M populations allow for near-purity enrichment, show stable color code expression over time, and consequently facilitate the study of competitive growth behavior of differentially marked populations in mixed cultures.
Single Color-Coded H9M Populations Show Comparable Growth Properties In Vivo
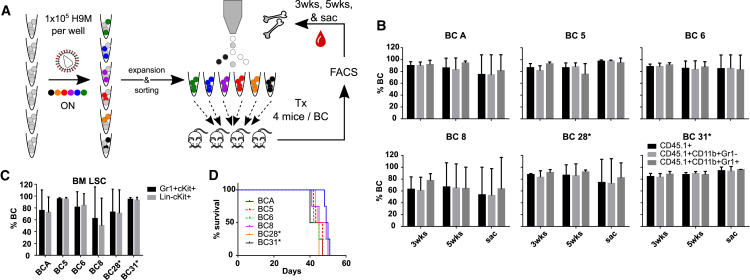
To extend the utilization of our 6xFGB system to in vivo leukemia studies, we next sought to verify the stable maintenance of fluorescent labels without influencing cell properties; e.g., disease latency and surface marker composition. Therefore, each of the six CD45.1Rep2-derived color-coded populations was individually transplanted into groups of four mice (Figure 2A). At 3 and 5 weeks post-transplantation, as well as at the time of sacrifice, the peripheral blood (PB) showed color code expression in ∼80% of donor-derived bulk CD45.1+ cells, as well as in myeloid cells (CD11b+Gr1− and CD11b+Gr1+) therein (Figures 2B; representative flow cytometry plots and gating strategies depicted in S2). Two (Gr1+cKit+ and lineage marker negative cKit+, Lin−cKit+) of the three BM-derived LSC subpopulations defined by Gibbs et al.34 showed similar gene marking rates (Figures 2C; representative flow cytometry plots and gating strategies depicted in S3). In contrast, color code expression in the third (lymphoid [Lym]+cKit+) LSC subpopulation could not be analyzed due to the small size of this population in the majority of mice. Nevertheless, sorting procedures enabled the enrichment of LSC subpopulations from all color codes (Figure S4). Notably, the slightly higher degree of variation seen in groups BCA and BC8 was due to the continuous expansion of transgene negative cells in single mice, potentially originating from non-gene marked cells or from vector silencing. Similarly, in single animals of groups BC6 and BC28*, intermediate bright meKO3+ populations emerged, which required exclusion from analyses, and potentially developed due to variegated vector expression (Figures S5A–S5C).35 Regardless, mice from all six groups succumbed with a comparable latency of 45 days (Figure 2D). Therefore, the application of stringent gating criteria allowed for specific immunophenotypic characterization of differently color-coded samples, which displayed comparable lineage composition (Figures S5D and S5E) and transforming potential as a prerequisite for subsequent functional multiplexing studies.
Figure 2.
Color-Coded H9M Cells Show Comparable Properties In Vivo
(A) Schematic experimental design. H9M cells were transduced in separate wells with either one of six FGB vectors, before sorting and transplantation of single color-coded populations per mouse. The PB was sampled over time, and the PB and BM status were assessed in moribund mice. (B) Determination of single color-coded cell fractions within donor-derived PB CD45.1+, CD45.1+CD11b+Gr1−, and CD45.1+CD11b+Gr1+ cells 3 and 5 weeks post-transplantation and at the time of sacrifice (sac). (C) Color code content in BM LSC subpopulations is shown. (D) Survival curves for all six single color code transplanted groups (4 mice per group). The number of mice analyzed per group and time point is indicated in Figures S5A and S5B. The bar graphs represent mean values ± SD.
Color-Coded H9M Cells Exhibit Heterogeneous Growth Behavior In Vivo
To assess the competitive behavior of H9M cells and to test the ability of FGB to resolve dynamic changes in H9M populations in vivo, we next transplanted cell mixes containing equal proportions of all six color-coded populations into lethally irradiated mice (Figure 3A). Longitudinal analyses of PB samples typically revealed the presence of all input color codes both 3 and 4 weeks after transplantation, although some mice displayed better preservation of the initial mixing ratios than others (representative mice shown in Figure 3B, left). However, between 4 and 5 weeks after transplantation, the mixing ratio of early detected color codes underwent rapid changes and led to the expansion of one or two dominant color codes. Due to this dynamic behavior, mice showed a steady decline from an average of four (at 3 weeks) to one (at 5 weeks) color-coded populations with contributions of ≥10% to PB lineages representative of the expansion of a single color code over time (Figure 3C). As expected, the bulk of the leukemic cell mass in the BM of moribund mice resembled the PB color code distribution from endpoint samples (Figure 3B, middle). However, when assessing color code distributions within the BM LSC subpopulations, some mice showed a good correlation between the overall PB and BM color code composition (e.g., mice 1 and 2), while others showed a stronger degree of variation (e.g., mice 3 and 4; Figures 3B, right, and S6). This observation suggests that separate cell populations with distinct differentiation preferences may form the three H9M LSC compartments.
Figure 3.
In Vivo Competition of Six Color-Coded H9M Populations Results in a Loss of Color Code Complexity over Time
(A) Experimental design. Purified color-coded cells were mixed in equal ratios and transplanted into lethally irradiated mice. The PB was sampled over time, and the PB and BM were analyzed in moribund mice. (B) Color code distribution within (left) donor-derived CD45+ PB cells over time, (middle) bulk BM cells, and (right) BM LSC subpopulations (subp.) of mice transplanted with 6xFGB transduced H9M (CD45.1 or CD45.2) cells. (C) Determination of color codes >10% contribution to the donor-derived PB cell fraction, from a total of 19 transplanted mice, 16, 18, 19, 12, and 18 were analyzed 3, 4, 5, and 6 weeks post-transplantation and at the time of sacrifice, respectively. The bar graphs from (C) represent mean values ± SD.
