Abstract
The genome of the fungal plant pathogen Fusarium graminearum harbors six catalases, one of which has the sequence characteristics of a fatty acid peroxide-metabolizing catalase. We cloned and expressed this hemoprotein (designated as Fg-cat) along with its immediate neighbor, a 13S-lipoxygenase (cf. Brodhun et al, PloS One, e64919, 2013) that we considered might supply a fatty acid hydroperoxide substrate. Indeed, Fg-cat reacts abruptly with the 13S-hydroperoxide of linoleic acid (13S-HPODE) with an initial rate of 700–1300 s−1. By comparison there was no reaction with 9R- or 9S-HPODEs and extremely weak reaction with 13R-HPODE (~0.5% of the rate with 13S-HPODE). Although we considered Fg-cat as a candidate for the allene oxide synthase of the jasmonate pathway in fungi, the main product formed from 13S-HPODE was identified by UV, MS, and NMR as 9-oxo-10E-12,13-cis-epoxy-octadecenoic acid (with no traces of AOS activity). The corresponding analog is formed from the 13S-hydroperoxide of α-linolenic acid along with novel diepoxy-ketones and two C13 aldehyde derivatives, the reaction mechanisms of which are proposed. In a peroxidase assay monitoring the oxidation of ABTS, Fg-cat exhibited robust activity (kcat 550 s−1) using the 13S-hydroperoxy-C18 fatty acids as the oxidizing co-substrate. There was no detectable peroxidase activity using the corresponding 9S-hydroperoxides, nor with t-butyl hydroperoxide, and very weak activity with H2O2 or cumene hydroperoxide at micromolar concentrations of Fg-cat. Fg-cat and the associated lipoxygenase gene are present together in fungal genera Fusarium, Metarhizium and Fonsecaea and appear to constitute a partnership for oxidations in fungal metabolism or defense.
Keywords: fusarium, catalase, lipoxygenase, peroxidase, HPODE, ABTS assay, NMR
1. Introduction
Although by far catalases are best known for their role in the metabolism of hydrogen peroxide, in recent years multiple examples have emerged of catalase-related hemoproteins that utilize fatty acid hydroperoxides as their natural substrate. The enzymes of this type described to date are involved in pathways of oxylipin biosynthesis. Examples are the fatty acid allene oxide synthases of corals and cyanobacteria [1–3], the fatty acid hydroperoxide lyase discovered recently in coral [4], and a cyanobacterial enzyme that forms fatty acid diols and a fatty acid containing a bicyclobutane ring [5, 6]. These enzymes all exist as the N-terminal domain of a natural fusion protein that has the lipoxygenase that forms their fatty acid hydroperoxide substrate on the C-terminus. The X-ray crystal structure of the catalase-related allene oxide synthase (cAOS) domain of the coral Plexaura homomalla confirms the close structural similarity to true catalases [7]. Indeed, each of these enzymes feature the most conserved elements of the heme environment of catalases, including the Tyr proximal heme ligand in the sequence context of R(x)3Y(x)6R and on the distal heme the His residue essential for catalysis [8] and the distal heme Asn that facilitates efficient enzymatic activity [9]. One key difference from the conserved sequence of true catalases is what at first appears an apparently nondescript change in the residue immediately preceding the distal heme catalytic His: this is conserved as Val in true catalases and is changed to Thr in all the catalase-related hemoproteins that metabolize fatty acid hydroperoxides [8]. It turns out that the change from Val to Thr is important in preventing any reaction with hydrogen peroxide [10], and indeed can be considered a hallmark of the catalase-related enzymes that react with fatty acid hydroperoxides [8].
The focus of the present study is a catalase-related enzyme from the fungal plant pathogen Fusarium graminearum (Gibberella zeae). F. graminearum causes head blight (scab) of wheat, barley, corn and rice and is listed among the Top 10 fungal pathogens as threats to the world’s food supply [11]. Not only does infestation cause huge losses in grain production, the scabby grain is contaminated with trichothecene and estrogenic mycotoxins, making it unsuitable for food or feed [12, 13]. The F. graminearum gene of interest, designated here as Fg-cat, has the characteristics of a fatty acid hydroperoxide-metabolizing enzyme with the distal heme signature sequence of Thr-His [8]. One of the immediate neighboring genes was annotated as a lipoxygenase (that we designate here as Fg-LOX), lending credence to the possibility that the two might cooperate in metabolism. We were cognizant of the fact that fungi form the hormone jasmonic acid from α-linolenic acid via an as-yet uncharacterized pathway [14], and initially we considered that Fg-cat was a candidate for the role of the “missing” AOS activity.
We decided to clone and express Fg-cat as well as the adjacent Fg-LOX gene and characterize their enzymatic activities. After this work was begun the cloning and characterization was reported of a homologous LOX gene from Fusarium oxysporum; it was characterized as a 13S-LOX [15], which agrees with our findings summarized briefly herein on the Fg-LOX. Linoleic is the major polyunsaturated fatty acid in Fusarium (30–60% of the fatty acid composition) with about one-tenth the amount of α-linolenic acid [15, 16], and undoubtedly these are the natural substrates for Fg-LOX, and as we report here, in turn for Fg-cat.
2. Materials and methods
2.1. Materials
Linoleic and α-linolenic acids were purchased from Nu-Chek Prep Inc. (Elysian, MN). Linoleate hydroperoxides were prepared by vitamin E-controlled autoxidation [17] followed by separation of the 9RS- and 13RS-hydroperoxides by SP-HPLC and chiral column resolution of the enantiomers [18] or using soybean lipoxygenase for preparation of the 13S-hydroperoxides [19], Anabaena 9R-LOX for preparation of 9R-hydroperoxides [20], and potato 9S-LOX for preparation of 9S-hydroperoxides [21]. [1-14C]Linoleic acid and [1-14C]arachidonic acid were from NEN (Dupont). [18O2]13S-HPODE was prepared by incubation of linoleic acid with soybean lipoxygenase under an atmosphere of 18O2 (Sigma-Aldrich, formerly Isotec). 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)diammonium salt (ABTS) was purchased from Sigma.
2.2. Cloning of Fg-cat and Fg-LOX
Genomic DNA for F. graminearum was kindly provided by Dr. Frances Trail of Michigan State University. Fg-cat, the Fusarium graminearum catalase-related gene (FGSG_02217), was cloned initially using two sets of primers, the first covering exon 1 with overlap of the downstream primer into the first 23 nucleotides of exon 2 (skipping the intron sequence), the other set covering exon 2 with overlap of the upstream primer to include 22 nucleotides of exon 1 (again by-passing the intron sequence). NdeI and EcoRI restriction sites were included at the N- and C-terminus respectively (in bold). Set 1: 5′ CAT ATG TCC ATC CAA GTT GTT CTT CAA AAC 3′ (upstream), and 5′ ATC TTC GGA AAA CAT CCT GGC TAC CGA ACA A INTRON CC CAT GCC AAA GGT ATT CTT GTG 3′ (downstream). Set 2: 5′ AAA CAT CCT GGC TAC CGA ACA A INTRON CC CAT GCC AAA GGT ATT CTT GTG GAA GGA 3′, and 5′ CGA CGA CAT GAA ACA CAG TCA GAG CAT CAC CAT CAC CAT CAC TAG GAA TTC 3′ (downstream). The initial three cycles used a lower annealing temperature (only the first half of the downstream primer could anneal) and then the temperature was raised for copying of the full-length primer. PCR was performed using 50 ng of genomic DNA and Easy A polymerase using the following PCR Protocol: 95°C for 2 min then 3 cycles of 55°C for 30 s, 72°C for 3 min, 95°C for 40 s, then 27 cycles of 60°C for 30 s, 72°C for 3 min, 95°C for 40 s, 72°C for 7 min, then hold at 4°C. The PCR products were extracted from a 1.2% Agarose/TAE gel and DNA recovered using Macherey-Nagel NucleoSpin Kit. Second round PCR was performed using a 1/50th dilution of the PCR products from sets 1 and 2. Easy A polymerase was used and the following PCR Protocol: 95°C 2 min then 3 cycles of 55°C 30 s, 72°C 2 min, 95°C 40s. This allowed annealing of the initial two PCR products and completion of the full-length cDNA. At this point 1 μl of 125 ng/μl of the upstream and downstream primers were added and the following PCR protocol run: 95°C 2 min, then 3 cycles of 55°C 30 s, 72°C 2 min, 95°C 40s, followed by 27 cycles of 60°C 30 s, 72°C 2 min, 95°C 40 s, 72°C 7 min, then hold at 4°C. The PCR product was extracted as above and ligated into pCR 2.1 vector using the Invitrogen protocol.
