Abstract
Group B streptococci (GBS) are serotyped according to capsular polysaccharide (CPS) type (Ia to VIII); an isolate is classified as nontypeable (NT) if no detectable CPS is found. Surface-localized protein antigens (α, β, R1, and R4) serve as additional markers to classify GBS isolates, which is particularly useful since NT isolates often express one or more of these proteins. To compare genetic resemblance among isolates with similar protein profiles, we studied 58 NT isolates digested with the SmaI macrorestriction enzyme prior to pulsed-field gel electrophoresis (PFGE). Of these 58, 15.5% expressed α only, 20.7% expressed α+β, 15.5% expressed R4, and 25.8% expressed R1,R4, while 22.4% of the isolates expressed no detectable proteins. The largest PFGE profile group, with 48% of the isolates, was group 4, composed primarily of isolates that expressed R1,R4 or no proteins. The second most common profiles were 3 and 32, each with 13.8% of the isolates. Since NT isolates in profile group 4 were highly related to type V isolates, as demonstrated by PFGE profiles, we investigated 45 type V isolates. Two-thirds of the type V isolates within profile group 4 were classified into subgroup 4a, compared to 28.2% of 39 NT isolates. Only 11% of the V/R1,R4 isolates were identical to the prototype group 4 profile, in contrast to 75% of the NT/R1,R4 isolates. A shift of type V isolates into profile 4 subgroups may be indicative of a genetic change over time. PFGE is a valuable approach for comparison of GBS isolate relatedness and for monitoring of NT and typeable GBS isolates for potential clonal divergence.
Group B streptococcal (GBS; Streptococcus agalactiae) isolates are classified according to their capsular polysaccharide (CPS) into one of nine types: Ia, Ib, and II to VIII (13, 19). However, when tested by routine typing methods, approximately 2.9% of colonizing isolates and 1.4% of invasive isolates lack a detectable CPS type and consequently are categorized as nontypeable (NT) (4, 9, 10).
In addition to the CPS type, protein markers can be useful for classification since most GBS isolates express either the c or the R surface-localized protein(s) (9, 10). The c protein is made of two components that are distinguished on the basis of their reaction with trypsin; α is trypsin resistant, while β is trypsin sensitive and binds immunoglobulin A. Isolates may possess only one or both of these components (14). The R proteins (R1, R2, R3, and R4) are the second group of surface proteins and are trypsin resistant; isolates may express one or more of them (10, 18).
Identification of surface-localized proteins is an important aspect of the classification of GBS isolates since some proteins are highly associated with specific CPS types. For example, α protein is commonly expressed by the majority of serotype Ia isolates, while more than half of serotype V isolates possess the R1 and R4 proteins (10). Thus, these CPS type-protein profile associations may permit one to compare similarities among NT and typeable isolates on the basis of their protein profiles.
Because pulsed-field gel electrophoresis (PFGE) is highly reproducible and yields well-resolved bacterial DNA, it is commonly used in molecular and epidemiological laboratories to classify and type bacterial isolates (2, 16). Using PFGE, our laboratory previously classified 78 NT GBS isolates into DNA profile groups on the basis of their DNA macrorestriction patterns (4). To continue validation of PFGE as an effective tool for classifying NT isolates, we recently studied 58 additional NT isolates. In addition, we investigated further the genetic relatedness between NT/R1,R4 and V/R1,R4 isolates and examined their DNA macrorestriction profiles for evidence of genomic divergence.
MATERIALS AND METHODS
Bacterial isolates.
All NT and type V isolates were received in our laboratory between 1998 and 2002 from a multicenter collaborative study (Pittsburgh, Houston, and Seattle). The 58 NT GBS clinical isolates studied included 1 invasive and 57 colonizing (34 vaginal, 23 rectal) isolates; the colonizing isolates were from 35 nonpregnant women. Most (n = 50) of the isolates were from Pittsburgh, while 7 were from Seattle and 1 was from Houston. The 45 type V GBS clinical isolates studied included 4 invasive and 41 colonizing (22 vaginal, 19 rectal) isolates; the colonizing isolates were from 39 nonpregnant women. Most (n = 42) of the isolates were from Pittsburgh, while 3 were from Houston.
