Abstract
Campylobacter curvus is a rarely encountered Campylobacter species in human, animal, and environmental samples. During the course of two investigations, one involving a search for possible bacterial agents causing bloody gastroenteritis and a second concerning a small outbreak of Brainerd's diarrhea in northern California, 20 strains of C. curvus or C. curvus-like organisms were isolated by a microfiltration technique and prolonged incubation. The results suggest that C. curvus may be an underappreciated Campylobacter that may be involved in sporadic and outbreak cases of bloody or chronic diarrhea in humans.
The genus Campylobacter consists of curved or S-shaped gram-negative, oxidase-positive microaerophilic bacilli with a respiratory type of metabolism (15). The genus has undergone several taxonomic revisions over the past decade, including the transfer of a number of species to either the genus Arcobacter or the genus Helicobacter (12). Sixteen species presently reside in the genus Campylobacter. Of these 16 species, Campylobacter jejuni, C. coli, and C. fetus are the major species of medical, public health, or veterinary interest involved in disease processes (12).
Of the 13 remaining Campylobacter species, very little information regarding their pathogenicity is available, although some species, such as C. upsaliensis, C. hyointestinalis, and C. lari, have been associated with various infectious syndromes (1, 3, 13). Even less information about campylobacters such as C. curvus is available. Originally described in 1984 (“Wolinella curva”) (14), only four strains of C. curvus were described in that report; two strains were associated with the oral cavity, one isolate was from blood, and the fourth was a clinical isolate of unknown origin. Since that initial report, few studies have reported on the isolation of C. curvus. C. curvus has been isolated from the stools of patients who subsequently presented with either Guillain-Barré or Fisher's syndrome, although no role in the development of these neurologic conditions could be associated with this species (8). In a 2000 study of more than 1,300 human stool specimens conducted to study the prevalence of campylobacters (other than C. jejuni and C. coli), only 3 of 48 campylobacters recovered were tentatively identified as C. curvus-like (4). A study from 2003 (11) reported that 320 consecutive liquid or semisolid fecal specimens submitted to a laboratory for enteric pathogen detection were culture negative for C. curvus, although one stool specimen was positive for C. curvus by 16S rRNA gene PCR. These cumulative results suggest that the frequency of C. curvus in the gastrointestinal tracts of symptomatic individuals is exceedingly low.
Over a 4-year period, we have isolated 20 strains of C. curvus from two separate and distinct clinical settings, namely, from a hospital survey of infectious causes of bloody diarrhea and from an outbreak of Brainerd's diarrhea in northern California. These isolations serve as the basis of this report.
MATERIALS AND METHODS
Settings.
The first setting from which C. curvus was isolated involved an analysis of 142 consecutive fecal specimens with a diagnosis of bloody diarrhea submitted to the Microbial Diseases Laboratory by a large regional microbiology laboratory in a major medical center in Oakland, Calif. These samples originated from individuals in all age groups, although the majority came from adults. None of these symptomatic patients had unusual risk factors. All 142 stool specimens were either grossly bloody or were positive for occult blood by the Hemoccult SENSA Fecal Occult Blood Test (Smith Kline Diagnostics, San Jose, Calif.). The period of sample analysis ran from October 1995 through February 1996. In the second setting, C. curvus was isolated in conjunction with a small outbreak of Brainerd's diarrhea in Humboldt County in northern California in March and April 1999. These cases of watery diarrhea involved individuals who had had symptoms for more than 4 weeks.
Isolation of C. curvus.
Liquid stools or emulsified suspensions of semisolid or solid feces in phosphate-buffered saline were inoculated onto 0.45-μm-pore-size filters placed on sheep blood agar plates (BAPs). The plates were incubated at room temperature (25°C) for 30 min, after which the filters and any excess fluid were removed and the BAPs were incubated under microaerophilic conditions for 7 days. The cultures were observed daily for evidence of growth.
Biochemical tests and 16S rRNA gene sequencing.