The Fusarium graminearum lipoxygenase (FGSG_02216) was cloned using the same strategy of individual primer sets for exon 1 and exon 2 followed by annealing and polymerization of the two sequences. Set 1: 5′ CAT ATG CAT CAC CAT CAC CAT CAC TCT GCA GTT GCC AGT CAC GTT GTC 3′ (upstream), and 5′ TTG ACT GAG ATC TAC AGC ATT CTG GAA GAT GC INTRON T GCT GTA TCC CAT TTT GAT AAG 3′; Set 2: 5′ ATC TAC AGC ATT CTG GAA GAT GC INTRON T GCT GTA TCC CAT TTT GAT AAG CGC GGC TAT 3′ and 5′ CC ACT GCC ATT TCA ATT TTA ATC TAG GAA TTC 3′ (downstream). PCR conditions were the same as for the Fusarium mini-catalase, including second round using 1/50th dilution of first round PCR products as template for the full length N- and C-terminal primers.
2.3. Expression and catalytic activity of Fg-LOX
Fg-LOX in the mammalian expression vector pCR 2.1 (Invitrogen) was expressed by transient transfection in HeLa cells (1 × 106 cells/35 mm well) using VTF-7, a recombinant vaccinia virus containing the T7 RNA polymerase gene [22, 23]. After 12 h the cell pellet from one well was resuspended in 150 μl Tris buffer pH 7.5 containing 150 mM NaCl, sonicated twice for 5 s, then 50 μl aliquots were transferred to separate Eppendorf tubes, and each diluted with 100 μl buffer. One tube was incubated with 20 μM [1-14C]linoleic acid (~ 330,000 DPM) and the second with 20 μM [1-14C]arachidonic acid for 45 min at 37 °C. The products were extracted using the Bligh and Dyer method [24] and one quarter analyzed by RP-HPLC using a solvent of MeOH/water/glacial acetic acid (90:10:0.01 by vol.), in each case indicating ~20% conversion of radiolabel to a mono-hydro(peroxy) product. The remaining three-quarters of each sample was reduced with triphenylphosphine and purified by RP-HPLC (MeOH/water/glacial acetic acid 80:20:0.01 by vol.), then SP-HPLC of the methyl ester derivative (hexane/isopropanol 100:2 by volume) in comparison to unlabeled authentic standards (9 and 13-HODEs, and 5, 8, 9, 11, 12, and 15-HETEs). The collected main radiolabeled product in each case was mixed with unlabeled racemic 15-HETE methyl ester or racemic 13-HODE methyl ester (to allow monitoring of the retention time of the R and S enantiomers by UV-diode array detection), and analyzed by chiral column HPLC using a Chiralpak AD column with a solvent of hexane/MeOH (100:2 by vol. for 15-HETE, 100:5 for 13-HODE [18]).
2.4. Expression of Fg-LOX and Fg-cat in E. coli
The His-tagged cDNAs in pET17b were expressed in E. coli BL21(DE3) cells in TB medium using a published protocol [25, 26]. Briefly, in a typical expression the transformed bacteria were plated overnight and a single colony inoculated into 1 ml LB medium containing 100 μg/ml ampicillin and grown at 37 °C. After 4 hours an aliquot of 200 μl was transferred into 10 ml TB medium containing 400 μg/ml ampicillin and grown at 37°C for 3 hours then added to 40 ml TB containing 100 μg/ml ampicillin in a 500 ml non-fluted flask and grown at 28°C, 250 RPM, for 21 hours. The harvested bacterial pellet was sonicated in Bugbuster HT (Novagen) containing 1 mM PMSF and 1 μl/5ml of benzonase and after centrifugation at 15,000 g for 15 min the Fg-LOX or Fg-cat were isolated via the His-tag by nickel affinity chromatography.
2.5. Incubations of Fg-cat with fatty acid hydroperoxides
Initially, Fg-cat (2 - 10 nM) was incubated in 0.5 mL 50 mM Tris pH 7.5 + 150 mM NaCl at room temperature with individual fatty acid substrates, 9R-, 9S-, 13R-, 13S-HPODE, 9S- and 13S-HPOTE (60 μM). Disappearance of the conjugated diene chromophore was monitored at 235 nm using a Lambda-35 UV-Vis spectrometer (PerkinElmer); to compensate for the absorbance of products at 235 nm (which slows the observed decrease), the rates of reaction are increased by a factor of 1.3 to correct to the loss of conjugated diene. To identify the profile of products and determine the yield, an incubation was conducted using [1-14C]13S-HPODE as substrate. To determine the fate of the 13-hydroperoxyl oxygens an incubation was conducted with [18O2]13S-HPODE as substrate with analysis of the profile of products by LC-MS. To prepare metabolites for structural analysis, subsequent reactions up to 10 ml in scale were conducted using 100 μM 13S-hydroperoxide incubated with Fg-cat for 2 min at room temperature.
For comparison with the activity of peroxidases that generate hypochlorous acid [27], 5 nM Fg-cat was incubated with 100 μM 13S-HPODE in the presence of 5 mM taurine in 50 mM Tris pH 7.5 + 150 mM NaCl buffer. Positive controls used 16 nM myeloperoxidase with 50 μM H2O2 as the oxidizing co-substrate, and also parallel incubations were conducted with “bleach” (a mixture of HOCl and HOBr solution) [27]. After 5 min, a quarter volume of developing buffer was added (0.4 M sodium acetate, pH 5.4, 2 mM tetramethylbenzidine, 10% dimethylformamide; or 100 μM sodium iodide for HOBr and HOCl detection), and the absorbance measured at 650 nm (blue color).
2.6. HPLC analysis
Products were acidified to pH 4.0 by addition of 1 M KH2PO4 and 1 N HCl and applied to a pre-conditioned 1-cc HLB Oasis cartridge (3-cc for larger incubations), the cartidge washed with water and the products eluted with MeOH. Initial analysis by RP-HPLC used a Waters Symmetry C18 5μ column (0.46 × 25 cm) with an isocratic solvent of acetonitrile/water/glacial acetic acid (60:40:0.01, by volume) and a flow rate at 1 mL/min. The UV signals at 205, 220, 235, 270 nm were recorded on an Agilent 1200 Series diode array detector. Further purification of individual peaks for NMR analysis was accomplished by SP-HPLC using a Beckman or Thomson 5μ silica column (25 × 0.46 cm) and solvent systems of hexane and 0.5% – 3% isopropanol and 0.1% glacial acetic acid.
2.7. Reaction of Fg-cat and 13S-HPOTE under anaerobic conditions
Fg-cat (5 nM) and 100 μM 13S-HPOTE substrate were incubated at room temperature in 0.5 mL oxygen-free 50 mM Tris pH 7.5 + 150 mM NaCl buffer (degassed with argon). The products were rapidly acidified with 1 M KH2PO4 and 1 N HCl down to pH 3.5–4.0, and extracted using a 1-cc HLB Oasis cartridge as described previously. Products were chromatographed on RP-HPLC (acetonitrile/water/glacial acetic acid, 60:40:0.01, by volume) by Waters Symmetry C18 column 5-μm column (0.46 × 25 cm) at a flow rate of 1 mL/min. About 0.5 mg of product formed in a scaled-up incubation were purified on RP-HPLC for 1H-NMR analysis.
2.8. Reaction of Fg-cat and 13S-HPOTE in the presence of ABTS
Peroxidase activity of Fg-cat was assayed as an increase in absorbance at 417 nm (green color) using a Lambda-35 UV-Vis spectrophotometer (Perkin-Elmer) of a Cary 60 UV-Vis spectrophotometer (Agilent) [28–30]. Reactions were conducted with 3 nM Fg-cat using 100 μM 9S- or 13S-HPOTE, 1 mM H2O2, 1 mM cumene hydroperoxide, and 1 mM t-butyl hydroperoxide substrates and ABTS (1 mM). Parallel incubations were conducted without the enzyme or substrate. The initial rate was expressed as nanomoles of ABTS oxidized per nanomole of Fg-cat per second. The reaction products from 13S-HPOTE were extracted with 2 volumes of dichloromethane in the absence of acid. The organic layer was transferred into a new vial, taken to dryness and redissolved in a RP-HPLC column solvent. Products were chromatographed initially on RP-HPLC as described above and subsequently 0.5–1 mg of products from scaled-up incubations were isolated, individually purified on SP-HPLC (hexane/ispropyl alcohol/glacial acetic acid; 100:3:0.1, by volume) using (0.46 × 25 cm) at a flow rate of 1 mL/min.