Growth conditions.
GBS isolates were stored frozen at −30°C in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) with 2% sheep blood until studied. They were grown overnight in Todd-Hewitt broth for CPS and protein typing and on 5% sheep blood agar plates (Remel, Lenexa, Kans.) for 2 days at 37°C for molecular studies (3).
Serotyping.
To determine the CPS type and surface-localized protein profile, isolates were typed by HCl extraction and Ouchterlony double immunodiffusion in agarose as previously described (10, 14). When necessary, the HCl extract was concentrated to enhance weak precipitin reactions and/or grown in broth with increased glucose and buffer to enhance CPS production (1). Isolates without detectable CPS were further analyzed by PFGE.
Molecular analysis by PFGE.
A rapid PFGE assay was performed on isolates with no detectable CPS in accordance with our published protocol (3), a modification of the method of Fasola et al. (8). Briefly, bacteria were embedded in agarose plugs and then treated with mutanolysin and proteinase K to lyse the cell wall and precipitate the protein. The bacterial DNA was digested in situ with the infrequently cutting restriction enzyme SmaI before the DNA fragments were resolved by PFGE. NT isolates with similar protein profiles were assembled into groups and studied together with the PFGE prototype typeable isolate for that particular protein. Each gel also contained a lambda DNA ladder (Bio-Rad) and our internal control, strain 89-022 (Ib/α+β).
DNA macrorestriction band pattern analysis.
For digital analyses, the gel was photographed under UV light with a DC 40 digital camera (Kodak, Rochester, N.Y.) and the image was imported into the 1D digital imaging software (Kodak) to gather information about the migration, intensity, size, and quantity of DNA bands from each isolate. Visual comparison of the PFGE profiles of the isolates to the DNA profiles of the prototypes was done by using our modification of the criteria of Tenover et al. (16). To achieve more precise analyses, we created the following method to compare the typeable and NT isolates for genotypic relatedness on the basis of their band patterns (4). Each DNA profile group had a prototype to which all similar isolates were compared. An isolate that did not have a band pattern identical to that of the prototype was assigned a letter to designate a profile subgroup. An isolate with a one- or two-band difference from the prototype DNA profile was assigned the letter a, one with a three-band difference was assigned the letter b, one with a four-band difference was assigned the letter c, and one with a five-band difference was assigned the letter d. Isolates with five or more band differences were not considered to be related to the PFGE profile group and were assigned a different profile number. Classification of isolates into PFGE profile subgroups ranging from a to d allowed one to identify immediately to what extent an isolate was related to the prototype. However, it was possible for isolates within a specific subgroup to have nonidentical band patterns, since band differences were defined as either the presence or absence of a band compared to the profile prototype.
RESULTS
Nontypeable GBS isolates.
A comprehensive summary of both protein expression patterns and PFGE profile data is shown in Table 1. The most prevalent protein profile among the 58 NT isolates was R1,R4, with 15 isolates (25.9%); the least common protein profiles were R4 and α only, each with 9 isolates (15.5%). Thus, 21 isolates expressed c protein (36.2%) and 24 isolates expressed R proteins (41.4%), while 13 isolates (22.4%) did not express a detectable protein.
TABLE 1.
Distribution of 58 NT GBS isolates into PFGE profile groups by protein expression patterna
| No. of isolates | Protein(s) | PFGE groupsb |
|---|---|---|
| 9 | c (α only) | 1a2, 1d1, 111, 13b1, 152, 161, 331 |
| 12 | c (α + β) | 2a2, 193, 323, 32b2, 341, 351 |
| 9 | R4 | 34, 3a1, 4a1, 221, 282, |
| 15 | R1, R4 | 49, 4a4, 4c2 |
| 13 | None | 44, 4a4, 4b1, 4d3, 161 |
Categorization of DNA profiles was done in accordance with our modifications of the criteria of Tenover et al. (16).