To demonstrate the successful color coding of cells with disease propagating potential, unfractionated BM from two mice was transplanted into two paired secondary recipients, each (mice 3 and 4; Figures 3B and S7), and assessed for the engraftment potential of color-coded cells. Although the BM of these two donor mice contained two populations with >20% contributions to the Lin−cKit+ LSC compartment, thought to contain the highest frequency of LSC,34 the BM of paired secondary moribund mice was dominated by a single color code (Figure S7). In the case of mouse 4, this color code may have originated from a Lin−cKit+-derived minor population (Figure S7C). The observed color code expansion indicates that the color code distribution in primary recipients may not represent a steady state and that major and minor populations continue to compete in secondary recipients. Furthermore, this suggests the successful color coding of functionally defined LSC.
In summary, multiplex transplantation assays allowed for the tracking of dynamic fluctuations of color-coded H9M subpopulations over time. Similar to in vitro experiments, early time points revealed multiple color-coded populations, while later time points reported a progressive loss of color code complexity.
FGB-Based Limiting Dilution Assays Show Differences in LIC Frequency
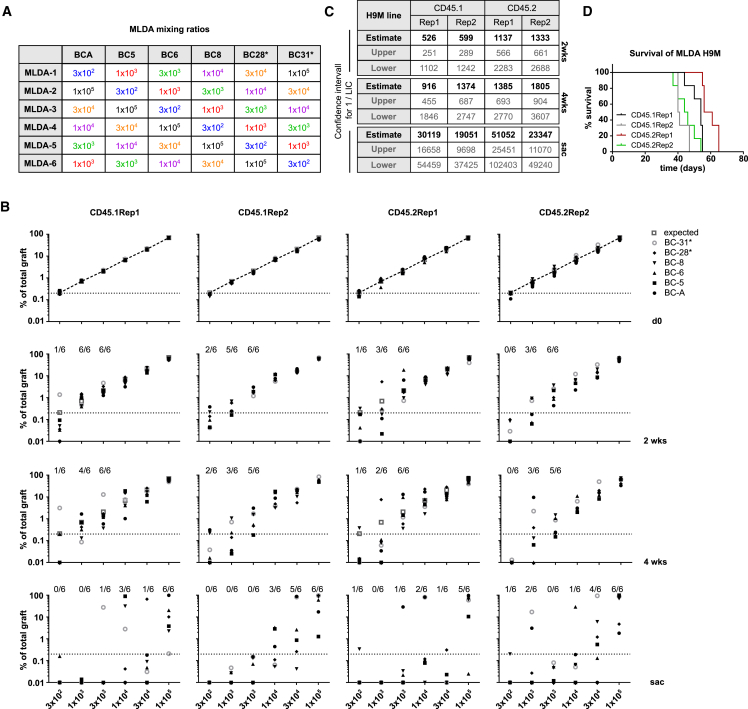
Based on the relatively stable color code distributions early after initiation of in vitro and in vivo multiplexing experiments, we next sought to harness this behavior for performing LDAs with multiple cell concentrations in a single mouse. We hypothesized that a group of six mice, each transplanted with a unique cell mix containing 3 × 102, 1 × 103, 3 × 103, 1 × 104, 3 × 104, and 1 × 105 differently color-coded input cells, would allow determination of the LIC frequency based on monitoring 36 populations in parallel (Figure 4A). Similar to conventional LDAs, color code contributions <0.2% (equal to the smallest input population) of the total PB donor cell fraction would be scored “negative” in these MLDAs. However, due to the competitive nature of MLDAs, LIC frequencies can be assessed in the PB and BM over time rather than using the conventional dose-dependent survival as the only readout. Assessment of MLDA color code mixing ratios in aliquots of the injected cell mixes from our four independently generated H9M lines yielded an almost perfect correlation between the expected and observed mixing ratios, demonstrating the potential of the FGB platform to report quantitative differences in mixed cell populations (Figure 4B, top panel). Assessment of the PB color code distributions 2 weeks after transplantation and subsequent LIC frequency calculation reproduced the ∼2-fold difference in self-renewing cell content already detected in in vitro colony-forming cell assays between the CD45.1- and the CD45.2-derived H9M lines (Figures S8A and S8B), although these differences evened out at the 4 week time point (Figures 4B, middle panels, and 4C).36 However, in the PB of moribund mice, the highest LIC frequency was detected in sample CD45.1Rep2 (1 in 19,051), the second highest in CD45.2Rep2 (1 in 23,347), the third highest in CD45.1Rep1 (1 in 30,119), and the lowest in CD45.2Rep1 (1 in 51,052) (Figures 4B, bottom panel, and 4C), which inversely correlated with median survival times of 40.5 days (CD45.1Rep2), 45 days (CD45.2Rep2), 54 days (CD45.1Rep1), and 58.5 days (CD45.2Rep1), respectively (Figure 4D). Importantly, and as expected, the PB and BM samples of moribund mice showed a similar distribution of color codes, which further underscores the utility of PB characterization for assessment of LIC frequencies in MLDAs (Figures S8C and S8D). Additionally, the detection of 62%, 19%, and 19% of dominant color codes in BM samples originating from the color codes with the highest (1 × 105), second highest (3 × 104), and third highest (1 × 104) input numbers, respectively, corroborates the quantitative nature of MLDAs (Figure S8E).
