Abstract
Single cell-laden three-dimensional (3D) microgels that can serve to mimic stem cell niches in vitro, and are therefore termed microniches, can be efficiently fabricated by droplet-based microfluidics. In this technique an aqueous polymer solution along with a highly diluted cell solution is injected into a microfluidic device to create monodisperse pre-microgel droplets that are then solidified by a polymer crosslinking reaction to obtain monodisperse single cell-laden microniches. However, problems limiting this approach studying the fate of single cells include Poisson encapsulation statistics that result in mostly empty microniches, and cells egressing from the microniches during subsequent cell culture. Here, we present a strategy to bypass Poisson encapsulation statistics in synthetic microniches by selective crosslinking of only cell-laden pre-microgel droplets. Furthermore, we show that we can position cells in the center of the microniches, and that even in protease-sensitive microniches this greatly reduces cell egress. Collectively, we present the development of a versatile protocol that allows for unprecedented efficiency in creation of synthetic protease-sensitive microniches for probing single stem cell fate in 3D.
Graphical abstract
We present a powerful strategy for evading Poisson encapsulation statistics and for cell centering in single cell-laden synthetic microniches to facilitate long-term culture in protease-sensitive 3D microenvironments.
Introduction
Combining droplet microfluidics and artificial extracellular matrices allows for fabrication of thousands of microgels per second that can serve as discrete cellular microenvironments for culturing cells in a close to physiological environment.1 Microgels are spherical hydrogel particles with a typical diameter of 10–1000 μm that can be designed to capture properties, such as elasticity, degradability and cell adhesion, of the three-dimensional (3D) natural environment.2 Cells have been encapsulated in microgels made out of a variety of materials based on synthetic and natural polymers.3 Cell-laden microgels have been used for multifaceted applications, e.g., as in-vitro 3D-cell culturing systems to assess tumor microenvironments 4, to study stem cells,5,6 and to test drugs,4 or for in-vivo studies to treat diabetes in animals7 and even in humans.8,9 A unique advantage of microgels as opposed to larger hydrogels is that they allow for efficient encapsulation and subsequent assessment of individual cells.10 Encapsulation of single cells is particularly relevant in the context of heterogeneous cell populations, such as stem cell populations, that need to be investigated on a single-cell level to appraise biological responses, which otherwise might be masked when assessing the average response of the complete population.11
Due to their therapeutic potential, mesenchymal stem cells (MSCs) are the focus of a great number of studies related to basic and applied research.12 Depending on external cues, individual MSCs can migrate, undergo planed cell death, remain quiescent, proliferate, or further differentiate into tissue lineages. To fully exploit their therapeutic potential, it is thus desirable to assess and control the fate of individual cells.13 What is more, in vivo, MSCs reside within unique tissue microenvironments, so called stem cell niches, which have a crucial role in influencing their fate by presenting a still not well understood set of biophysical and biochemical cues.14 Microfluidic encapsulation of single stem cells in microgels mimicking such stem cell niches (here termed microniches) presents, therefore, an ideal method to assess the fate of individual MSCs with simultaneous consideration of their surrounding three-dimensional (3D) microenvironment.
To encapsulate single cells in microniches, highly diluted cell solutions along with aqueous polymer solutions need to be injected into a microfluidic device to create microdroplets that are then solidified. The following distribution of cells in these artificial microniches follows Poisson distribution statistics. This means that for typical cell dilutions used for single cell encapsulation only every 10–20th droplet will be loaded with a cell, which makes further processing and analysis a laborious venture.15 For alginate microniches an elegant approach to beat Poisson distribution statistics was recently presented by exclusively gelating microdroplets loaded with cells.16 However, due to the lack of degradation sites, the typical nanoporous alginate network does not allow for cells to actively remodel their surrounding. Within their in-vivo environment, however, cells do so extensively by degrading the existing matrix components through secretion of proteases in interplay with secretion of freshly synthesized matrix components, such as proteins and sugars.17 Engineering an artificial extracellular matrix that is susceptible to cell-induced degradation is often necessary to assess and manipulate cellular processes, such as migration,18 morphogenesis19 or differentiation20. What is more, due to the natural origin of alginate, batch-to-batch variations might exist and unintended biological activity might be introduced. It would thus be desirable for strategies to beat Poisson encapsulation statistics to be available for a fully synthetic hydrogel platform that is permissive to cellular remodeling. Importantly, though, remodeling can lead to cells rapidly escaping the microniche,21 which limits ones ability to probe stem cell fate over a time course of days. If cell egress can be delayed within protease-sensitive microniches, and long-term cultures of stem cells become possible, new paths will be opened to address, in a high throughput manner, questions related to stem cell biology and their niches.
Here, we report on development of a strategy for beating Poisson encapsulation statistics during fabrication of synthetic single cell-laden microniches. Moreover, we demonstrate that positioning of single cells within the center of protease-sensitive microniches can delay cell egress and allows for their long-term in-vitro assessment in a close to physiological microenvironment. We show that this approach allows for efficiently probing differentiation of single MSCs in 3D, in dependence of the mechanical properties of their surrounding microniches.