A number of biochemical tests were performed with all isolates, based on the tests recommended for use in the differentiation of aerobic, microaerophilic, motile, or helical vibrios or gram-negative bacteria (6). These included tests for catalase, oxidase, and nitrate reductase activities; H2S production from triple sugar iron agar (TSI) and lead acetate paper strips; growth in 1% glycine and 3.5% NaCl; growth at 25, 35, and 42°C; hydrolysis of indoxyl acetate; and sensitivity to cephalothin (30-μg disk) and nalidixic acid (30-μg disk). The cellular fatty acid (CFA) profiles for all isolates were determined by fatty acid methyl ester analysis (MIS system software, version 3.2C; Microbial Identification Systems, Newark, Del.) (10).
Phylogenetic relatedness studies were performed with all campylobacter-like isolates by 16S rRNA gene sequencing (500-bp alignment), as described previously (7). Sequence assembly and analysis were accomplished with MicroSeq Identification software (Applied Biosystems, Foster City, Calif.). An isolate was defined as belonging to a particular species if it exhibited ≤1.2% sequence divergence from the type strain of a given species.
RESULTS
Sporadic cases of bloody diarrhea.
Of 142 consecutive stool specimens containing macroscopic amounts of blood or occult blood, 15 (10.6%) yielded a campylobacter or a campylobacter-like isolate upon subculture on BAPs. These campylobacter-like isolates typically took between 3 and 7 days of incubation under microaerophilic conditions before they first appeared as pinpoint colonies on BAPs. Of the 15 patients from which campylobacter isolates were detected, 3 presented with frankly bloody stools, while the remainder were occult blood positive. No other bacterial pathogens were identified in 12 patients, including Salmonella, Shigella, Escherichia coli (seven pathogenic groups), Yersinia enterocolitica, Vibrio spp., other Campylobacter species, Aeromonas, Plesiomonas, and Edwardsiella tarda. The stool of one person who was occult blood positive contained Cryptosporidium, as determined by enzyme immunoassay, while the stools of two other individuals (one of whom presented with frankly bloody diarrhea) also harbored diffusely adhering E. coli. The average age of a person from whom a campylobacter isolate was recovered was 42.8 years (age range, 18 months to 78 years). The ratio of males to females was 1:2.
Biochemically, these 15 strains met the definition of members of the genus Campylobacter, as they were nonfermentative, microaerophilic gram-negative rods that were oxidase positive and that grew at both 35 and 42°C (9). Consistent with the biochemical data was the result of CFA analysis: a lack of branched-chain fatty acids but the presence of hydroxy acids, with the majority of CFAs present as straight-chained saturated and monounsaturated fatty acids. However, neither the biochemical nor the CFA profiles exactly matched the phenotypic and CFA profiles described for existing taxa in the genus Campylobacter or in the genera Helicobacter or Arcobacter (Table 1). C. curvus strains have been described as catalase negative and indoxyl acetate positive. However, in our study 50% of the C. curvus strains were catalase positive, while only 15% were indoxyl acetate positive. On TSI, only 40% of our strains produced H2S, although all were positive by the lead acetate paper method.
TABLE 1.
Phenotypic features of C. curvus and C. curvus-like strains
| Characteristic | No. (%) of strains (n = 20) positive | C. curvus phenotypea |
|---|---|---|
| Oxidase | 20 (100) | + |
| Catalase | 10 (50) | − |
| Growth at: | ||
| 25°C | 5 (25) | − |
| 35°C | 20 (100) | + |
| 42°C | 20 (100) | + |
| Growth in: | ||
| 1% Glycine | 20 (100) | + |
| 3.5% NaCl | 1 (5) | − |
| H2S production on: | ||
| TSI | 8 (40) | + |
| PbAcb | 20 (100) | NA |
| Indoxyl acetate hydrolysis | 3 (15) | + |
| Sensitive to: | ||
| Nalidixic acid (30-μg disk) | 20 (100) | + |
| Cephalothin (30-μg disk) | 20 (100) | + |
Data are from reference 6. NA, not available.
PbAc, lead acetate.
Brainerd's diarrhea outbreak.
During the early part of 1999 an outbreak of Brainerd's diarrhea, characterized by three or more loose bowel movements in a 24-h period for ≥4 consecutive weeks, occurred in northern California. At least 23 cases were identified in this outbreak, and stool samples from 6 of the symptomatic individuals were submitted for analysis. Of these six stool samples, five (83.3%) yielded a campylobacter-like organism with phenotypic properties similar to those associated with bloody diarrhea described above. No other enteric bacterial or enteric viruses (some samples) were isolated from these 23 samples, although only some samples were tested for enteric viruses. Five fecal samples obtained from asymptomatic household or close contacts of these individuals were negative for all enteric pathogens, including Campylobacter species.