Figure 4.
Multiplex Limiting Dilution Assays Detect Differences in LIC Frequency of Individual H9M Lines
(A) Mixing ratios of individual MLDAs with color-coded input cells of indicated doses. (B) Assessment of color code contributions to the total MLDA cell mix at the time of transplantation (d0) and within the donor-derived PB CD45.1+ or CD45.2+ cells 2 and 4 weeks after transplantation as well as at sacrifice (sac). The dotted line indicates 0.2% graft content as a cutoff for successful engraftment. The numbers at the top of each graph indicate the fraction of mice that showed successful engraftment. (C) Estimation of leukemia-initiating cell (LIC) frequency for each of the four H9M lines from (B). (D) Comparison of survival times of mice transplanted with MLDA H9M cell mixes. There were six mice that were transplanted per H9M line.
Taken together, this suggests that in contrast to conventional LDAs, MLDAs facilitate a quantitative and longitudinal assessment of LIC frequencies with unparalleled efficiency in a 6-fold reduced number of mice.
FGB-Based Multiplex Assays Reveal an Immediate Effect of In Vitro Entinostat Treatment on In Vivo Cell Properties
Given the quantitative nature of the FGB multiplex transplantation assays, we hypothesized that the FGB approach could also enable the characterization of in vivo LSC properties in response to in vitro drug treatment in a reduced number of mice. H9M cells were thus subjected to a series of functional in vitro and in vivo assays after challenge with the histone deacetylase (HDAC) inhibitor Entinostat (Figure 5A). Growth curves showed that all four H9M lines similarly tolerated up to 1 μM Entinostat exposure for 24 hr (Figure 5B). Likewise, mixing of color-coded Entinostat-exposed H9M cells in equal ratios revealed the disappearance of the 3 μM-treated sample within the first week of observation, whereas contributions of all other populations remained relatively stable for up to 28 days in culture (Figure 5C).
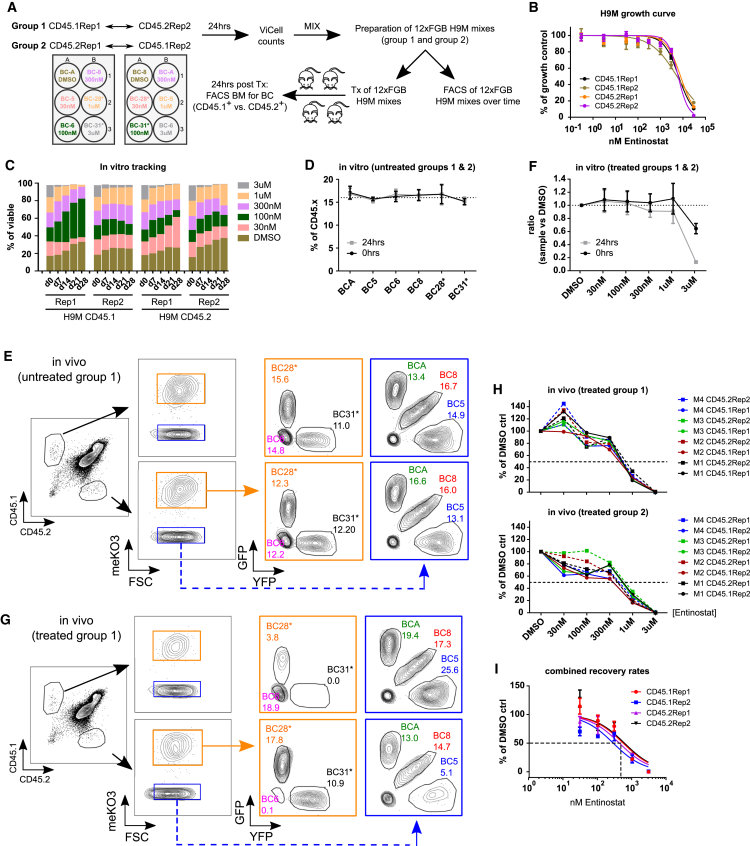
Figure 5.
Multiplex Drug Assay
(A) Experimental design for short-term Entinostat assays. CD45.1- and CD45.2-derived color-coded H9M cells were treated with Entinostat for 24 hr before assessment of cell growth by Vi-Cell counts. Treated cells from a CD45.1- and a CD45.2-derived H9M line were mixed in equal ratios to yield the “group 1” and “group 2” 12xFGB mixes for longitudinal in vitro tracking and injection into four CD45.1 × CD45.2 F1 mice per group. For comparison, non-treated 12xFGB cell mixes were prepared in parallel. (B) AlamarBlue-dependent assessment of dose-dependent cell growth of four different H9M lines after 24 hr of Entinostat exposure (n = 3–4 per data point). (C) Each color-coded population was exposed to a specific Entinostat concentration for 24 hr before the preparation of cell mixes and subsequent longitudinal assessment of growth properties. (D) Assessment of color code distribution within non-treated 12xFGB H9M cell mixes at the time of transplantation (0 hr) and after 24 hr of in vitro cultivation. Cells were stained for CD45.1 and CD45.2 to distinguish all six color codes within the parental CD45 (CD45.x = CD45.1 and CD45.2) populations (n = 4 per color code). (E) Exemplified flow cytometry profiles of non-treated 12xFGB cell mixes recovered from the BM of lethally irradiated mice 24 hr after transplantation. The BM cells were first analyzed for expression of CD45.1 and CD45.2 markers before detecting color codes within these populations. (F) Assessment of color code distribution within Entinostat-treated 12xFGB H9M cell mixes at the time of transplantation (0 hr) and after 24 hr of in vitro cultivation (combined data from group 1 and group 2 cells). Cells were stained for CD45.1 and CD45.2 to distinguish all six color codes within the parental CD45 (CD45.x = CD45.1 and CD45.2) populations and normalized to the size of the DMSO control-treated samples (n = 4 per color code). (G) Exemplified flow cytometry profiles of Entinostat-treated 12xFGB H9M cell mixes 24 hr after transplantation into lethally irradiated CD45.1 × CD45.2 F1-derived mice. The gating strategy is as in (E). (H) Entinostat-dependent color code sizes normalized to DMSO control-treated cells from 12xFGB cell mixes recovered from the BM of recipient mice 24 hr after transplantation. M1–M4 indicate the individual mice transplanted for each group. (I) IC50 (nM) determination for Entinostat-dependent homing potential from 96 data points from all 8 mice depicted in (H). The error bars represent mean values ± SD.