2.9. LC-MS and NMR analysis
Products formed from 13S-HPODE or 13S-HPOTE by Fg-cat were run on LC-MS using a Kinetex C18 2.6-μm column (3 × 100 mm) with acetonitrile/water/glacial acetic acid (60:40:0.01, by volume) column solvent at 0.4 mL/min, and molecular weights were established from the M-H anions measured by negative ion electrospray LTQ1 ion trap LC-MS instrument.
1H-NMR and 1H,1H COSY NMR spectra were obtained on a Bruker 600 MHz spectrometer at 298 K. The parts/million values are reported relative to residual nondeuterated benzene (C6D6; δ = 7.15 ppm) and methanol (MeOD; δ = 3.31 and 4.78 ppm). All spectra were analyzed on Bruker TopSpin 3.0 software.
3. Results
3.1. Identification of Fg-cat and Fg-LOX
Fg-cat is annotated on the Superfamily website (http://supfam.org/SUPERFAMILY/) as one of six catalase-related genes in F. graminearum, although the only one with the tell-tale sequence of Thr next to the distal heme His residue (Table 1), potentially signifying reaction with fatty acid hydroperoxides [8]. In addition to the Thr-His sequence, Fg-cat also retains the conserved catalase-related residues around the heme (notably the Tyr proximal heme ligand in the sequence context of R(X)3Y(X)6R), and like the other cAOS-related proteins it is smaller in size than classic catalases (cf. ref [31]), Table 1. Unlike other cAOS-type enzymes, Fg-cat is present in the genome as a stand-alone open-reading frame. Nonetheless the immediate neighboring genes were annotated as a putative esterase and a putative lipoxygenase (LOX), Fig. 1.
Table 1.
Catalase genes in Fusarium graminearum
| F. graminearum Gene designation | Number of amino acids in monomer | Sequence around the distal heme Histidine | Name or annotation |
|---|---|---|---|
| FGSG_02217 | 378 | RTTHAK | “Fg-cat” |
| FGSG_02881 | 533 | RSVHAK | Catalase |
| FGSG_05695 | 550 | RVVHAK | Catalase |
| FGSG_06554 | 738 | RVVHAR | Catalase-peroxidase |
| FGSG_06596 | 584 | RVVHSC | Catalase |
| FGSG_06733 | 716 | RAVHAR | Catalase-peroxidase |
Fig. 1. Region of the Fusarium graminearum gene map with Fg-cat and Fg-LOX.
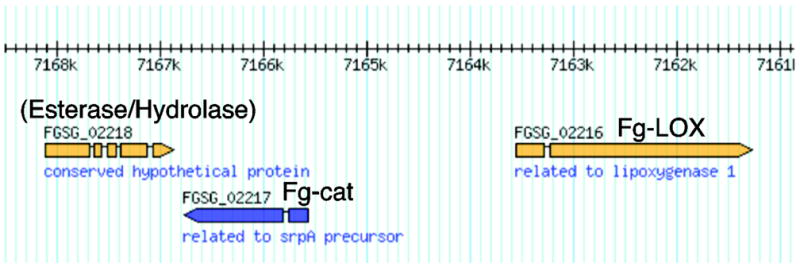
Fg-cat (FGSG_02217) is labeled “related to srpA precursor”, the gene of a 36 kD catalase in Synechococcus [49]. FGSG_02216 is the single LOX gene in F. graminearum, and BLAST searches indicate FGSG_02218 is a putative esterase/hydrolase.
3.2. Cloning and expression of Fg-LOX and Fg-cat
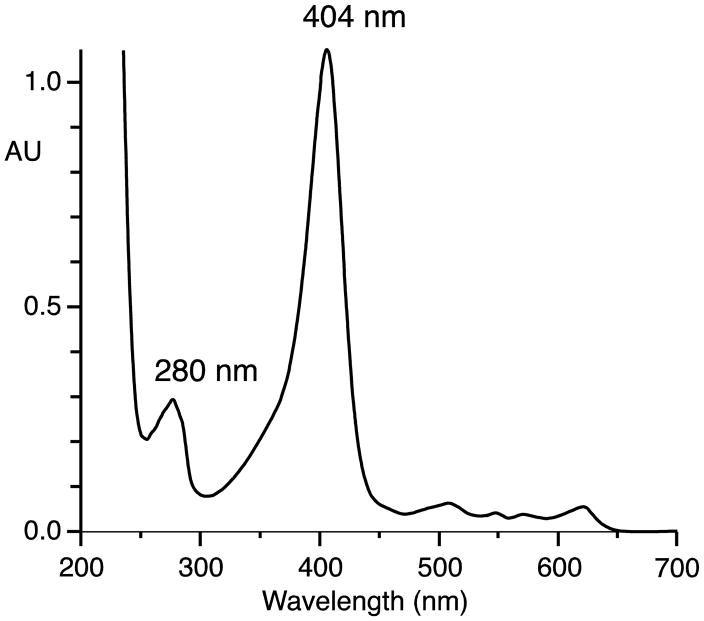
The single intron in each gene was removed by PCR of the two exons and the exons recombined to give cDNAs of the open reading frames, and after transfer to pET plasmids the proteins were expressed in E. coli. Characterization of the activity of the LOX domain by HPLC and chiral HPLC versus authentic standards proved it to convert linoleic acid almost exclusively to the 13S-hydroperoxide (13S-HPODE), and arachidonic acid to 15S-HPETE, (Supplement, Fig. S1, S2), in agreement with the reported activity of the F. oxysporum homologue [15]. The Fg-cat expresses particularly well in E. coli, turning the bacterial protein pellets to an obvious green color. The UV-Vis spectrum of the purified protein exhibits a strong Soret band at 404 nm with a relatively weak signal for the protein at 280 nm, as predicted from the low content of aromatic amino acids (Figure 2). Fg-cat was quantified by UV-Vis spectroscopy assuming a molar extinction coefficient of 100,000 M−1 cm−1 at 404 nm.
Fig. 2.
UV-Vis spectrum of Fg-cat
3.3. Specificity in metabolism of fatty acid hydroperoxides
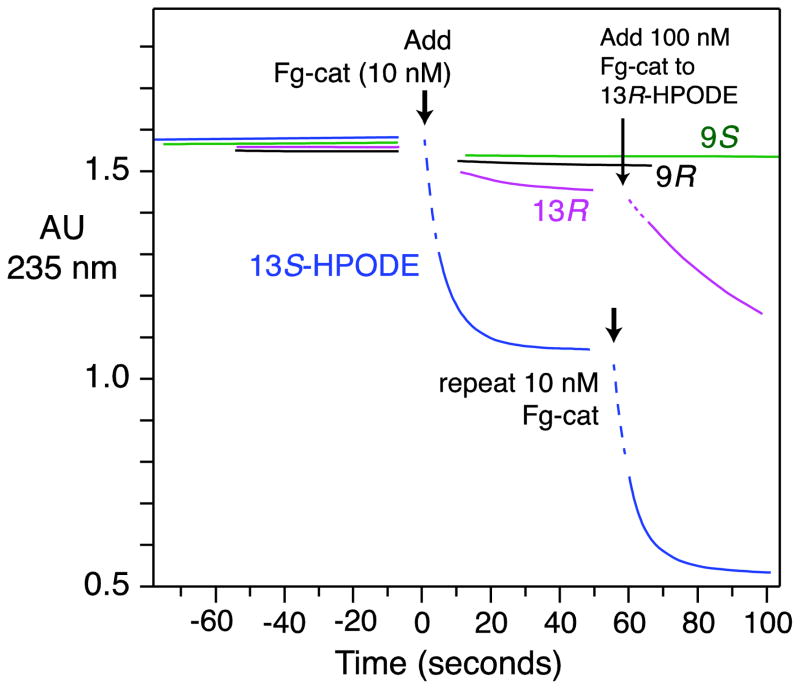
Assays of transformation of fatty acid hydroperoxides by Fg-cat were performed by observing changes in the UV absorbance of the conjugated diene chromophore of HPODE isomers recorded over time at 235 nm. These experiments indicated the selective and rapid reaction with 13S-HPODE with an initial rate in the order of 700–1300 turnovers s−1 (Figure 3). By comparison there was only extremely weak reaction (~0.5% of the rate) with 13R-HPODE (which is not a natural product), and no reaction with 9R- and 9S-HPODEs at the Fg-cat concentrations employed (Figure 3). Fg-cat is inactivated in the course of the transformation of 13S-HPODE and reaction is reinitiated by further addition of enzyme (Figure 3). The selective metabolism of 13S-HPODE matches with the specificity of the adjacent Fg-LOX gene in forming 13S-HPODE as product from linoleic acid and likely has functional relevance.