Each superscript number is the number of isolates with that DNA macrorestriction profile.
Analysis of the PFGE band profile patterns resulted in assignment of the 58 NT isolates to 15 of 35 DNA macrorestriction profile groups. Among these 35 groups were profiles 32 to 35, which we added recently to enable classification of an additional five isolates. As shown in Table 1, 9 α-only isolates were distributed among six different PFGE profiles (1, 11, 13, 15, 16, and 33), 12 α+β isolates were distributed among five PFGE profiles (2, 19, 32, 34, and 35), and 9 R4 isolates were distributed among four PFGE profiles (3, 4, 22, and 28), while all 15 R1,R4 NT isolates were classified into DNA profile group 4. Of 13 isolates lacking a detectable protein, 12 were classified into profile 4, while 1 isolate was assigned to profile group 16.
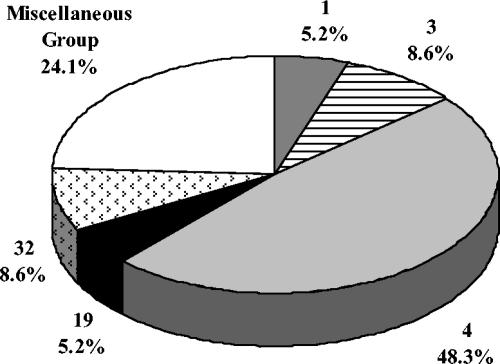
Profile 4 was the most prevalent PFGE group, with 28 isolates (48.3%), and included isolates with three different protein profiles: 15 R1,R4 isolates, 1 R4 isolate, and 13 isolates with no detectable protein (Fig. 1). Profiles 3 and 32, each with five isolates, were the second most common PFGE profiles. All isolates within newly created profile 32 were α+β and included the only invasive NT isolate. All PFGE profile groups with fewer than three isolates were consolidated into a miscellaneous group for Fig. 1. This group of NT isolates was composed of two isolates each from PFGE profile groups 2, 15, 16, and 28 and one isolate each from groups 11, 13, 22, 33, 34, and 35.
FIG. 1.
PFGE profile group distribution among 58 nontypeable GBS isolates. PFGE profile groups with less than 5% were consolidated into a miscellaneous group. This group included two isolates each from PFGE profile groups 2, 15, 16, and 28 and one isolate each from profile groups 11, 13, 22, 33, 34, and 35.
Clonal divergence of type V isolates.
Since almost half of the 58 NT isolates were classified within profile group 4 and the prototype for this profile group is a V/R1,R4 isolate, it seemed warranted to study V/R1,R4 isolates more extensively and to compare their PFGE band patterns with those of the NT isolates in this profile group. Of the 84 PFGE profile group 4 isolates used in this analysis (Table 2), 45 were type V and 39 were NT (15 R1,R4 isolates and 13 isolates without protein from the 58 NT isolates were included, plus an additional 11 from the same time period).
TABLE 2.