To further resolve potential in vitro effects of Entinostat on in vivo cell properties, we next investigated the Entinostat-dependent engraftment rate of transplanted H9M cells in short-term assays. These assays serve as surrogates for immediate drug effects on survival, differentiation, and migration capacity to the BM, which together reflect the cellular homing potential. For increasing the complexity of traceable populations and for concomitantly reducing the number of required mice, drug treatment experiments of single color-coded H9M populations were repeated before mixing of CD45.1Rep1 with CD45.2Rep2 (group 1) and CD45.1Rep2 with CD45.2Rep1- (group 2) derived color-coded cells, respectively. The antibody-mediated detection of CD45.1- and CD45.2-derived input cells in these assays enables monitoring of 12 color-coded populations in a single culture well or CD45.1 × CD45.2 F1 animal (Figure 5A). Longitudinal in vitro tracking of treated and untreated 12xFGB cell mixes showed the same growth behavior as correlating 6xFGB cell mixes (Figures 1G, 5C, and S9). In addition, non-treated 12xFGB mixes showed similar color code ratios in aliquots of transplanted cell mixes, as well as in in vitro cultures, and in the BM extracted cells 24 hr later (Figures 5D, 5E, S9A, and S9B). In contrast, analysis of Entinostat-treated 12xFGB cell mixes 24 hr after injection revealed a dose-dependent decrease in homing potential at even lower concentrations than required for impairing in vitro cell growth (Figures 5F–5H, S9C, and S9D). Subsequent calculations of half maximal inhibitory concentration (IC50) values predicted a 50% decreased engraftment rate at Entinostat concentrations of 548.5 nM (CD45.1Rep1), 282.1 nM (CD45.1Rep2), 425.6 nM (CD45.2Rep1), and 599 nM (CD45.2Rep2) for our four H9M lines, respectively (Figure 5I). Together, results from monitoring 96 non-treated and 96 Entinostat-treated populations in a total of 16 mice indicated a severe reduction in H9M cell homing potential shortly after in vitro Entinostat treatment. These observations underscore the importance of in vivo readouts for assessing in vitro drug responses and further illustrate the power of FGB to enable such readouts in an efficient manner with decreased mouse utilization.
Discussion
Our study establishes the potential of lentiviral FGB for the multiplex characterization of leukemia samples. Despite growing interest in the molecular mechanisms of the development, maintenance, and therapy of leukemia, these studies either monitor a single sample per experimental animal or utilize vector libraries that do not permit the real-time tracking of cells with similar properties.37, 38, 39, 40 Our newly developed FGB vector system employs a different strategy by providing a small number of color codes for tracking unique cell populations in multiplex flow cytometry-driven real-time assays. This allows properties of cellular populations; e.g., differentiation potential, size and dynamic expansion, or contraction processes, to become readily accessible and make the FGB platform a versatile tool for small-scale screening approaches.
The initial development of the FGB vector platform sought to allow for color coding and tracking of mixed target cell populations independent of a priori sorting steps.32 In particular, abbreviated lentiviral transduction and transplantation protocols offered enhanced gene marking and preservation of stemness for cell types that would benefit from short-term in vitro cultivation times, such as HSCs.41, 42, 43 In addition, we recognized that this tool could be adapted to yield improved readouts in multiplexing experiments with leukemic cells if initiated with controlled ratios of color-coded cells per cell mix. For this purpose, stringent sorting procedures were established that routinely yielded color-coded H9M cell purities >95%, allowed for longitudinal tracking of single color-coded populations, and showed stable in vitro and in vivo FGB vector-mediated expression of all six color codes without altering cell properties (Figures 1 and 2). The advantage of in vitro multiplexing became apparent, when single color codes occasionally outcompeted the other five populations (Figure 1F), despite stable maintenance of all color codes when monitored in isolation. Since DNA barcoding of cell lines and mesenchymal stem cells previously detected clonal restriction, color coding may represent a simple method to investigate heterogeneous growth properties in vitro, albeit requiring integration site analyses for the assessment of clonal complexities.44, 45, 46
To extend these in vitro studies on population dynamics, we employed FGB for the visualization of hematopoietic reconstitution and leukemogenesis dynamics in an in vivo H9M transplantation model (Figures 3 and S6). At 3 weeks post-transplantation, recipients of 6xFGB H9M cell mixes displayed an average of 4.4 color codes with contributions of >10% to donor-derived CD45+ PB. Color code frequencies subsequently decreased so that the majority of moribund mice only contained a single dominant color code in bulk CD45+ PB cells (Figure 3C). Nevertheless, as shown by secondary transplantation experiments, minor populations from primary BM samples were still capable of expanding in paired secondary recipients (Figure S7B). This argues against the development of dominant color codes purely by the stochastic distribution of limited color-coded LSC numbers in the graft. It rather appears more likely, that stochastic events, such as the acquisition of secondary mutations, insertional mutagenesis, epigenetic alterations, or pre-determined cell autonomous characteristics within a mixed cell population,40, 47, 48 may have influenced the competitive expansion of a color-coded population over time. Although the investigation of the underlying molecular aberrations exceeded the scope of this manuscript, we successfully established sorting strategies for the enrichment of color-coded LSC subpopulations (Figure S4), which may aid comparative gene expression studies and clonality assessments. In light of the previous reported clonal nature of H9M leukemia,33, 48 it would, however, be challenging to dissect, if our FGB approach suggested clonal restriction because of the transplantation of limited LSC numbers or as a result of competitive clonal outgrowth.