Fig. 3. UV assay of the metabolism of HPODE isomers by Fg-cat.
UV absorbance at 235 nm was monitored over time as equivalent concentrations of 9R-, 9S-, 13R- and 13S-HPODE (~ 70 μM) were reacted with Fg-cat (10 nM). The assays were conducted at room temperature in 25 mM phosphate pH 7.5 containing 150 mM NaCl. The dotted lines were not recorded and are added to help visualize the connection to the starting absorbance of 13S-HPODE. The results are representative of two separate experiments comparing the four HPODE isomers and over ten experiments with 13S-HPODE.
3.4. Metabolism of 13S-HPODE by Fg-cat: analysis of the products
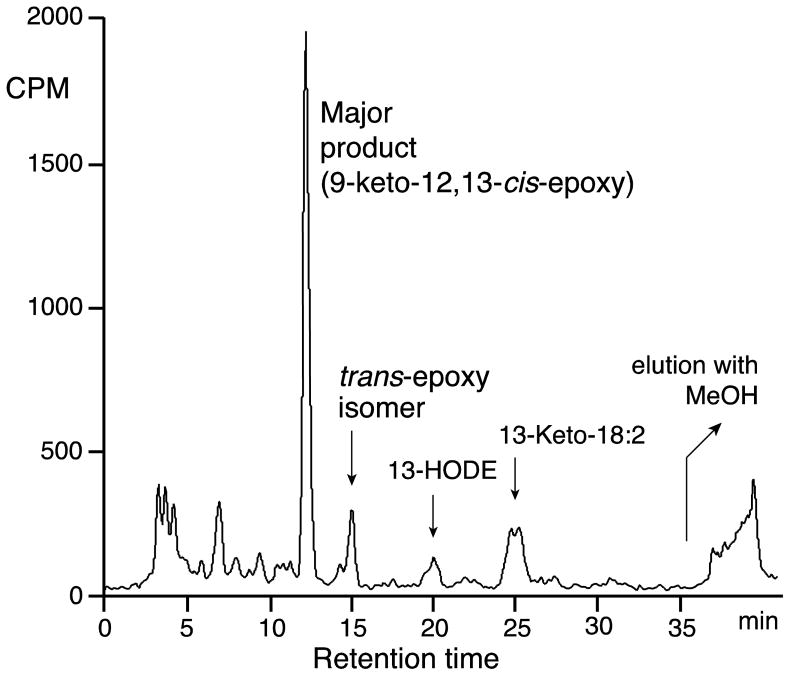
Using [1-14C]13S-HPODE as substrate for Fg-cat, the product profiles as monitored by RP-HPLC with on-line radioactive and UV detection showed formation of many minor peaks along with one major product that comprised about 60% of the recovered material (Figure 4). After scaling up the incubations the main peak was purified and analyzed by HPLC, UV, LC-MS and 1H-NMR. The UV spectrum exhibited a smooth chromophore typical of a conjugated enone with lambda-max at 235 nm in the 60:40 CH3CN/water proportions of the RP-HPLC solvent. LC-MS (negative ion) gave the major M-1 ion at m/z 309, establishing a molecular weight of 310. 1H-NMR with COSY analysis (Supplement, Table S1) indicated a covalent structure of 9-oxo-10E-12,13-epoxy with the designation as a 12,13-cis-epoxide made from the chemical shifts of the epoxide protons (H12 at 3.49 ppm and H13 at 3.18 ppm) and the J12,13 coupling constant (~5 Hz), a value typical for cis epoxides and clearly distinguished from the ~2 Hz value in trans epoxides (ref [32] and many subsequent precedents). In linoleic acid products, NMR cannot readily distinguish between 9-oxo-10E-12,13-epoxy and the reverse orientation, 9,10-epoxy-11E-13-oxo. The former was established by formation of the methoxime derivative followed by acid hydrolysis of the epoxide moiety and GC-MS analysis of the methyl ester methoxime trimethylsilyl ether derivative: the electron impact mass spectrum gave m/z 173 as the base peak (indicating a 13-hydroxyl, CH(OTMS)C5H11), and establishing the parent epoxide as 12,13-epoxy. Accordingly, the structure of the main product from 13S-HPODE was determined as 9-oxo-12,13-cis-epoxy-octadec-10E-enoic acid. One of the minor peaks, eluting just after the cis epoxide on RP-HPLC and in ~10-fold smaller amounts (Figure 4), was identified as the analogous 9-oxo-10E-12,13-trans-epoxide (Supplement, Table S2).
Fig. 4. RP-HPLC analysis of [1-14C]13S-HPODE metabolism by Fg-cat.
The analysis used a Waters C18 Symmetry column (25 × 0.46 cm), with a solvent of acetonitrile/water/glacial acetic acid (60:40:0.01 by vol.) and a flow rate of 1 ml/min. In addition to the main epoxy-ketone, the trans-epoxy-ketone, 13-HODE and 13-keto-18:2 were identifed as minor peaks on the chromatogram.
3.5. Mechanism of formation of the 9-oxo-12,13-epoxide
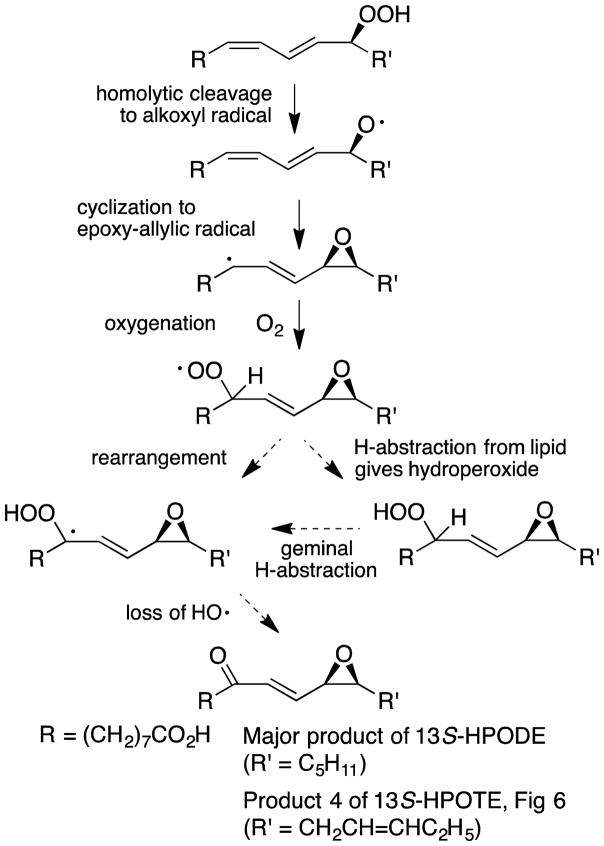
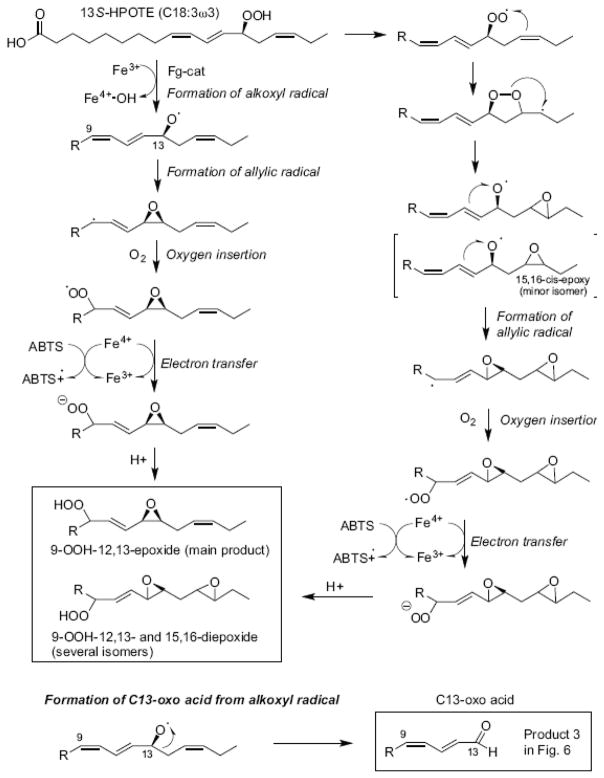
[18O2]13S-HPODE with labels in the hydroperoxide group was prepared by incubation of linoleic acid with soybean LOX-1 under an atmosphere of 18O2. After incubation with Fg-cat, LC-MS analysis of the resulting products indicated that the major 9-oxo-12,13-cis-epoxide retained only one of the two hydroperoxide oxygens (data not shown). This fits with a scheme whereby the 13S-hydroperoxide is homolytically cleaved to an alkoxyl radical which mainly reacts to form a 12,13-epoxy allylic radical which itself then reacts with a new molecule of molecular oxygen that eventually becomes the 9-oxo moiety (Scheme 1). Two possibilities are illustrated for conversion of the oxygenated intermediate to the epoxy-ketone: either a rearrangement of the epoxy-hydroperoxy radical followed by loss of a hydroxy radical (left hand pathway) or an intermediate formation of the epoxy-hydroperoxide (right-hand pathway). The latter is a covalently complete molecule and was not detected in these incubations; it was, however, isolated as a main product from incubations in the presence of ABTS (see below), which provides a means to trap the hydroperoxy radical as the hydroperoxide.