Distribution of 39 NT and 45 serotype V GBS isolates classified within PFGE profile group 4
| Serotype | Total no. of isolates | No. of isolates in DNA macrorestriction profile group or subgroup:
|
||||
|---|---|---|---|---|---|---|
| 4 | 4a | 4b | 4c | 4d | ||
| NT/R1, R4 | 24 | 18 | 4 | 2 | ||
| NT/none | 14 | 4 | 6 | 1 | 3 | |
| NT/R4 | 1 | 1 | ||||
| V/R1, R4 | 27 | 3 | 18 | 6 | ||
| V/none | 13 | 5 | 8 | |||
| V/R4 | 5 | 4 | 1 | |||
| Total | 84 | 30 | 41 | 8 | 2 | 3 |
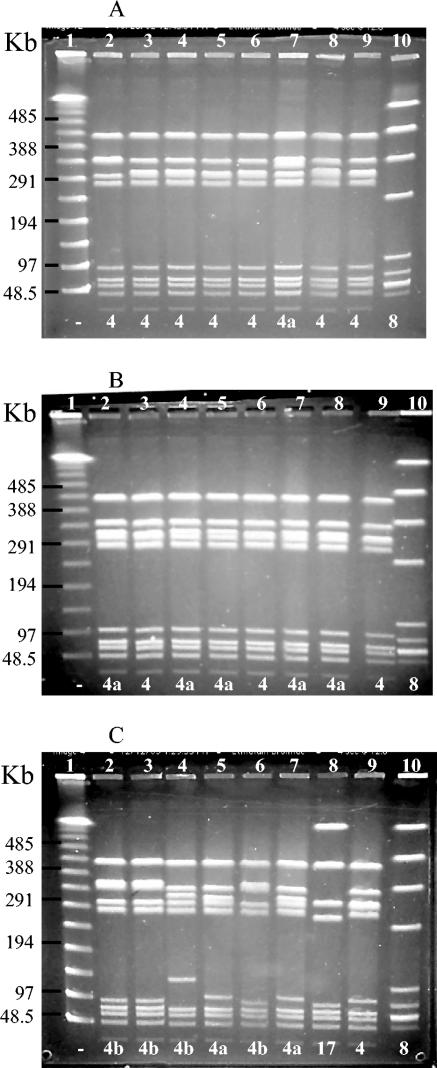
Three representative gels are shown in Fig. 2, with NT and V isolates compared to the profile group 4 prototype in lane 9 of each panel. Panel A shows the high degree of homogeneity found in NT/R1,R4 isolates, with six of the seven isolates displayed in this gel classified into profile group 4 and only one (lane 7) classified into subgroup 4a. In contrast, the V/R1,R4 isolates in panel B show some genetic divergence from the classic V/R1,R4 profile group 4 prototype (lane 9), since more than half of the isolates in this gel were classified into subgroup 4a. Likewise, the PFGE profiles of the isolates in panel C were representative of recent V/R1,R4 isolates and all were profiled as subgroup 4a or 4b. The markedly different restriction pattern in lane 8 of panel C was that of a type VII/R1,R4 isolate that was classified into profile group 17.
FIG. 2.
SmaI macrorestriction analysis by PFGE of NT/R1,R4 and V/R1,R4 GBS isolates. Numbers 1 to 10 at the top of each gel are lane designations; the numbers at the bottom of the gel represent PFGE profile groups (−, NT). (A) Lanes: 1, lambda molecular size standard with sizes in kilobases on the left; 2 to 8, NT/R1,R4 isolates; 9, V/R1R4 profile 4 control; 10, internal control Ib/α+β isolate. (B) Lanes: 1, lambda molecular size standard; 2 to 8, V/R1,R4 isolates; 9, profile 4 control; 10, internal control. (C) Lanes: 1, lambda molecular size standard; 2 to 7, V/R1,R4 isolates; 8, VII/R1,R4; 9, profile 4 control; 10, internal control.
The distribution of the NT and serotype V isolates within PFGE profile group 4 or its subgroups is summarized in Table 2. Whereas 22 (56.4%) of the 39 NT isolates had profiles that were identical to that of the profile 4 prototype, only 8 (17.8%) of the 45 serotype V isolates were classified into profile 4. When the isolates were analyzed by their protein profiles, it was found that 18 (75.0%) of the 24 NT/R1,R4 isolates were identical to the prototype isolate of profile group 4, as shown in Fig. 2A. However, in contrast to the NT/R1,R4 isolates, the NT/none group was heterogeneous, with only 4 (28.6%) isolates classified into profile 4 and the remaining 10 of the 14 isolates classified into subgroups 4a to 4d. Similar analysis of the typeable isolates showed that the majority (88.9%) of the V/R1,R4 isolates were in profile subgroup 4a (18 isolates) or 4b (6 isolates). The same trend was observed with V/R4 and V/none isolates, with the majority in subgroup 4a rather than group 4.