Experimental Section
TG-PEG hydrogel precursors preparation
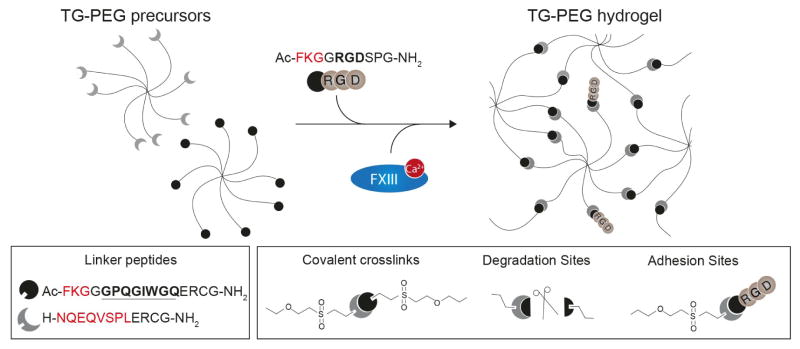
TG-PEG hydrogel precursors are fabricated as described previously.22 In brief, star shaped eight-arm PEG-vinylsulfone (mol. wt. 40000 Da) is functionalized with two kinds of pending FXIII substrate peptides serving either as glutamine acceptor (Gln-PEG; Substrate peptide: H-NQEQVSPL-ERCG-NH2) or as lysine donor additionally containing an MMP-sensitive linker (Lys-MMP-PEG; Substrate peptide: Ac-FKGG-GPQGIWGQ-ERCG-NH2). To make TG-PEG precursor solution, Gln-PEG and Lys-MMP-PEG are mixed stoichiometrically in 50 mM Tris (Sigma-Aldrich, cat. no. T5941) pH 7.6 to a final concentration of 30% (v/w). The peptides Ac-FKGG-RGDSPG-NH2 (Lys-RGD) and Ac-FKGG-K-FITC (Lys-FITC) are introduced as adhesion ligand and for FITC labeling of the matrix, respectively. For stock solutions, Lys-RGD and Lys-FITC are diluted in 50 mM Tris pH 7.6 to 13.2 mM and 1.32 mM, respectively. All peptides are purchased from PolyPeptide Group, France.
Cell culture
D1 cells (ATCC, cat. no. CRL-12424, passage 22 and verified to be mycoplasma-free by manufacturer) in maintenance culture (maintained at sub-confluency) and when encapsulated as single cells in TG-PEG microniches are cultured at 37 °C and 5% CO2 in a humidified atmosphere. D1 cells between passages 24 and 26 are used for all experiments. As culture medium Dulbecco’s Modified Eagle Medium (DMEM; Gibco, cat. no. 12800017) supplemented with 10% (v/v) fetal bovine serum (FBS; Atlanta Biologicals, cat. no. S11150) and 1% (v/v) Penicillin-Streptomycin solution (P/S; Gibco, cat. no. 15140122) is used.
Preparation of CaCO3 nanoparticles
10 mg CaCO3 particles (CalEssence, cat. no. 70 PCC) are placed in 1 mL DMEM supplemented with 10% (v/v) FCS and 1% (v/v) P/S (termed ‘DMEM-complete’ in all following paragraphs) and sonicated for 20 seconds at 70% using a tip sonicator (SONICS vibra cell) before an additional 19 ml DMEM-complete is added. For removal of super-micron particles, the mixture is centrifuged (5 min; 150 × g; room temperature) and supernatant containing sub-micron particles is recovered. Supernatant is then sharply centrifuged (5 min at 1000 × g at room temperature), the pellet now containing the sub-micron particles is re-suspended in 1 mL DMEM-complete and sonicated for 1 min using a sonicator bath (Branson 3510).
Preparation of MSCs prior to encapsulation
To minimize Ca2+ concentration and at the same time prevent CaCO3 nanoparticles from dissolving, CaCl2-free but CaCO3 saturated DMEM is fabricated (termed ‘DMEM-CaCO3’ in all following paragraphs). To do so, 0.5 L CaCl2-free DMEM (Gibco, cat. no. 21068028) supplemented with 10% (v/v) FCS is incubated for 30 min at room temperature with 20 g CaCO3 powder (Sigma-Aldrich, cat. no. C5929). Next, the mixture is filtered through a 0.22 μm cellulose nitrate membrane (Corning, cat. no. 430756) to remove undissolved CaCO3.
D1 cells in maintenance culture are detached for 5 min using a Trypsin-EDTA (0.05%) solution (Gibco, cat. no. 25300054). Cell suspension is washed with DMEM-complete by centrifugation (5 min at 250 × g at room temperature) and is adjusted to 107 cells mL−1 using DMEM-complete. 0.5 mL of the cell suspension is mixed with the prior prepared 1 mL of CaCO3 nanoparticles in DMEM-complete. Mixture of D1 cells and CaCO3 nanoparticles is centrifuged (5 min at 50 × g at 4 °C), agitated by pipetting and again centrifuged (5 min at 50 × g at 4 °C). Pellet, which now contains D1 cells loaded with CaCO3 nanoparticles is washed twice with 10 mL DMEM-CaCO3 by centrifugation (10 min at 30 × g at 4 °C). Pellet is re-suspended in 500 μL DMEM-CaCO3. Final cell concentration is 107 cells mL−1.