Interestingly, the Scadden group30 recently published the “HUe” mouse model that utilizes stochastic recombination for the random creation of >1 × 103 color codes. Tracking of up to 15 color-coded HSCs in multiplex transplantation assays by flow cytometry revealed dynamic changes in the color code composition early after transplantation, before the manifestation of a stable pattern, mainly consisting of 1–3 color codes at later time points. Since these results compare to our observations in the H9M AML model, they further underline the feasibility of color coding approaches for the investigation of dynamic cell behavior in vivo. However, while the HUe mouse model provides a greater selection of color codes, due to subtle differences in fluorescent properties, not all of them might be suitable for flow cytometry-driven cell tracking. In contrast, the lentiviral FGB approach clearly benefits from the generation of a priori defined color codes in a variety of cell types without the requirement for transgenic animals.
Studies involving in vitro drug treatment of leukemic cells typically focus on the assessment of bulk cell survival, due to a lack of markers allowing for the prospective isolation of pure LSC population.2, 7, 17 Stringent drug screens, therefore, require in vivo assessment of drug efficacy.17, 18, 49 FGB thus presents a potent tool to efficiently facilitate these assays through the ability to subject in vitro drug-exposed cells to monitoring in a single animal or culture well. The importance of this in vivo verification step became apparent when in vitro treatment of H9M cells with the HDAC-inhibitor Entinostat only impaired competitive in vitro cell growth at concentrations >1 μM (Figures 5B and 5C), but led to a 50% reduced homing rate at 463.8 nM ± 141.3 in short-term in vivo assays (Figures 5G–5I). Importantly, these data are comparable to results from the Thompson group,18 who exposed primary H9M leukemia cells to 300 nM Entinostat and reported a reduction in H9M leukemogenicity and extended survival of recipient mice. This suggests that the multiplex FGB screening platform may be especially applicable to drug screening approaches in the context of leukemia research and potentially in regenerative medicine.12, 50, 51 In addition to detecting qualitative differences; e.g., resistance to drug treatment, adaptation of competitive transplantation assays also facilitated quantification of the competitive fitness of cells, when initiated with different cell concentrations per color code. Therefore, MLDA experiments allowed for estimation of LIC frequencies in a limited number of mice (Figure 4), and similar studies may be feasible for assessing drug-dependent changes in LIC numbers.
Given that the current FGB platform in comparison to conventional transplantation assays permits a 6- to 12-fold reduction in mouse numbers, FGB promises accelerated data acquisition, as well as increased reproducibility of results from transplantation studies. However, the potential of the FGB vector platform extends further, as evidenced by our recently reported applications for marking and tracking of HSC, CRISPR/Cas9-mediated genome modifications, DNA barcoding, and miRNA screening.32 With the anticipated continuing expansion in the repertoire of fluorescent marker proteins, as well as in the power of flow cytometers to separate fluorophores with similar emission spectra, the complexity of the FGB platform will also continue to expand making FGB amenable to even more ambitious multiplexing experimentations.
Materials and Methods
Cell Culture
293T cells for the production of lentiviral particles were maintained in DMEM with 4,500 mg/L glucose (STEMCELL Technologies) supplemented with 10% Performance Plus fetal calf serum (PP-FCS), 100 U/mL penicillin + 100 μg/mL streptomycin (Pen/Strep), and 0.1 mg/mL sodium pyruvate (all Gibco; DMEM+++). H9M cells were maintained in 36SF medium consisting of DMEM supplemented with 15% FCS (STEMCELL Technologies, product #06250), Pen/Strep, glutamine and 100 ng/mL murine stem cell factor (mSCF; R&D Systems), 10 ng/mL human interleukin-6 (hIL-6), and 6 ng/mL murine IL-3 (mIL-3) (both STEMCELL Technologies).
Generation of Hoxa9 and Meis1 Overexpressing BM Cell Lines
H9M overexpressing BM cells were generated as previously described.33, 52 In brief, C57BL/6:Pep3b (Pep3b; CD45.1+) and C57BL/6 (CD45.2+) mice were injected with 150 mg/kg 5-fluorouracil. BM cells were harvested 4 days later and were then pre-stimulated for 2 days in 36SF medium. Cells were subsequently seeded onto irradiated (40 Gy) GP+E−86 MSCV-Meis1-ires-Hoxa9-pgk-neo retroviral producer cells. At 2 days later, transduced cells were subjected to in vitro selection with 750 μg/mL G418 for 7–14 days depending on non-transduced control cells survival. Selected cells were subsequently expanded and frozen for later use. In total, four independent H9M lines (CD45.1Rep1, CD45.1Rep2, CD45.2Rep1, and CD45.2Rep2) were generated through independent transduction and selection processes.