DISCUSSION
This work not only verified the value of grouping isolates according to expressed surface proteins in studying NT isolates but also reaffirmed the validity of classifying NT GBS isolates according to DNA macrorestriction PFGE profiles. Furthermore, since we analyzed all of the isolates from a 5-year period, the results of this epidemiological study will assist researchers in monitoring GBS trends with regard to the serotyping of NT isolates and monitoring divergence among type V GBS isolates.
The importance of continuous monitoring of NT GBS isolates by PFGE was emphasized by our discovery of PFGE band patterns not seen previously in our laboratory. In 2002, we reported that 135 isolates were classified into 26 different PFGE profile groups (4); currently, our laboratory has identified 35 profiles. These additional, unique band profile groups will permit the classification of more isolates for future reference.
The distribution of NT isolates expressing the same protein profile among PFGE profile groups was consistent with trends that we have observed previously with typeable GBS isolates. We had reported that isolates of the historical serotypes (Ia, Ib, II, and III) were distributed among a number of PFGE band patterns (8), whereas most isolates of the more recently emerging serotypes were in a very limited number of profile groups, as exemplified by our finding that all of the type V isolates studied were classified into one PFGE profile group (4). In addition, we found that certain protein profiles were highly associated with specific CPS types, such as α with type Ia, α+β with type Ib, R4 with type III, and R1,R4 with type V (10). We observed similar trends in our NT isolates. Specifically, NT isolates with an α protein profile such as α only that would be associated with serotype Ia were distributed among more profile groups than the NT/R1,R4 isolates, which, like the V/R1,R4 isolates, were all in the same profile group, reflecting perhaps less genetic diversity (7).
It was of interest that NT isolates classified into PFGE profile group 4 or its subgroups, and therefore considered to be highly related to V/R1,R4 isolates, accounted for 48.3%, nearly half, of the NT isolates studied. This reflected a slight increase since 2002, when we found that of 78 NT isolates, 30 (38.5%) were in this profile group (4). We do not know why profile group 4 constituted such a significant percentage of the NT isolates. One can speculate that it may reflect their advantage in colonization or perhaps that poorly encapsulated or nonencapsulated variants or variants related to type V were more likely to be produced than by isolates of other serotypes. PFGE profile group 4, described here, is identical to PFGE subtype O, described by Elliott et al. (7), which made up the majority of isolates of GBS type V from 1986 to 1996.
Among typeable GBS isolates, whether invasive or colonizing, CPS type V has increased in prominence over the past 15 years. In the early 1990s, the percentage of type V isolates causing invasive disease in all patient groups increased from approximately 3% to about 20% (5, 7) and to 29% for nonpregnant adults (12). We reported that type V was responsible for 14% of invasive infections in neonates and for 22.6% of those in pregnant women (19) and that this serotype accounted for approximately 12% of the colonizing GBS isolates from neonates or parturient women at the time of delivery (13). However, the percentage may be even higher since in a study of vaginal and rectal colonization in 102 nonpregnant women, we found that if multiple colonies were picked from the initial culture plate for serotyping, the relative percentage of type V isolates was 14.3%, versus 13.6% when only one colony was processed, as is customary in most laboratories (10, 11). Furthermore, our recent data from a much larger number of women with paired vaginal and rectal cultures taken at up to four culture visits, indicated that 18.4% of the women were colonized with type V, underscoring the importance of this CPS type (unpublished observations).