Fabrication of microfluidic devices
The microfluidic devices are produced using soft lithography23 by pouring poly(dimethylsiloxane) (PDMS) along with crosslinker (Sylgard 184 elastomer kit, Dow Corning, base : crosslinker = 10 : 1) onto a silicon wafer patterned with SU-8 photoresist. After solidifying the material for 1 h at 65 °C, devices are fabricated by oxygen plasma bonding of the PDMS replicas onto glass slides. To render the devices hydrophobic, and hence, suitable for water-in-oil emulsification, they are treated with Aquapel® (PPG, Pittsburgh, PA, USA.), a commercial windshield treatment. Therefore, Aquapel® is injected into the channels, allowed to sit for a few minutes, and then removed by thorough airdrying and incubation at 65 °C for 10 min.
Microfluidic fabrication of single cell-laden TG-PEG microniches
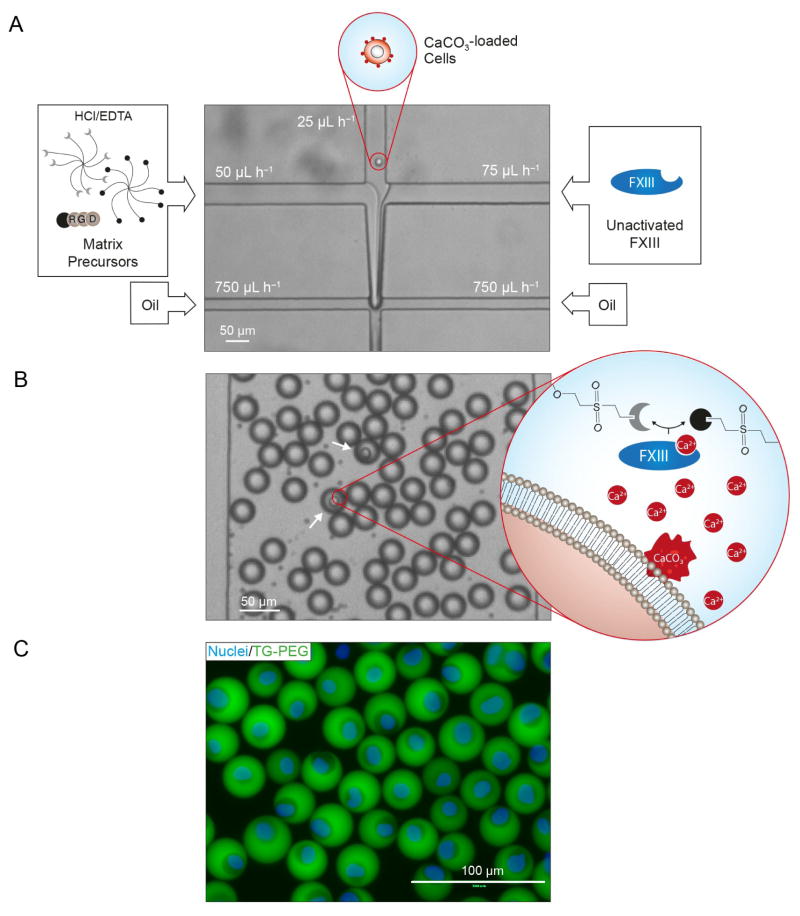
To fabricate single cell-laden microniches, a microfluidic device with a channel geometry as depicted in Figure 2A is produced. The channel width is 50 μm at the first cross-junction. The channel width at the second cross-junction as well as the uniform channel height is 20 or 30 μm depending on whether resultant microniches should have a diameter of 20 or 30 μm, respectively. A wide basin channel is patterned downstream of the second cross-junction to allow for observation of the resultant droplets, as also illustrated in Figure 2B.
Figure 2. Strategy for on-demand gelation of synthetic microniches.
A) Matrix precursors supplemented with hydrochloric acid (HCl) and ethylenediaminetetraacetic acid (EDTA), a diluted solution of cells loaded with CaCO3 nanoparticles and unactivated FXIII are separately injected into a microfluidic chip. Reagents are joined in a laminar flow and subsequently sheared by oil at an X-junction resulting in an emulsion of oil and droplets containing precursors for hydrogel formation. Channel dimensions at X-junction define droplet diameter. Here, a 20 μm × 20 μm X-junction resulted in a droplet diameter of 22.4±0.7 μm before microniche swelling. B) Emulsion is shown in the collection channel of the microfluidic chip. The number of cells per droplet follows Poisson distribution statistics. In the event that a cell is present in the droplet (white arrows), the HCl dissolves CaCO3 nanoparticles on the cell leading to Ca2+-induced activation of FXIII and thereby on-demand microniche formation. Supplied EDTA ensures prevention of background gelation occurring in absence of a cell. C) Fluorescence image of cells stained with Hoechst 33342 (Nuclei) encapsulated in FITC labeled TG-PEG microniches after microniche retrieval from the emulsion and transfer to cell culture.
The following 3 fluids containing matrix components, cells and unactivated FXIII are prepared at 4°C and all concentrations are given as initial concentrations:
To supply matrix components to the microfluidic device, 250 μL of a solution containing 15 or 22.5% (w/v) TG-PEG precursor solution, 750 μM Lys-RGD, 18.75 μM Lys-FITC and three times the denoted EDTA and HCl concentrations in CaCl2-free DMEM (no supplements) is prepared.
To supply D1 cells, the prior prepared 500 μL D1 cell suspension containing 107 mL−1 cells in DMEM-CaCO3 is supplemented with 15% (v/v) OptiPrep™ (Sigma-Aldrich, cat. no. D1556) to prevent cell sedimentation.