Viral Vectors, Virus Production, and Gene Transfer
The lentiviral 6xFGB platform was described previously.32 In brief, a silencing resistant spleen focus forming virus promoter-derivative (CSF) drives the expression of either one of six fluorescent color codes. Color codes consist of GFP, YFP, or mKO2 derivatives or co-expressed fluorescent protein pairs. meKO3 constitutes an mKO2 variant, and YFPe is a codon-diversified YFP. Each color code is linked to a BC for PCR-based detection as previously described.32 The production of concentrated FGB lentiviral supernatants was performed as previously described.32, 53 For transduction, 1 × 105 H9M cells were seeded in 100 μL 36SF medium supplemented with 4 μg/mL protamine sulfate into 96-well round bottom plates before the addition of a single concentrated FGB vector supernatant per well. Cells were washed after overnight transduction, and samples with comparable gene transfer rates (typically between 40%–80%) were subsequently expanded for purification of color-coded populations by fluorescent-activated cell sorting (FACS). The sorting purity was checked before the initiation of tracking experiments, and the day of the first flow cytometric analysis with subsequent sample mixing was considered d0.
Entinostat Treatment for Dose-Response Assessment and Transplantation
Entinostat (Selleckchem) was dissolved in DMSO and added to 1 × 105 H9M cells seeded into 96-well plates (flat bottom) in 100 μL 36SF, while keeping the DMSO concentration at 0.3%. Cell growth was assayed 24 hr later by the addition of 10% v/v alamarBlue (Invitrogen) and detection on a Varioskan plate reader (Varioskan; Thermo Fisher Scientific) 2–4 hr later. Data were normalized to untreated DMSO controls, and for each Entinostat concentration, 3–4 data points were acquired in parallel. H9M cells destined for longitudinal in vitro assays or transplantation were cultured with Entinostat for 24 hr, followed by extensive washing, mixing of cells in equal ratios, and assessment of initial mixing ratios by flow cytometry.
Transplantation of H9M Cells
FACS-purified color-coded H9M cells were either transplanted as single color-coded cells or as cell mixes containing all six color codes. For studies with a leukemia endpoint, 5 × 104 - × 105 cells per color-coded population were transplanted in combination with 2 × 105 radioprotective BM cells of syngeneic mice by tail vein injection into lethally irradiated (810 cGy) C57BL/6 or Pep3b recipients. Mice were sacrificed when showing signs of leukemia. For quantification of LICs by MLDA, color-coded cell mixes were prepared with 3 × 102, 1 × 103, 3 × 103, 1 × 104, 3 × 104, and 1 × 105 cells for injection with 2 × 105 syngeneic helper cells into lethally irradiated (810 cGy) C57BL/6 or Pep3b recipients. For short-term assays, cell mixes were prepared with 5 × 105 cells for each of the six CD45.1-derived color-coded populations and each of the six CD45.2-derived color-coded populations. Cell mixes were subsequently injected into lethally irradiated CD45.1 × CD45.2 F1 mice, and mice were sacrificed 24 hr after injection. All mice were monitored daily, and experimental protocols were approved by the University of British Columbia Animal Care Committee.
Flow Cytometry
H9M suspension cells were pelleted and resuspended in PBS (Gibco) supplemented with 2% FBS and 1 μM 4’,6-diamidino-2-phenylindole (DAPI; Sigma Aldrich) before flow cytometric analysis. Aliquots of BM cells from moribund mice were stained with CD11b-APC/Cy7 (M1/70), Gr1-Alexa Fluor 700 (RB6-8C5), cKit-PE/Cy7 (2B8), B220-biotin (RA3-6B2), CD3e-biotin (145-2C11), and optionally CD45.1-APC (A20) or CD45.2-APC (104) (all BioLegend), prior to staining with Streptavidin-eFluor450 (eBioscience). Data acquisition was performed in the presence of 0.5 μg/mL propidium iodide (PI). PB nucleated cells were stained with CD11b-APC/Cy7, Gr1-Alexa Fluor 700, cKit-PE/Cy7, and either CD45.1-APC or CD45.2-APC antibodies. Cells were resuspended in 1 μM DAPI for flow cytometric analysis. BM cells from short-term homing assays were flushed from the tibia, femurs, and iliac crests of recipient mice before staining with CD45.1-Pacific Blue (A20; BioLegend) and CD45.2-APC and 0.5 μg/mL PI for dead cell exclusion. 12xFGB H9M cell mixes were stained likewise and tracked over time. All data were recorded on an LSRFortessa flow cytometer (BD Biosciences), and data analyses were performed with FlowJo (Tristar).
Author Contributions
T.M. performed experiments, coordinated research activities, analyzed data, designed the study, and wrote the manuscript; R.K.H. analyzed data, designed the study, and wrote the manuscript; C.K.L. performed research and wrote the manuscript; and J.R., L.S.M., and N.v.K. performed research.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by a Terry Fox Foundation Program Project grant (TFF-122869) and a Stem Cell Network Global Research grant (9/5264(CT10)). T.M. was supported by a Dr. Mildred Scheel Postdoctoral Research Fellowship by the German Cancer Aid. C.K.L. received studentships from the Canadian Institute of Health Research (http://www.cihr-irsc.gc.ca/e/193.html#), the University of British Columbia (https://www.grad.ubc.ca/awards/four-year-doctoral-fellowship-4yf), and the UBC Faculty of Medicine, as well as from the BC Cancer Agency (http://www.bccancer.bc.ca/default.htm). Furthermore, we would like to thank the staff of the Flow Core and of the Animal Resource Center of the British Columbia Cancer Agency Research Center for excellent technical support.