16S rRNA gene sequencing.
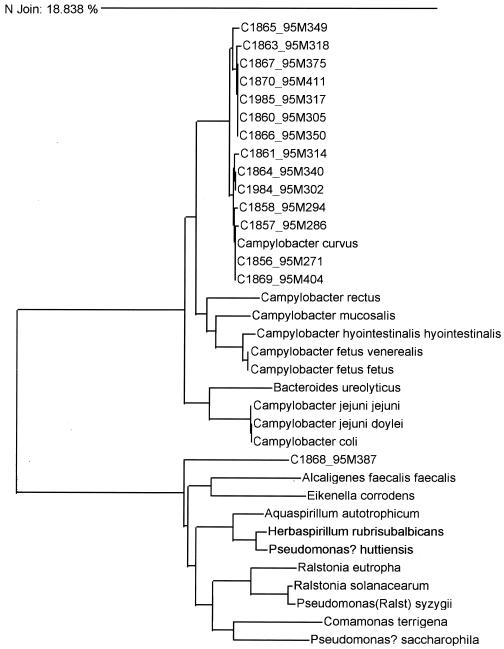
Because phenotypic tests, including CFA analysis, failed to yield a definitive answer regarding species identification, the 20 campylobacter-like strains were subjected to 16S rRNA gene sequencing. Overall, 18 of these 20 strains were identified as C. curvus by 16S rRNA gene sequencing analysis. These 18 strains had a sequence divergence from the type strain of C. curvus (ATCC 35224) ranging from 0 to 1.2% (average divergence, 0.64%). One additional strain (MDL 99A2133) had a nonexpressed ∼50-bp intervening sequence in the rRNA gene, which complicated the analysis, but the strain was also subsequently identified as C. curvus by 16S rRNA gene sequencing analysis. A final isolate (MDL 95M387) was unrelated to the genus Campylobacter and had the closest match to Aquaspirillum autotrophicum (10.88% divergence) upon alignment of a 500-bp sequence. Strain MDL 95M387 had properties identical or very similar to those of the other strains, with the exception that it was indoxyl acetate positive. The results of an unweighted pair group method analysis with the 15 strains recovered from patients with bloody diarrhea is depicted in Fig. 1.
FIG. 1.
Dendrogram depicting the linkage of 16 C. curvus or C. curvus-like strains by 16S rRNA gene sequencing by the neighbor-joining method.
DISCUSSION
Since the original description of C. curvus, which included only four strains, there have been only a couple of citations in the literature regarding the isolation of a single strain of this bacterium (4, 8) or the detection of its genetic material (11) from clinical samples. The present report describes for the first time a large collection of C. curvus strains associated with two infectious processes, namely, bloody diarrhea (n = 14) and Brainerd's diarrhea (n = 5). Although it cannot be conclusively proven that C. curvus was the etiologic agent of either disease, there is compelling evidence that suggests that this was, in fact, the case. With the exception of diffusely adhering E. coli as a potential but still controversial copathogen in two patients and a Cryptosporidium isolate that infected one individual, no other enteric pathogens were identified in the feces of symptomatic individuals in either group containing C. curvus. The fact that >10% of persons who presented with frankly bloody stools or who were positive for occult blood were positive for C. curvus and the fact that this species is rarely isolated from gastrointestinal samples in large-scale studies (4, 8) strongly suggest a causal role in this setting. Furthermore, in the Brainerd's diarrhea outbreak, C. curvus was isolated from five of six sick patients but not from any of five symptomless household or close contacts of the infected individuals.