Scheme 1.
Transformation of 13S-HPODE to the epoxy-ketone
3.6. Metabolism of 13S-HPOTE by Fg-cat
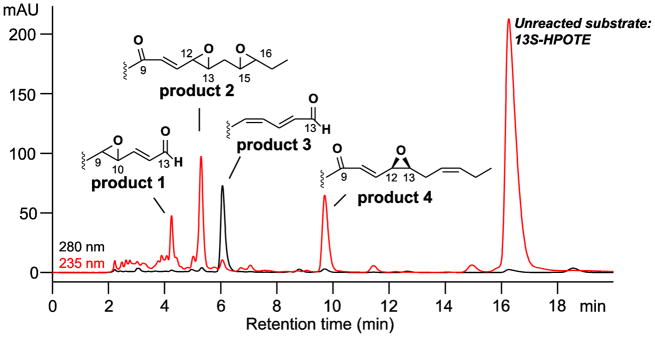
When the 13S-hydroperoxide of α-linolenic acid was used as substrate (13S-HPOTE), the analogous 9-oxo-10E,15Z-12,13-cis-epoxide remained as one of the main products although it was accompanied by several others of similar prominence (Figure 5). The corresponding NMR data are given in Supplement, Tables S3–S6, Figure S3.
Fig. 5. RP-HPLC analysis of products formed from 13S-HPOTE by Fg-cat.
Products were chromatographed using a Waters Symmetry C18 column (0.46 × 25 cm), a solvent of CH3CN/water/glacial acetic acid (60:40:0.01 by volume) at a flow rate of 1 mL/min, with the UV profiles depicted at 235 nm (red) and 280 nm (black). The main peaks were further purified by SP-HPLC and the structures established by LC-MS and 1H-NMR (Supplement Fig. S3, Tables S3–S6).
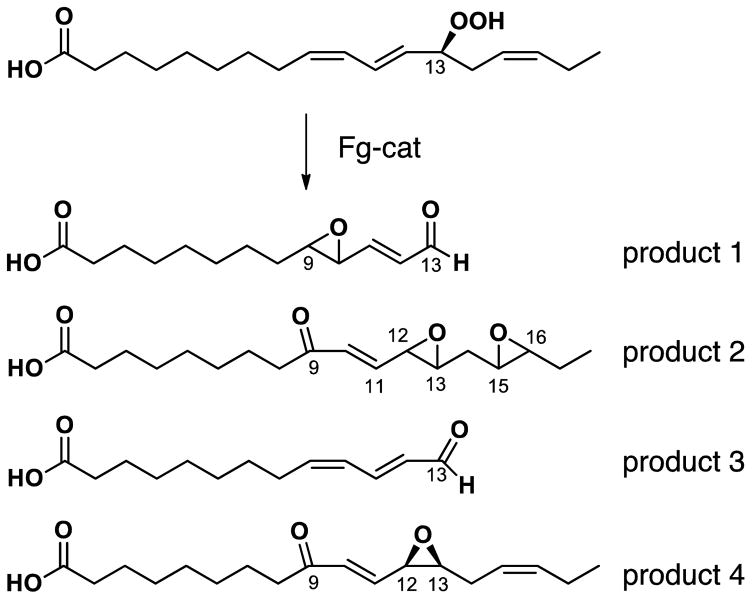
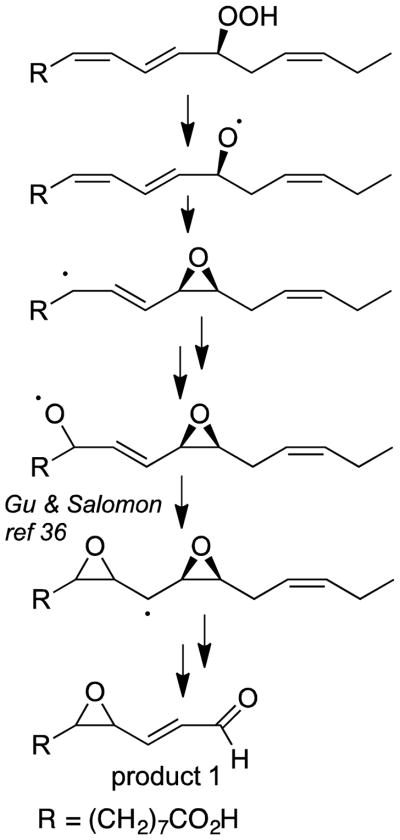
The products were analyzed by LC-MS and NMR. The structures included two fatty acid C13 aldehydes and what appear to be doubly oxidized derivatives (Figure 6). The appearance of several isomers of product 2, the unusual diepoxides, mimics a well characterized mechanism [33] whereby initial conversion of the 13-hydroperoxide to a peroxy radical is followed by formation of an endoperoxide bridge to C15, whereupon the associated carbon radical at C-16 induces formation of the 15,16-epoxide while the nascent C-13 alkoxyl radical forms an epoxyallylic radical that is oxygenated at C-9, Scheme 2. A mechanistically related sequence of reactions was characterized in human cyclooxygenase-2 when the normal carbon ring cyclization was disturbed by active site mutations [34]. A possible route to product 1, the fatty acid 9,10-epoxy C13 aldehyde is via conversion of an epoxyallyic radical to a pseudosymmetrical diepoxycarbinyl radical as proposed by Gu and Salomon [35], Scheme 3.
Fig. 6.
Structures of the main products formed from 13S-HPOTE
Scheme 2.
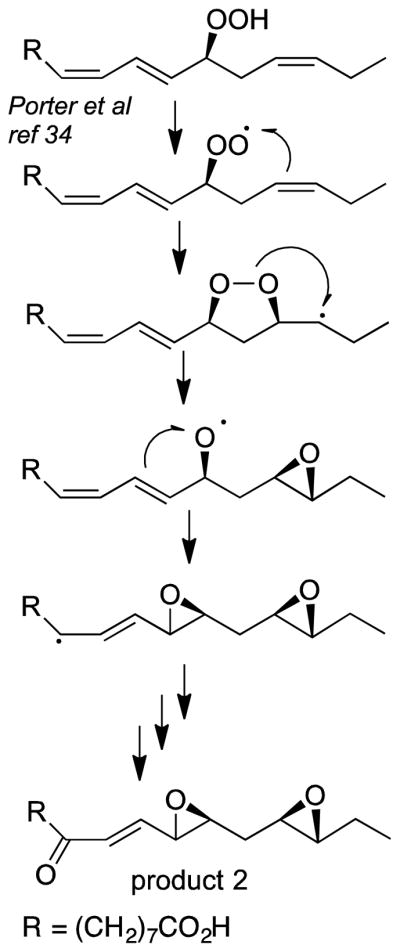
Mechanism of formation of fatty acid diepoxy ketone
Scheme 3.