To supply unactivated FXIII, 90 μL of 60 U mL−1 FXIII (CSL Behring, cat. no. 1250E FXIII N) in distilled water and 90 μL of 60 U mL−1 thrombin (Sigma-Aldrich, cat. no. T6884-250UN) in 50 mM Tris pH 7.6 and 120 μL CaCl2-free DMEM (no supplements) are mixed. Thrombin is required for additional proteolytic activation of FXIII.
All three fluids are kept on ice while being supplied to the microfluidic device using analytical plastic syringes and polyethylene tubing (Scientific Commodities, cat. no. BB31695-PE/5). The flow rates are adjusted by syringe pumps (New Era Pumps).
To monitor the droplet formation, all devices are operated on a horizontally oriented microscope mounted on an optical table and equipped with a digital camera (Sony XCD-V50). The cell suspension is injected through the center inlet and the matrix precursor solution and the unactivated FXIII solution are injected through the two outer inlets. Varying the flow rates of the three phases adjusts their respective volume fractions in the laminar co-flowing stream that forms downstream of the first cross-junction. Subsequent flow focusing by the continuous phase, a mixture of HFE-7500 (3M Novec, cat. no. 98-0212-2928-5) and a 0.5% (w/w) triblock copolymer surfactant (Krytox–Jeffamine–Krytox; synthesized as described earlier24), leads to droplet formation. The following volume flow rates are used for matrix precursor solution, cell phase, unactivated FXIII phase and continuous oil phase, respectively: 50:25:75:750 μL h−1. This results in a dilution of matrix components, cell solution and unactivated FXIII in microdroplets by a factor of 3, 6 and 2, respectively. The resultant pre-microgel emulsion is collected on ice. To initiate polymer crosslinking to form microniches, the emulsion is incubated in a thermo shaker (Eppendorf) set to 37 °C and as denoted either 0 or 1000 rpm.
Recovery of cell-selective gelled microniches
For microniche recovery, first a DMEM emulsion is created by vortexing 200 μL CaCl2-free DMEM with 200 μL HFE-7500 containing 0.5% (w/w) triblock copolymer surfactant. Next, microniche and DMEM emulsions are washed twice with 400 μL HFE-7500 by inverting the tube, waiting for phase-separation and removal of lower oily phase. Washed microniche and DMEM emulsions are mixed together and washed again with 400 μL HFE-7500 in the same way as before. Next, 200 μL CaCl2-free DMEM supplemented with 10% (v/v) FCS is added to the emulsion mix and the emulsion is broken by adding 200 μL 50% (v/v) 1H,1H,2H,2H-perfluoro-1-octanol (PFO; Sigma-Aldrich, 370533) in HFE-7100 and vigorous pipetting. The broken emulsion is immediately washed twice by adding 400 μL HFE-7500 and removal of complete lower oily phase. To remove remaining traces of HFE-7500 and PFO, aqueous phase containing microniches is transferred to a new tube, 1 ml DMEM-complete supplemented with 50 mM EDTA is added, microniches are densified by centrifugation (1 min at 100 g at room temperature) and upper aqueous phase is again transferred to a new tube. To remove undissolved CaCO3 nanoparticles on cells, aqueous phase containing microniches is centrifuged (5 min at 500 × g at room temperature), all but the lowest 100 μL of the supernatant is discarded, 1 ml DMEM-complete supplemented with 50 mM EDTA is added and microniches are incubated for 20 min at room temperature. For removal of EDTA, microniches are washed twice by adding 1 ml DMEM-complete, centrifugation (5 min at 500 × g at room temperature) and discarding of all but lowest 100 μL of the supernatant. Finally, retrieved microniches are transferred to cell culture well plates.
Statistical analysis
Data is depicted as mean ± standard deviation (SD). Typically, the number of analyzed microniches is > 200. All measurements have been repeated three times. Mean values are compared by a student’s t-test using MATLAB. Statistical significance is accepted for p < 0.05. Further details on experimental methods and materials are provided in Supplementary Information.
Results and discussion
We use a recently developed synthetic hydrogel platform (TG-PEG) for fabrication of microniches.22 TG-PEG hydrogels have been widely used for studying stem cells from various origins when they are encapsulated in bulk hydrogels25–28 or microgels.21 TG-PEG is based on two polyethylene glycol precursors that are crosslinked by the transglutaminase factor XIII (FXIII) resulting in a biocompatible nanoporous matrix (Figure 1). To make the peptide bond that links the two PEG precursors, FXIII needs to be activated by binding a calcium ion (Ca2+) as cofactor.29 To render the matrix responsive to cell secreted proteases, matrix-metalloproteinase (MMP)-sensitive sequences are introduced into the network. Moreover, adhesion sites are added at a concentration of 250 μM to allow MSCs to interact with the surrounding matrix, which is necessary for anchorage dependent cells to prevent them from undergoing anoikis.30 To beat Poisson encapsulation statistics, we load MSCs with calcium carbonate (CaCO3) nanoparticles and rinse away unbound nanoparticles resulting in an average of 300±10 nanoparticles per cell, as calculated by measuring the Ca2+ concentration after completely dissolving bound CaCO3 nanoparticles using 7.4% (v/v) HCl. The used CaCO3 nanoparticles have a diameter of 770±30 nm (intensity-average), a zeta potential of −14.0±0.2 mV and a polydispersity index of 0.22±0.02 (Figure S1A and B). CaCO3 nanoparticles efficiently bind to the membrane of the MSCs (Figure S1C). Of note, once MSCs are loaded with CaCO3 nanoparticles all handling is performed at 4 °C, which effectively prevents internalization of CaCO3 nanoparticles as shown recently by measuring intracellular calcium levels.16 Also, loading of MSCs with CaCO3 nanoparticles neither affects cell viability nor the MSCs’ ability to adhere to culture plates (Figure S1D). Next, we inject the cells into a microfluidic device with two consecutive cross-junctions along with matrix precursors and unactivated FXIII (Figure 2A). At the first cross-junction, the three solutions are joined in a laminar flow. At the second cross-junction, the fluid stream is sheared with fluorinated oil containing 0.5% (w/w) fluorinated triblock copolymer surfactant, which makes monodisperse droplets. Subsequently, mixing of the three solutions within the droplets occurs (Figure 2B) and hydrochloric acid (HCl) dissolves bound CaCO3 nanoparticles, which leads to the activation of FXIII by Ca2+ ions and to the crosslinking of the polymeric precursors; droplets that contain no cell and as a result no Ca2+ ions do not jellify.