The phenotypes of the strains identified as C. curvus in this study deviated from the ideal phenotype in several characteristics (Table 1). These deviations are because of the small subset (n = 4) of C. curvus strains used to establish the biochemical profile of this species in the original publication (14) or because of methodological differences. The latter possibility can be seen in the test results for H2S production by two different methods. Sequencing of the 16S rRNA gene indicates that several strains generated 100% sequence matches (no divergence) to the type strain of C. curvus, further supporting their identification. While there are no criteria or established values for 16S rRNA gene sequence similarity for species identification, one recent study established a similarity score of ≥99% (2), which is very similar to that used in the present study. Whether strains with divergence values of ∼1% are in fact C. curvus or a phylogenetically related and biochemically similar but distinct taxon await DNA-DNA hybridization studies. One strain of C. curvus (MDL 99A2133) was difficult to identify because of an intervening sequence. However, this phenomenon has previously been reported in Campylobacter species, including C. rectus, C. sputorum, and C. curvus (5). The exact reason why these intron-like DNA sequences are inserted within ribosomal genes is unknown.
The large number of C. curvus strains identified in the present study appears to result from two laboratory procedures, namely, inoculation of stool suspensions onto a 0.45-μm-pore-size filter and prolonged incubation (>3 days) of BAPs in a microaerophilic environment. The significance of the isolation of C. curvus from gastrointestinal specimens will require additional prospective studies evaluating the frequency and occurrence of this species in distinct diarrheal disease syndromes.
REFERENCES
- 1.Chiu, C.-H., C.-Y. Kuo, and J. T. Ou. 1995. Chronic diarrhea and bacteremia caused by Campylobacter lari in a neonate. Clin. Infect. Dis. 21:700-701. [DOI] [PubMed] [Google Scholar]
- 2.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J.-P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmonds, P., C. M. Patton, P. M. Griffin, T. J. Barrett, G. P. Schmid, C. N. Baker, M. A. Lambert, and D. J. Brenner. 1987. Campylobacter hyointestinalis associated with human gastrointestinal disease in the United States. J. Clin. Microbiol. 25:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etoh, Y., A. Yamamoto, and N. Goto. 1998. Intervening sequences in 16S rRNA genes of Campylobacter sp.: diversity of nucleotide sequences and uniformity of location. Microbiol. Immunol. 42:241-243. [DOI] [PubMed] [Google Scholar]
- 6.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Aerobic/microaerophilic, motile, helical/vibroid gram-negative bacteria, p. 58-63. In Bergey's manual of determinative bacteriology, 9th ed. The Williams & Wilkins Co., Baltimore, Md.
- 7.Janda, J. M., S. L. Abbott, S. Khashe, and W. Probert. 2002. Phenotypic and genotypic properties of the genus Hafnia. J. Med. Microbiol. 51:575-580. [DOI] [PubMed] [Google Scholar]
- 8.Koga, M., N. Yuki, M. Takahashi, K. Saito, and K. Hirata. 1999. Are Campylobacter curvus and Campylobacter upsaliensis antecedent infectious agents in Guillain-Barre and Fisher's syndrome? J. Neurol. Sci. 163:53-57. [DOI] [PubMed] [Google Scholar]
- 9.Lawson, A. J., S. L. W. On, J. M. J. Logan, and J. Stanley. 2001. Campylobacter hominis sp. nov., from the human gastrointestinal tract. Int. J. Syst. Evol. Microbiol. 51:651-660. [DOI] [PubMed] [Google Scholar]
- 10.Lindquist, D., D. Murrill, W. P. Burran, G. Winans, J. M. Janda, and W. Probert. 2003. Characteristics of Massilia timonae and Massilia timonae-like isolates from human patients, with an emended description of the species. J. Clin. Microbiol. 41:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher, M., C. Finnegan, E. Collins, B. Ward, C. Carroll, and M. Cormican. 2003. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 41:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.On, S. L. W. 2001. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: current status, future prospects and immediate concerns. J. Appl. Microbiol. 90:1S-15S. [DOI] [PubMed] [Google Scholar]
- 13.Patton, C. M., N. Shaffer, P. Edmonds, T. J. Barrett, M. A. Lambert, C. Baker, D. M. Perlman, and D. J. Brenner. 1989. Human disease associated with “Campylobacter upsaliensis” (catalase-negative or weakly positive Campylobacter species) in the United States. J. Clin. Microbiol. 27:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanner, A. C. R., M. A. Listgarten, and J. L. Ebersole. 1984. Wolinella curva sp. nov.: “Vibrio succinogenes” of human origin. Int. J. Syst. Bacteriol. 34:275-282. [Google Scholar]
- 15.Vandamme, P., and J. De Ley. 1991. Proposal for a new family, Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451-455. [Google Scholar]