Mechanism of formation of the fatty acid epoxy C13 aldehyde
3.7. Fg-cat activity as a peroxidase
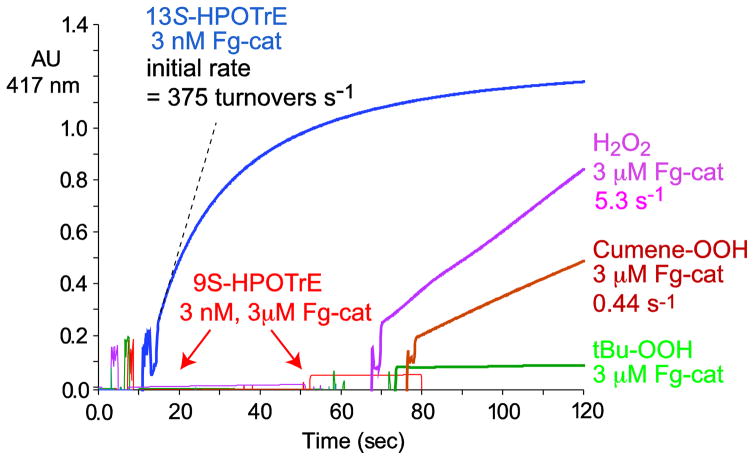
The potential peroxidase activity of Fg-cat was assessed using ABTS as the oxidizable substrate and using a number of different peroxides as co-substrate. The results, Figure 7, show a remarkable activity that is dependent on the availability of fatty acid 13S-hydroperoxides as the peroxide co-substrate. Using 60 μM 13S-HPOTE in the spectrophotometric assay and monitoring the oxidation of ABTS at 417 nm, the reaction consumed the ABTS using only 3 nM Fg-cat and with an initial rate of 375 s−1 (Figure 7). By contrast, there is no reaction using 9S-HPOTE as co-substrate, even with the Fg-cat concentration increased 1000-fold to 3 μM. Similarly, H2O2, cumene hydroperoxide and t-butyl hydroperoxide are ineffective, showing weak reaction only with a 1000-fold increase in the enzyme concentration (Figure 7). A kinetic analysis of the ABTS oxidation using 13S-HPOTE gave a kcat of 552 s−1, a Km value of 25 μM, and kcat/Km = 22 × 106 M−1 s−1 (Supplement, Figure S4).
Fig. 7. Effect of different peroxide co-substrates on the peroxidase activity of Fg-cat.
Activity was measured as the rate of oxidation of ABTS (increased absorbance at 417 nm). Reactions were conducted at room temperature in 1 ml 50 mM Tris containing 150 mM NaCl using 3 nM for reaction with 95 μM 13S-HPOTE, or 3 μM Fg-cat for reaction with 9S-HPOTE (95 μM), or H2O2, cumene hydroperoxide or t-butyl hydroperoxide (each 1 mM).
In an assay in comparison to mammalian myeloperoxidase for production of hypochlorous acid from hydrogen peroxide or 13S-HPOTE and chloride ion (Methods), Fg-cat showed no blue color development and thus no activity as a haloperoxidase.
3.8. Metabolic fate of the fatty acid hydroperoxide in the presence of ABTS
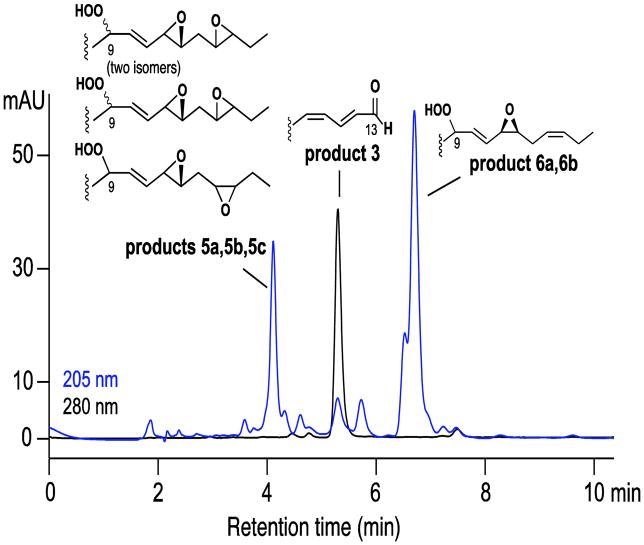
The peroxidase activity of Fg-cat with oxidation of ABTS is associated with conversion of the 13S-HPOTE co-substrate to three classes of product, Figure 8. The individual products were purified by SP-HPLC and identified by NMR (Supplement, Figures S5 – S9; Tables S7–S10). They are related to the products formed in Fg-cat reactions in the absence of ABTS, except that the peroxyl radical intermediates at C-9 are reduced to the corresponding hydroperoxides by electron transfer via ABTS, Scheme 4.
Fig. 8. RP-HPLC analysis of products formed from 13S-HPOTE by Fg-cat in the presence of ABTS.
The products were chromatographed using a Waters Symmetry C18 column (0.46 × 25 cm), a solvent of CH3CN/water/glacial acetic acid (60:40:0.01 by volume) at a flow rate of 1 mL/min, with the UV profiles depicted at 205 nm (blue) and 280 nm (black). The earlier and later peaks were further resolved by SP-HPLC and the structures established by 1H-NMR as illustrated (Supplement, Figures S5 – S9; Tables S7–S10).
Scheme 4.
Proposed mechanisms of transformation of 13S-HPOTE by Fg-cat in the presence of ABTS
4. Discussion
There are six catalase-related genes in F. graminearum (Table 1) presumably in some cases with overlapping functions [36]. However only Fg-cat (FGSG_02217) has the characteristics predicted for a fatty acid peroxidizing heme catalase (with an RXTH sequence on the distal heme [8]) and its immediate neighbor is a 13S-LOX gene (FGSG_02216) that could supply 13S-HPODE or 13S-HPOTE as co-substrate for the peroxidase activity we have detected. This same combination of LOX and catalase with the signature RTTH sequence on the distal heme occurs in over a dozen annotated fungal genomes including in the genera Fusarium, Metarhizium and Fonsecaea. Previously reported heme catalases that metabolize fatty acid hydroperoxides occur as the N-terminal domain of a fusion protein with a C-terminal LOX domain. Fg-cat and its homologues are the first described as stand-alone genes, albeit side-by-side to a lipoxygenase.
4.1. Proposed mechanisms of transformations of 13S-HPODE and 13S-HPOTE
A mechanistic issue raised by our findings is the reaction equation in the transformation of 13-hydroperoxides to the main epoxy-ketone product does not balance. Comparing the structure of 13-HPODE (the central 5 carbons being C5H6O2) and the epoxy-ketone product (C5H4O2), there is an overall loss of 2H in the course of the transformation. Given the imbalance in the reaction producing the main epoxy-ketone product, for catalysis to continue through multiple turnovers there must be side-reactions that return the enzyme to the resting ferric state (see following discussion for details). This may explain why the epoxy-ketone is formed in only ~ 60% yield from 13S-HPODE and is one of multiple products from 13S-HPOTE. The fact that several reactions are going on concurrently may also contribute to the catalysis-dependent enzyme inactivation. In Figure 3, 10 nM Fg-cat reduced the hydroperoxide absorbance at 235 nm by 0.5 AU, which (after allowing for the residual 30% absorbance of products) equates to a change in concentration of 28 μM and calculates as 2,800 turnovers per enzyme molecule prior to Fg-cat inactivation.
The UV-Vis spectrum of Fg-cat is typical of a ferric hemoprotein, and Fe3+ heme is a suitable initiating catalyst of hydroperoxide metabolism. We suggest a reaction pathway beginning with homolytic cleavage of the 13-hydroperoxide to give a fatty acid alkoxyl radical, a species that coexists as an epoxyallylic carbon radical [37] (Scheme 1). The latter reacts with O2 to form the 12,13-epoxy-9-hydroperoxyl radical that abstracts hydrogen from another lipid molecule to form the corresponding hydroperoxide. Subsequent dehydration (loss of H2O) of the hydroperoxy moiety is probably initiated via the facile loss of the geminal C-9 hydrogen, producing the epoxy-ketone. Similar events leading to the 12,13-epoxy-9-hydroperoxyl radical were proposed in the reaction of metmyoglobin with 13-HPODE [38].
The reaction with the α-linolenic acid analog, 13S-HPOTE, produces several products of similar abundance, the formation of which we rationalize as in Scheme 2. In addition to the epoxy-ketone analog are the unusual diepoxy ketone products and the chain cleavage aldehydes, the C13-oxo acid and its epoxy derivative. The diepoxy ketones are formed through a well precedented route involving an endoperoxide intermediate, as illustrated in Scheme 2. This reaction was characterized originally in transformations of fatty acid endoperoxyl radicals [33], and identified subsequently as a significant product outcome associated with mutation of two active site residues of human COX-2 [34]. That the C13-aldehyde acid product is more prominent than from the linoleic acid hydroperoxide is rationalized by the extra 15,16-double bond of 13S-HPOTE stabilizing the alkoxyl radical intermediate and promoting a β-scission reaction resulting in cleavage of the carbon chain [39]. When we tested the reaction of Fg-cat with 13S-HPOTE under anaerobic conditions, the C13 oxo acid was the sole product (data not shown), with the lack of molecular oxygen curtailing the other pathways.