Figure 1. Synthetic PEG-based hydrogel for microniche formation.
8-arm PEG were functionalized with two kinds of pending linker peptides to create TG-PEG polymer precursors. Amino acids in red denote complimentary transglutaminase factor XIII (FXIII) substrate sequences. Amino acids in bold denote a matrix metalloproteinase (MMP) sensitive sequence. TG-PEG precursors are crosslinked by FXIII in a calcium dependent manner to form a covalently crosslinked synthetic matrix (TG-PEG hydrogel) that can be degraded by cell-secreted MMPs (scissors). Peptides containing amino acids serving as integrin binding site (RGD) adjacent to a FXIII substrate sequence are admixed to the reaction mixture to introduce cell adhesion sites.
Ethylenediaminetetraacetic acid (EDTA) is added to reduce background gelation of empty droplets. Of note, due to the temperature dependence of FXIII activity, which has a maximum activity at 37 °C, formation of microniches is temperature dependent. An increase in temperature from 4 °C to 37 °C is therefore used to induce gelation of microniches; droplets are collected at 4 °C and are then transferred to 37 °C. After breaking the subsequent droplet in oil emulsion using 1H,1H,2H,2H-perfluoro-1-octanol, on-demand gelled microniches are washed to remove unpolymerized TG-PEG precursors and exposed to 50 mM EDTA to remove undissolved cell-bound CaCO3 nanoparticles. Finally, they are transferred to cell culture medium (Figure 2C).
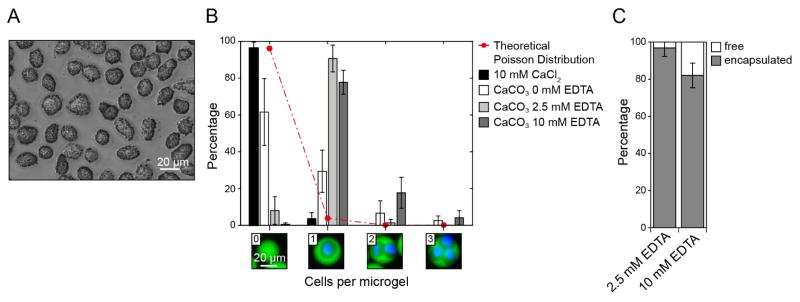
To define the EDTA concentration required to minimize background gelation, we load MSCs with CaCO3 nanoparticles (Figure 3A) and vary EDTA concentrations in the reaction mixtures in microdroplets. Microniches are formed using 15% (w/v) matrix precursor solution and 60 U mL−1 unactivated FXIII. MSCs are then injected into the microfluidic device, which has channel dimensions of 20 μm × 20 μm at the second cross-junction. Formed droplets are monodisperse and have a diameter of 22.4±0.7 μm. The dilution of injected solutions in droplets is controlled by the flow rates, and here results in a final polymer concentration of 5% (w/v) and 30 U mL−1 unactivated FXIII in individual droplets. After transfer to cell culture medium, the microniches swell by a factor of 1.6 to a diameter of 36±2 μm. To test the influence of varying EDTA concentrations, the distribution of cells among fabricated microniches is assessed (Figure 3B). As a positive control for gelation, we add 10 mM CaCl2 to the cell solution in the absence of EDTA, which results in a cell distribution that follows Poisson distribution statistics, with merely 4±3% single cell-laden microniches. The use of the cell bound CaCO3 nanoparticles as the sole calcium source, in the absence of EDTA, allows one to increase the fraction of single cell-laden microniches to 29±2%. To test if the high number of empty hydrogels results from CaCO3 nanoparticles that are not associated with cells, in spite of the rinse step, we add EDTA to the input solution containing the matrix components. Indeed, the presence of 2.5 mM or 10 mM EDTA in the droplets reduces the number of empty microniches, leading to 91±7% and 78±6% single cell-laden microniches, respectively. Although in both cases the majority of injected cells are encapsulated in microniches, 10 mM EDTA leads to significantly more free cells suggesting that it may lower calcium levels below that required for Factor FXIII activation (Figure 3C).
Figure 3. Loading of cells with CaCO3 nanoparticles and tuning of EDTA concentration in microdroplets allows for favored fabrication of single cell-laden microniches.