Analyses of the V/R1,R4 isolates suggested a genotypic change since the majority of recent isolates were classified as having profile 4a, unlike our past isolates that had a PFGE band pattern indistinguishable from that of the prototype isolate for profile group 4. Genetic diversity among type V isolates has also been observed by Thomas-Bories et al. (17), as they found 11 distinct patterns in 64 isolates. In contrast to our V/R1,R4 isolates, the NT/R1,R4 isolates appeared to have retained genetic homogeneity, with three-quarters still classified within PFGE profile 4. This change in the distribution of NT and type V isolates among subgroups of profile 4 was of interest, since our previous publication demonstrated that most of the NT and type V isolates producing proteins R1 and R4 or no proteins were in profile group 4 (4).
Because of recent interest in vaccine development for prevention of GBS disease, precise identification of GBS isolates is of importance. Consequently, DNA dot blot hybridization and PCR methods have been developed as an alternative to conventional serotyping and have proven to be highly sensitive techniques (6, 15). However, PFGE is an excellent adjunct to these methods since the analysis of DNA band patterns enables one to compare isolates and assess the degree of relatedness among them, providing an overall view of the extent of homogeneity or clonal divergence within groups of isolates over time.
Acknowledgments
This study was supported in part by contract N01-AI-75326 from the National Institutes of Health, Bethesda, Md.
REFERENCES
- 1.Baker, C. J., and D. L. Kasper. 1976. Microcapsule of type III strains of group B Streptococcus: production and morphology. Infect. Immun. 13:189-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, J. A., and P. Ferrieri. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, J. A., A. E. Flores, C. J. Baker, S. L. Hillier, and P. Ferrieri. 2002. Improved methods for typing nontypeable isolates of group B streptococci. Int. J. Med. Microbiol. 292:37-42. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 6.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2004. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasola, E., C. Livdahl, and P. Ferrieri. 1993. Molecular analysis of multiple isolates of the major serotypes of group B streptococci. J. Clin. Microbiol. 31:2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrieri, P., D. S. Cho, C. Livdahl, C. E. Rubens, and A. E. Flores. 1997. DNA restriction profiles of nontypeable group B streptococcal clinical isolates. In T. Horaud, A. Bouvet, R. Leclercq, H. de Montclos, and M. Sicard (ed.), Streptococci and the host. Proceedings of the XIIIth Lancefield International Symposium on Streptococci and Streptococcal Diseases (Paris). Adv. Exp. Med. Biol. 418:343-346. [DOI] [PubMed] [Google Scholar]
- 10.Ferrieri, P., and A. E. Flores. 1997. Surface protein expression in group B streptococcal invasive isolates. In T. Horaud, A. Bouvet, R. Leclercq, H. de Montclos, and M. Sicard (ed.), Streptococci and the host. Proceedings of the XIIIth Lancefield International Symposium on Streptococci and Streptococcal Diseases (Paris). Adv. Exp. Med. Biol. 418:635-637. [DOI] [PubMed] [Google Scholar]
- 11.Ferrieri, P., S. L. Hillier, M. A. Krohn, D. Moore, L. C. Paoletti, and A. E. Flores. 2004. Characterization of vaginal and rectal colonization with multiple serotypes of group B streptococci using multiple colony picks. Indian J. Med. Res. 119(Suppl.):208-212. [PubMed] [Google Scholar]
- 12.Harrison, L. H., J. A. Elliot, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, and the Maryland Emerging Infections Program. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 13.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-208. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas-Bories, I. L., F. Fitoussi, P. Mariani-Kurkdjian, J. Raymond, N. Brahimi, P. Bidet, V. Lefranc, and E. Bingen. 2001. Clonal relationship between U.S. and French serotype V group B streptococcus isolates. J. Clin. Microbiol. 39:4526-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson, H. W. 1972. Comparison of streptococcal R antigens. Appl. Microbiol. 24:669-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M.-L. T. Lee, A. E. Flores, P. Ferrieri, and C. J. Baker. 2000. Invasive disease due to group B streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]