The C13 aldehyde and the 9-oxo-10E-trans-12,13-epoxide were identified earlier as metabolites of 13-HPOTE in partially purified extracts of the green alga Chlorella pyrenoidosa [40]. Furthermore, epoxyallylic hydroperoxides analogous to those we isolated from incubations containing ABTS were shown to be products of the activation steps in lipoxygenase catalysis [41].
Overall, our impression is that the reaction of Fg-cat with 13S-HPODE and 13S-HPOTE is not well controlled by the enzyme, allowing multiple outcomes. Nonetheless, there is some enzymatic control because in non-enzymatic systems, due to steric hindrance in formation of the corresponding cis-epoxy isomer, the trans epoxy-hydroxy and epoxy-keto isomers are the more prominent [42].
4.2 Functional role of Fg-cat?
Association of the peroxidase activity specifically with 13S-hydroperoxides is remarkable and fits with the 13S-LOX being the neighboring gene. Values for the peroxidase activity of Fg-cat are directly comparable to the reported peroxidase activity of mammalian cyclooxygenase enzymes in oxidation of ABTS [29]: with COX-1 and COX-2 using 15S-HPETE as the oxidizing co-substrate, the enzymes exhibit turnover numbers of 51 – 120 s−1, a Km for 15-HPETE of 8.5 – 9.9 μM, and kcat/Km values of 6 – 12 μM−1.s−1 [29]. In comparison, the kcat/Km for Fg-cat in oxidizing ABTS using 13-HPOTE as co-substrate gave the slightly higher value of 22 μM−1.s−1 (Figure 8). This kcat/Km for Fg-cat in the ABTS assay is about 200-fold higher than we determined previously for a small (33 kD) catalase-related peroxidase from Mycobacterium avium paratuberculosis [30].
Several catalase hemoproteins that metabolize fatty acid hydroperoxides function as an allene oxide synthase (AOS), an activity associated with metabolic pathways of cyclopentenone biosynthesis [8, 43]. Thus, marine corals synthesize the prostanoid-related clavulones via 8R-LOX and a catalase-related AOS (cAOS) [8, 44]. By contrast, the jasmonic acid pathway in plants involves a 13S-LOX acting on α-linolenic acid and a cytochrome P450 (CYP74A) as the AOS [43]. We had considered Fg-cat as a candidate for the role of the “missing” AOS activity in fungi, in which case the adjacent 13S-LOX gene in F. graminearum could provide the appropriate AOS substrate, as suggested as a potential role for the F. oxysporum 13S-LOX homologue [15]). Many fungi including Fusarium species synthesize jasmonates [45–48] (although we find no evidence concerning F. graminearum). Notwithstanding the apparent logic, in the course of our extensive analyses we found no evidence of allene oxide synthesis nor cyclopentenone formation from interaction of Fg-cat with either 13S-HPODE or 13S-HPOTE, likely dismissing the possibility of its involvement in jasmonate synthesis in F. graminearum. Instead, our data suggest Fg-cat has an oxidative function as a peroxidase, possibly in metabolism, defense or fungal infection of plants.
Supplementary Material
Highlights.
F. graminearum and related fungi have adjacent 13S-lipoxygenase and catalase genes
The catalase reacts specifically with 13S fatty acid hydroperoxides (and not with H2O2)
A main product from 13S-HPODE and 13S-HPOTE is a 9-keto-10E-12,13-cis-epoxide
Specifically 13S-hydroperoxides support peroxidase activity with ABTS as substrate
The LOX and catalase may partner in oxidations for fungal metabolism or defense
Acknowledgments
This work was supported in part by National Institutes of Health Grants GM-074888 and GM-15431 (to A. R. B.), GM-076592 (to C. S.) and by the Institutional Research Funding IUT19-9 of the Estonian Ministry of Education and Research and the Estonian Science Foundation Grant 9410 (to N. S.). We thank Dr. M. Wade Calcutt for help with the LC-MS analyses and Dr. Scott McCall for assistance with the myeloperoxidase assay.
Abbreviations
- ABTS
2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)
- cAOS
catalase-related allene oxide synthase
- Fg-cat and Fg-LOX
Fusarium graminearum products of the genes FGSG_02217 and FGSG_02216, respectively
- HPODE
hydroperoxyoctadecadienoic acid
- HPOTE
hydroperoxyoctadecatrienoic acid
- HPETE
hydroperoxyeicosatetraenoic acid
- LOX
lipoxygenase
- RP-HPLC
reversed phase high pressure liquid chromatography
- SP-HPLC
straight phase high pressure liquid chromatography
References
- 1.Koljak R, Boutaud O, Shieh BH, Samel N, Brash AR. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 2.Lõhelaid H, Järving R, Valmsen K, Varvas K, Kreen M, Järving I, Samel N. Identification of a functional allene oxide synthase-lipoxygenase fusion protein in the soft coral Gersemia fruticosa suggests the generality of this pathway in octocorals. Biochim Biophys Acta. 2008;1780:315–321. doi: 10.1016/j.bbagen.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Gao B, Boeglin WE, Zheng Y, Schneider C, Brash AR. Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the cyanobacterium Acaryochloris marina. J Biol Chem. 2009;284:22087–22098. doi: 10.1074/jbc.M109.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teder T, Lohelaid H, Boeglin WE, Calcutt WM, Brash AR, Samel N. A catalase-related hemoprotein in coral Is specialized for synthesis of short-chain aldehydes: DISCOVERY OF P450-TYPE HYDROPEROXIDE LYASE ACTIVITY IN A CATALASE. J Biol Chem. 2015;290:19823–19832. doi: 10.1074/jbc.M115.660282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang I, Göbel C, Porzel A, Heilmann I, Feussner I. A lipoxygenase with linoleate diol synthase activity from Nostoc sp. PCC 7120. Biochem J. 2008;410:347–357. doi: 10.1042/BJ20071277. [DOI] [PubMed] [Google Scholar]
- 6.Schneider C, Niisuke K, Boeglin WE, Voehler M, Stec DF, Porter NA, Brash AR. Enzymatic synthesis of a bicyclobutane fatty acid by a hemoprotein-lipoxygenase fusion protein from the cyanobacterium Anabaena PCC 7120. Proc Natl Acad Sci USA. 2007;104:18941–18945. doi: 10.1073/pnas.0707148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oldham ML, Brash AR, Newcomer ME. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc Natl Acad Sci USA. 2005;102:297–302. doi: 10.1073/pnas.0406352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mashhadi Z, Newcomer ME, Brash AR. The Thr-His connection on the distal heme of catalase-related hemoproteins: a hallmark of reaction with fatty acid hydroperoxides. ChemBioChem. 2016;17:2000–2006. doi: 10.1002/cbic.201600345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao B, Boeglin WE, Brash AR. Role of the conserved distal heme asparagine of coral allene oxide synthase (Asn137) and human catalase (Asn148): mutations affect the rate but not the essential chemistry of the enzymatic transformations. Arch Biochem Biophys. 2008;477:285–290. doi: 10.1016/j.abb.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosha T, Uchida T, Brash AR, Kitagawa T. On the relationship of coral allene oxide synthase to catalase. A single active site mutation that induces catalase activity in coral allene oxide synthase. J Biol Chem. 2006;281:12610–12617. doi: 10.1074/jbc.M600061200. [DOI] [PubMed] [Google Scholar]
- 11.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Jacobsen S, Jorgensen HJ, Collinge DB, Svensson B, Finnie C. Fusarium graminearum and its interactions with cereal heads: Studies in the proteomics era. Front Plant Sci. 2013;4:37. doi: 10.3389/fpls.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richard JL. Some major mycotoxins and their mycotoxicoses - An overview. Int J Food Microbiol. 2007;119:3–10. doi: 10.1016/j.ijfoodmicro.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Tsukada K, Takahashi K, Nabeta K. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry. 2010;71:2019–2023. doi: 10.1016/j.phytochem.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Brodhun F, Cristobal-Sarramian A, Zabel S, Newie J, Hamberg M, Feussner I. An iron 13S-lipoxygenase with an α-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PloS one. 2013;8:e64919. doi: 10.1371/journal.pone.0064919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther JC, Hallen-Adams HE, Bucking H, Shachar-Hill Y, Trail F. Triacylglyceride metabolism by Fusarium graminearum during colonization and sexual development on wheat. Mol Plant-Microbe Interact. 2009;22:1492–1503. doi: 10.1094/MPMI-22-12-1492. [DOI] [PubMed] [Google Scholar]