A) Phase contrast image of mesenchymal stem cells (MSCs) loaded with CaCO3 nanoparticles before injection into the microfluidic chip. B) Theoretical Poisson distribution (red line) and experimental distribution (bars) of MSCs in TG-PEG microniches (5% (w/v)) after their recovery from the emulsion and transfer to cell culture conditions. Channel dimensions at X-junction were 20 μm × 20 μm. Data is depicted as mean ± standard deviation (SD; number of analyzed microniches > 200). C) Percentage of MSCs encapsulated in microniches as a function of alterations in initial EDTA concentration. Data is depicted as mean ± SD (number of analyzed cells > 200).
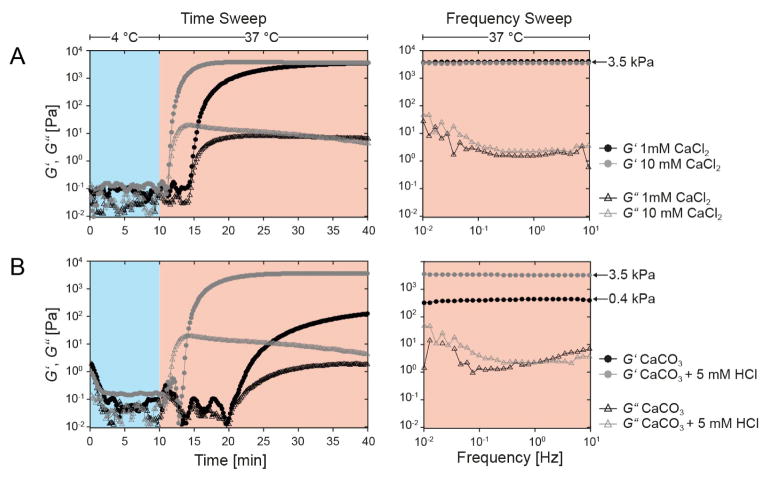
We next test if the elasticity of fabricated microniches is dependent on calcium levels present during the gelation process. To do so, we perform oscillatory rheology measurements of 5 and 7.5% (w/v) bulk hydrogels in the presence of 1 mM CaCl2 and 10 mM CaCl2 (Figure 4A and S2). Gelation in the rheology experiments is induced using an increase in temperature from 4 °C to 37 °C. Intriguingly, the 10-fold increase in calcium concentration results in accelerated kinetics, but the final storage moduli remain the same at 3.5 kPa and 6.5 kPa for 5 and 7.5% polymer content gels (w/v), respectively. This indicates that Ca2+ concentrations above 1 mM lead to complete gelation of microniches. Based on a measured nanoparticle diameter of 770±30 nm and a density of CaCO3 of 2.71 g cm−3 this corresponds to only 2.1 fully dissolved CaCO3 nanoparticles, which is significantly below the calculated number of 300±10 nanoparticles bound per cell. To assess the mechanical properties of microniches created using the on-demand gelation method, we repeat the same rheology measurement with CaCO3 nanoparticles as calcium source. At a normal reaction pH of 7.6, the resulting storage modulus is 0.4 kPa and gelation speed is also dramatically reduced (Figure 4B). Lowering of the reaction pH can efficiently dissolve CaCO3 nanoparticles. Thus, we add 5 mM HCl to the reaction to lower the pH to 6.5, and indeed the additionally available Ca2+ increases reaction kinetics and results again in a storage modulus of 3.5 kPa, equal to that observed with CaCl2 as the calcium source. Due to the pH dependency of FXIII activity,31 with an optimal activity at pH 7.6, the addition of HCl needs to be well tuned since overshooting should lead to prevention of microniche formation. Indeed, increasing HCl concentrations to 40 mM completely prevents microniche formation, resulting in 100% free cells (Figure S3A). For HCl concentrations from 5–20 mM, though, cell encapsulation yield and distribution remain at similar levels as in absence of HCl (Figure S3A and S3B). We select 2.5 mM EDTA and 5 mM HCl as input concentrations for fabrication of microniches when using channel dimension of 20 μm × 20 μm, which allows for efficiently beating Poisson encapsulation statistics and favored encapsulation of single MSCs.
Figure 4. Investigation of parameters regulating gelation kinetics and final mechanics in TGPEG microniches.
Rheology data is shown for TG-PEG precursor solutions (5% (w/v)) that are covalently crosslinked by FXIII enzymatic activity with CaCl2 (A) and CaCO3 nanoparticles (B) as calcium source. Gelation is induced by an increase in temperature from 4 °C (blue background) to 37 °C (red background). Plots show the elastic (circle, G′) and viscous (triangle, G″) parts of the complex shear modulus as a function of time (left panel) and test frequency (right panel).