Footnotes
Supplemental Information includes nine figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.05.007.
Supplemental Information
References
- 1.Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C., Wartman L.D., Lamprecht T.L., Liu F., Xia J. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klco J.M., Spencer D.H., Miller C.A., Griffith M., Lamprecht T.L., O’Laughlin M., Fronick C., Magrini V., Demeter R.T., Fulton R.S. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paguirigan A.L., Smith J., Meshinchi S., Carroll M., Maley C., Radich J.P. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci. Transl. Med. 2015;7:281re2. doi: 10.1126/scitranslmed.aaa0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somervaille T.C., Matheny C.J., Spencer G.J., Iwasaki M., Rinn J.L., Witten D.M., Chang H.Y., Shurtleff S.A., Downing J.R., Cleary M.L. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong S.H., Goode D.L., Iwasaki M., Wei M.C., Kuo H.P., Zhu L., Schneidawind D., Duque-Afonso J., Weng Z., Cleary M.L. The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell. 2015;28:198–209. doi: 10.1016/j.ccell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 8.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 9.Kreso A., Dick J.E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imren S., Heuser M., Gasparetto M., Beer P.A., Norddahl G.L., Xiang P., Chen L., Berg T., Rhyasen G.W., Rosten P. Modeling de novo leukemogenesis from human cord blood with MN1 and NUP98HOXD13. Blood. 2014;124:3608–3612. doi: 10.1182/blood-2014-04-564666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pabst C., Krosl J., Fares I., Boucher G., Ruel R., Marinier A., Lemieux S., Hébert J., Sauvageau G. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat. Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 13.Wei J., Wunderlich M., Fox C., Alvarez S., Cigudosa J.C., Wilhelm J.S., Zheng Y., Cancelas J.A., Gu Y., Jansen M. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buechele C., Breese E.H., Schneidawind D., Lin C.H., Jeong J., Duque-Afonso J., Wong S.H., Smith K.S., Negrin R.S., Porteus M., Cleary M.L. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood. 2015;126:1683–1694. doi: 10.1182/blood-2015-05-646398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellegast J.M., Rauch P.J., Kovtonyuk L.V., Müller R., Wagner U., Saito Y., Wildner-Verhey van Wijk N., Fritz C., Rafiei A., Lysenko V. inv(16) and NPM1mut AMLs engraft human cytokine knock-in mice. Blood. 2016;128:2130–2134. doi: 10.1182/blood-2015-12-689356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wunderlich M., Chou F.S., Link K.A., Mizukawa B., Perry R.L., Carroll M., Mulloy J.C. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24:1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDermott S.P., Eppert K., Notta F., Isaac M., Datti A., Al-Awar R., Wrana J., Minden M.D., Dick J.E. A small molecule screening strategy with validation on human leukemia stem cells uncovers the therapeutic efficacy of kinetin riboside. Blood. 2012;119:1200–1207. doi: 10.1182/blood-2011-01-330019. [DOI] [PubMed] [Google Scholar]
- 18.Ramsey J.M., Kettyle L.M., Sharpe D.J., Mulgrew N.M., Dickson G.J., Bijl J.J., Austin P., Mayotte N., Cellot S., Lappin T.R. Entinostat prevents leukemia maintenance in a collaborating oncogene-dependent model of cytogenetically normal acute myeloid leukemia. Stem Cells. 2013;31:1434–1445. doi: 10.1002/stem.1398. [DOI] [PubMed] [Google Scholar]
- 19.Sykes M., Eisenthal A., Sachs D.H. Mechanism of protection from graft-vs-host disease in murine mixed allogeneic chimeras. I. Development of a null cell population suppressive of cell-mediated lympholysis responses and derived from the syngeneic bone marrow component. J. Immunol. 1988;140:2903–2911. [PubMed] [Google Scholar]
- 20.Abkowitz J.L., Persik M.T., Shelton G.H., Ott R.L., Kiklevich J.V., Catlin S.N., Guttorp P. Behavior of hematopoietic stem cells in a large animal. Proc. Natl. Acad. Sci. USA. 1995;92:2031–2035. doi: 10.1073/pnas.92.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spangrude G.J., Heimfeld S., Weissman I.L. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 22.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt M., Hoffmann G., Wissler M., Lemke N., Müssig A., Glimm H., Williams D.A., Ragg S., Hesemann C.U., von Kalle C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- 24.Lu R., Neff N.F., Quake S.R., Weissman I.L. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerrits A., Dykstra B., Kalmykowa O.J., Klauke K., Verovskaya E., Broekhuis M.J., de Haan G., Bystrykh L.V. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 26.Livet J., Weissman T.A., Kang H., Draft R.W., Lu J., Bennis R.A., Sanes J.R., Lichtman J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 27.Weber K., Thomaschewski M., Warlich M., Volz T., Cornils K., Niebuhr B., Täger M., Lütgehetmann M., Pollok J.M., Stocking C. RGB marking facilitates multicolor clonal cell tracking. Nat. Med. 2011;17:504–509. doi: 10.1038/nm.2338. [DOI] [PubMed] [Google Scholar]