- 17.Peers KE, Coxon DT. Controlled synthesis of monohydroperoxides by α-tocopherol inhibited autoxidation of polyunsaturated lipids. Chem Phys Lipids. 1983;32:49–56. [Google Scholar]
- 18.Schneider C, Boeglin WE, Brash AR. Enantiomeric separation of hydroxy eicosanoids by chiral column chromatography: effect of the alcohol modifier. Anal Biochem. 2000;287:186–189. doi: 10.1006/abio.2000.4847. [DOI] [PubMed] [Google Scholar]
- 19.Brash AR, Song WC. Detection, assay, and isolation of allene oxide synthase. Methods Enzymol. 1996;272:250–259. doi: 10.1016/s0076-6879(96)72030-x. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Y, Boeglin WE, Schneider C, Brash AR. A 49-kDa mini-lipoxygenase from Anabaena sp. PCC 7120 retains catalytically complete functionality. J Biol Chem. 2008;283:5138–5147. doi: 10.1074/jbc.M705780200. [DOI] [PubMed] [Google Scholar]
- 21.Thomas CP, Boeglin WE, Garcia-Diaz Y, O’Donnell VB, Brash AR. Steric analysis of epoxyalcohol and trihydroxy derivatives of 9-hydroperoxy-linoleic acid from hematin and enzymatic synthesis. Chem Phys Lipids. 2013;167–168:21–32. doi: 10.1016/j.chemphyslip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blakely RD, Clark JA, Rudnick G, Amara SG. Vaccinia-T7 RNA polymerase expression system: evaluation for the expression cloning of plasma membrane transporters. Anal Biochem. 1991;194:302–308. doi: 10.1016/0003-2697(91)90233-j. [DOI] [PubMed] [Google Scholar]
- 23.Jisaka M, Kim RB, Boeglin WE, Nanney LB, Brash AR. Molecular cloning and functional expression of a phorbol ester-inducible 8S-lipoxygenase from mouse skin. J Biol Chem. 1997;272:24410–24416. doi: 10.1074/jbc.272.39.24410. [DOI] [PubMed] [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR. Lactose fed-batch overexpression of recombinant metalloproteins in Escherichia coli BL21 (DE3): Process control yielding high levels of metal-incorporated soluble protein. Prot Express Purific. 1995;6:646–654. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 26.Boutaud O, Brash AR. Purification and catalytic activities of the two domains of the allene oxide synthase-lipoxygenase fusion protein of the coral Plexaura homomalla. J Biol Chem. 1999;274:33764–33770. doi: 10.1074/jbc.274.47.33764. [DOI] [PubMed] [Google Scholar]
- 27.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nature chemical biology. 2012;8:784–790. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Childs RE, Bardsley WG. The steady-state kinetics of peroxidase with 2, 2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem J. 1975;145:93–103. doi: 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duggan KC, Musee J, Marnett LJ. Peroxidase active site activity assay. Methods Mol Biol. 2010;644:55–65. doi: 10.1007/978-1-59745-364-6_6. [DOI] [PubMed] [Google Scholar]
- 30.Pakhomova S, Gao B, Boeglin WE, Brash AR, Newcomer ME. The structure and peroxidase activity of a 33-kDa catalase-related protein from Mycobacterium avium ssp. paratuberculosis. Protein Sci. 2009;18:2559–2568. doi: 10.1002/pro.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klotz MG, Loewen PC. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol Biol Evol. 2003;20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- 32.Mercier J, Agoh B. Comportement d’hydroperoxydes allyliques a longue chaine en presence de complexes de certains metaux de transition. II. Structure des époxy-alcools formés à partir d’hydroperoxydes d’octadécène-9 oates de méthyle cis et trans en présence d’acétylacétonate de vanadyle. Chem Phys Lipids. 1974;12:239–248. [Google Scholar]
- 33.Porter NA, Zuraw PJ, Sullivan JA. Peroxymercuration-demercuration of lipid hydroperoxides. Tetrahed Lett. 1984;25:807–810. [Google Scholar]
- 34.Schneider C, Boeglin WE, Brash AR. Identification of two cyclooxygenase active site residues, leucine-384 and glycine-526, that control carbon ring cyclization in prostaglandin biosynthesis. J Biol Chem. 2004;279:4404–4414. doi: 10.1074/jbc.M307431200. [DOI] [PubMed] [Google Scholar]
- 35.Gu X, Salomon RG. Fragmentation of a linoleate-derived γ-hydroperoxy-α,β-unsaturated epoxide to γ-hydroxy- and γ-oxo-alkenals involves a unique pseudo-symmetrical diepoxycarbinyl radical. Free Radic Biol Med. 2012;52:601–606. doi: 10.1016/j.freeradbiomed.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansberg W, Salas-Lizana R, Dominguez L. Fungal catalases: function, phylogenetic origin and structure. Arch Biochem Biophys. 2012;525:170–180. doi: 10.1016/j.abb.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Wilcox AL, Marnett LJ. Polyunsaturated fatty acid alkoxyl radicals exist as carbon-centered epoxyallylic radicals: a key step in hydroperoxide-amplified lipid peroxidation. Chemical research in toxicology. 1993;6:413–416. doi: 10.1021/tx00034a003. [DOI] [PubMed] [Google Scholar]
- 38.Reeder BJ, Wilson MT. Mechanism of reaction of myoglobin with the lipid hydroperoxide hydroperoxyoctadecadienoic acid. Biochem J. 1998;330(Pt 3):1317–1323. doi: 10.1042/bj3301317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labeque R, Marnett LJ. Reaction of hematin with allylic fatty acid hydroperoxides: identification of products and implications for pathways of hydroperoxide-dependent epoxidation of 7,8-dihydroxy-7, 8-dihydrobenzo[a]pyrene. Biochemistry. 1988;27:7060–7070. doi: 10.1021/bi00418a059. [DOI] [PubMed] [Google Scholar]
- 40.Vick BA, Zimmerman DC. Metabolism of fatty acid hydroperoxides by Chlorella pyrenoidosa. Plant Physiol. 1989;90:125–132. doi: 10.1104/pp.90.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y, Brash AR. On the role of molecular oxygen in lipoxygenase activation: comparison and contrast of epidermal lipoxygenase-3 with soybean lipoxygenase-1. J Biol Chem. 2010;285:39876–39887. doi: 10.1074/jbc.M110.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner HW, Kleiman R. Degradation of linoleic acid hydroperoxides by a cysteine. FeCl3 catalyst as a model for similar biochemical reactions. 2. Specificity in formation of fatty acid epoxides. Biochim Biophys Acta. 1981;665:113–125. doi: 10.1016/0005-2760(81)90239-3. [DOI] [PubMed] [Google Scholar]
- 43.Tijet N, Brash AR. Allene oxide synthases and allene oxides. Prostaglandins Other Lipid Mediators. 2002;68–69:423–431. doi: 10.1016/s0090-6980(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 44.Corey EJ, d’Alarcao M, Matsuda SPT, Lansbury PT., Jr Intermediacy of 8-(R)-HPETE in the conversion of arachidonic acid to pre-clavulone A by Clavularia viridis. Implications for the biosynthesis of marine prostanoids. J Am Chem Soc. 1987;109:289–290. [Google Scholar]
- 45.Miersch O, Günther T, Fritsche W, Sembdner G. Jasmonates from different fungal species. Nat Prod Lett. 1993;2:293–299. [Google Scholar]
- 46.Miersch O, Bruckner B, Schmidt J, Sembdner G. Cyclopentane fatty acids from Gibberella fujikuroi. Phytochemistry. 1992;31:3835–3837. [Google Scholar]
- 47.Miersch O, Bohlmann H, Wasternack C. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry. 1999;50:517–523. [Google Scholar]
- 48.Eng F, Haroth S, Feussner K, Meldau D, Rekhter D, Ischebeck T, Brodhun F, Feussner I. Optimized jasmonic acid production by Lasiodiplodia theobromae reveals formation of valuable plant secondary metabolites. PloS one. 2016;11:e0167627. doi: 10.1371/journal.pone.0167627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicholson ML, Laudenbach DE. Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J Bacteriol. 1995;177:2143–2150. doi: 10.1128/jb.177.8.2143-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.