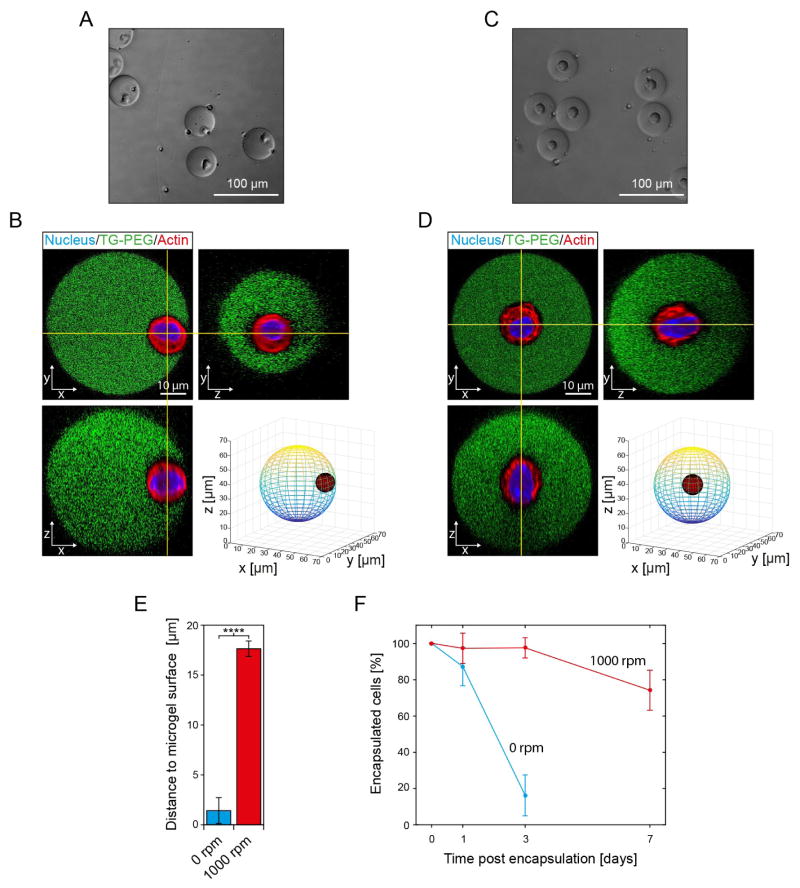
MSCs encapsulated within TG-PEG microniches have the potential to degrade the surrounding synthetic matrix via secretion of MMPs, allowing them to proliferate, spread and migrate as they would in their natural environment. Cell shape changes, presumably MMP-dependent, were observed to occur within hours after encapsulating fibroblasts in MMP-sensitive PEG gels.32 However, when staining nuclei of MSCs encapsulated in FITC-labeled TG-PEG microniches 24 hours after encapsulation we find that no overlapping signal from nuclei can be detected for 51±13% of microniches. This indicates that encapsulated MSCs have escaped from microniches within this time frame, leaving completely empty microniches behind and making efficient long-term cultures difficult. Of note, this problem has also been observed to an equal extent in microniches lacking degradation sites.21 To test if an increase in microniche diameter leads to reduced cell egress we increase the channel dimensions at the second cross-junction to 30 μm × 30 μm, which leads to fabrication of microniches with a diameter of 50.7±1.4 μm after hydrogel swelling. Furthermore, we also correct for the increase in microniche volume and adjust EDTA and HCl concentrations to 1 mM and 1.5 mM, respectively, which allows for efficient on-demand gelation (Figure S4A and S4B). MSC egress is initially reduced to 87±11% after 24 hours, but despite the increase in microniche size, only 16±11% of cells remain in their microniches after 3 days in culture. We hypothesize that this is due to the fact that initially MSCs are predominantly positioned at the edge of microniches (Figure 5A). Indeed, analysis of confocal stacks of microniches reveals that the average minimal distance from the outer edge of the cell nuclei to the microniche surface is only 1.4±1.3 μm (Figure 5B). To test if increasing the distance to the microniche surface would lead to a reduction of cell egress, we investigated techniques to influence cell position within microniches. Intriguingly, we find that orbital shaking at 1000 rpm during the gelation step at 37 °C allows for positioning of cells in the center of their microniches (Figure 5C and D). Orbital shaking at 1000 rpm significantly increases the minimal distance to the microniche surface to 17.6±0.8 μm, indicating that MSCs are indeed robustly positioned at the center of microniches (Figure 5E). What is more, positioning of MSCs in the center of microniches almost fully prevents them from escape over 3 days in culture, with 98±6% of cells encapsulated; even after 7 days 74±11% of MSCs remain encapsulated (Figure 5F). Of note, cells that escape between day 3 and 7 appear to do so by degradation of a micrometer sized channel to the microniche surface, through which they squeeze (Figure S5).
Figure 5. Positioning of cells in the center of microniches by orbital shaking the droplet in oil emulsion during gelation delays cell egress.
Single MSCs are encapsulated in microniches (5% (w/v)) using the on-demand gelation method. Channel dimensions at X-junction are 30 μm × 30 μm and 1 mM EDTA and 1.5 mM HCl are used as input concentrations. Phase contrast images of microniches 2 hours after fabrication that were gelled at 37 °C on an orbital shaker set at 0 rpm (A) or 1000 rpm (C). Corresponding confocal sections in x–y, x–z, and y–z planes of FITC-labeled TG-PEG microniches and encapsulated cells stained with Hoechst 33342 (Nucleus) and Rhodamine-phalloidin (Actin), as well as a 3D model of cell placement within the microniches for gelation at 0 rpm (B) and 1000 rpm (D). E) 3D model-based quantification of shortest distance from outer edge of cell (actin signal) to the hydrogel surface (exterior of FITC signal). Data are shown as mean ± SD (n=10) and **** indicates p < 0.0001 (Student’s t-test). F) Assessment of percentage of cells, which remain in microniches over time. Data is depicted as mean ± SD (number of analyzed cells > 100).