- 28.Malide D., Métais J.Y., Dunbar C.E. Dynamic clonal analysis of murine hematopoietic stem and progenitor cells marked by 5 fluorescent proteins using confocal and multiphoton microscopy. Blood. 2012;120:e105–e116. doi: 10.1182/blood-2012-06-440636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohme M., Maire C.L., Riecken K., Zapf S., Aranyossy T., Westphal M., Lamszus K., Fehse B. Optical barcoding for single-clone tracking to study tumor heterogeneity. Mol. Ther. 2017;25:621–633. doi: 10.1016/j.ymthe.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu V.W., Yusuf R.Z., Oki T., Wu J., Saez B., Wang X., Cook C., Baryawno N., Ziller M.J., Lee E. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell. 2016;168:944–945. doi: 10.1016/j.cell.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smurthwaite C.A., Hilton B.J., O’Hanlon R., Stolp Z.D., Hancock B.M., Abbadessa D., Stotland A., Sklar L.A., Wolkowicz R. Fluorescent genetic barcoding in mammalian cells for enhanced multiplexing capabilities in flow cytometry. Cytometry A. 2014;85:105–113. doi: 10.1002/cyto.a.22406. [DOI] [PubMed] [Google Scholar]
- 32.Maetzig T., Ruschmann J., Lai C.K., Ngom M., Imren S., Rosten P., Norddahl G.L., von Krosigk N., Sanchez Milde L., May C. A lentiviral fluorescent genetic barcoding system for flow cytometry-based multiplex tracking. Mol. Ther. 2017;25:606–620. doi: 10.1016/j.ymthe.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibbs K.D., Jr., Jager A., Crespo O., Goltsev Y., Trejo A., Richard C.E., Nolan G.P. Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell. 2012;10:210–217. doi: 10.1016/j.stem.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 36.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Miller P.G., Al-Shahrour F., Hartwell K.A., Chu L.P., Järås M., Puram R.V., Puissant A., Callahan K.P., Ashton J., McConkey M.E. In vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell. 2013;24:45–58. doi: 10.1016/j.ccr.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puram R.V., Kowalczyk M.S., de Boer C.G., Schneider R.K., Miller P.G., McConkey M., Tothova Z., Tejero H., Heckl D., Järås M. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165:303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J., Wang E., Milazzo J.P., Wang Z., Kinney J.B., Vakoc C.R. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhang H.E., Ruddy D.A., Krishnamurthy Radhakrishna V., Caushi J.X., Zhao R., Hims M.M., Singh A.P., Kao I., Rakiec D., Shaw P. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015;21:440–448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 41.Kurre P., Anandakumar P., Kiem H.P. Rapid 1-hour transduction of whole bone marrow leads to long-term repopulation of murine recipients with lentivirus-modified hematopoietic stem cells. Gene Ther. 2006;13:369–373. doi: 10.1038/sj.gt.3302659. [DOI] [PubMed] [Google Scholar]
- 42.Kurre P., Anandakumar P., Harkey M.A., Thomasson B., Kiem H.P. Efficient marking of murine long-term repopulating stem cells targeting unseparated marrow cells at low lentiviral vector particle concentration. Mol. Ther. 2004;9:914–922. doi: 10.1016/j.ymthe.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Mostoslavsky G., Kotton D.N., Fabian A.J., Gray J.T., Lee J.S., Mulligan R.C. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 2005;11:932–940. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Nolan-Stevaux O., Tedesco D., Ragan S., Makhanov M., Chenchik A., Ruefli-Brasse A., Quon K., Kassner P.D. Measurement of cancer cell growth heterogeneity through lentiviral barcoding identifies clonal dominance as a characteristic of in vivo tumor engraftment. PLoS ONE. 2013;8:e67316. doi: 10.1371/journal.pone.0067316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter S.N., Baker L.C., Mittelman D., Porteus M.H. Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 2014;15:R75. doi: 10.1186/gb-2014-15-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selich A., Daudert J., Hass R., Philipp F., von Kaisenberg C., Paul G., Cornils K., Fehse B., Rittinghausen S., Schambach A., Rothe M. Massive clonal selection and transiently contributing clones during expansion of mesenchymal stem cell cultures revealed by lentiviral RGB-barcode technology. Stem Cells Transl. Med. 2016;5:591–601. doi: 10.5966/sctm.2015-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung N., Dai B., Gentles A.J., Majeti R., Feinberg A.P. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat. Commun. 2015;6:8489. doi: 10.1038/ncomms9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin G., Yamazaki Y., Takuwa M., Takahara T., Kaneko K., Kuwata T., Miyata S., Nakamura T. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 49.Mulgrew N.M., Kettyle L.M., Ramsey J.M., Cull S., Smyth L.J., Mervyn D.M., Bijl J.J., Thompson A. c-Met inhibition in a HOXA9/Meis1 model of CN-AML. Dev. Dyn. 2014;243:172–181. doi: 10.1002/dvdy.24070. [DOI] [PubMed] [Google Scholar]
- 50.Fares I., Chagraoui J., Gareau Y., Gingras S., Ruel R., Mayotte N., Csaszar E., Knapp D.J., Miller P., Ngom M. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345:1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boitano A.E., Wang J., Romeo R., Bouchez L.C., Parker A.E., Sutton S.E., Walker J.R., Flaveny C.A., Perdew G.H., Denison M.S. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorsteinsdottir U., Mamo A., Kroon E., Jerome L., Bijl J., Lawrence H.J., Humphries K., Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 53.Maetzig T., Brugman M.H., Bartels S., Heinz N., Kustikova O.S., Modlich U., Li Z., Galla M., Schiedlmeier B., Schambach A., Baum C. Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells. Blood. 2011;117:3053–3064. doi: 10.1182/blood-2010-08-303222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.