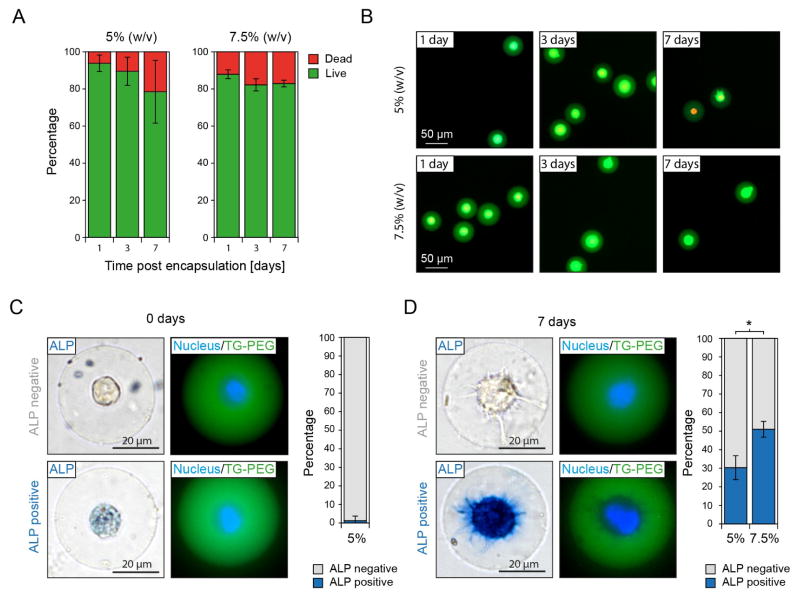
To evaluate if the protocols developed here can be used for long-term 3D culture of single cells, we encapsulate MSCs and assess cell viability in microniches. 79±17 and 83±2% of encapsulated cells are viable after 7 days in culture within 5 and 7.5% (w/v) TG-PEG microniches, respectively (Figure 6A and B). As a side note, the observed drop in percentage of encapsulated viable cells from day 3 to day 7 is largely due to viable cells being able to escape their microniches whereas all dead cells stay encapsulated. Next, we investigate if MSCs retain their inherent potential to differentiate down an osteogenic path. To ensure that loading of MSCs with CaCO3 nanoparticles does not interfere with the course of osteogenic differentiation, we load MSCs with CaCO3 nanoparticles and culture them in differentiation medium in 2D cell culture plates before assessing alkaline phosphatase (ALP) expression as an early marker for osteogenic differentiation. When comparing the number of ALP positive cells to untreated control cells we do not detect a difference after 1 or 7 days in culture (Figure S6). Next, we encapsulate single MSCs in microniches using the herein presented strategy and assess ALP expression after culture in differentiation medium. Two hours after encapsulation, only 1.3±2.5% of cells stain slightly positive for ALP expression (Figure 6C). However, after 7 days in culture within microniches, 30±6% and 51±4% of MSCs stain strongly positive for ALP in 5 and 7.5% (w/v) TG-PEG microniches, respectively (Figure 6D). This is in agreement with earlier data reporting enhanced osteogenic differentiation of mouse and human MSCs with increasing microenvironment stiffness in non-degradable 3D bulk hydrogels33 as well as the percentage of human MSCs differentiating down the osteogenic path observed in non-degradable microgels with similar stiffness.6 Taken together, this data indicates that on-demand gelation via loading of cells with CaCO3 nanoparticles, as well as cell centering during microniche formation, do not interfere with long-term cell culture and thus allow for efficiently probing single cell fate in 3D.
Figure 6. On-demand gelation is compatible with long-term culture and allows for probing single stem cell fate in 3D.
Single MSCs are encapsulated in microniches (5% and 7.5% (w/v)) applying the on-demand gelation method and positioned in the microniche center by orbital shaking (1000 rpm) of the emulsion during gelation at 37 °C. Channel dimensions at X-junction are 30 μm × 30 μm and 1 mM EDTA and 1.5 mM HCl are used as input concentrations. A) Percentage of viable encapsulated cells as function of polymer concentration at indicated time points post microniche formation. B) Representative fluorescent microscopy images of live (green) and dead (red) stained cells in microniches at indicated time points. C) Single cell-laden microniches are cultured in differentiation media for 7 days. Representative images and quantification of alkaline phosphatase expression as marker for osteogenic differentiation are shown after 2 hours (C) and 7 days (D) of culture. For all panels data is depicted as mean ± SD (number of analyzed cells > 100) and * indicates p < 0.05 (Student’s t-test).
Conclusions
In conclusion, we report a strategy based on droplet microfluidics for selective fabrication of single cell-laden synthetic microniches. The presented strategy provides a straightforward alternative to removal of empty waste droplets by sorting or to deterministic encapsulation by cell ordering or on-demand encapsulation.15 The interaction of herein used CaCO3 nanoparticles with cells is unspecific and cell type independent,16 allowing this strategy to be applied for encapsulating various cells and addressing a variety of biological questions related to single cells and microniches. To our knowledge the approach for centering cells in microniches is the first attempt to control the position of encapsulated cells within microgels and should be adaptable for other cell types. The resulting reduction of cell egress presents a simple alternative to embedding microniches in larger non-degradable hydrogels.21 We show that the presented strategy can be applied for formation of single cell-laden microniches with varying diameter and elastic moduli. With increasing elastic moduli, we observe enhanced osteogenic differentiation on a single cell level in correspondence to multiple cell-laden bulk gels. In this regard, in future work our approach can serve for studying the influence of a broad set of mechanical and biochemical matrix properties on single cells as well as the effects of a lack of cell–cell contact on cellular processes in 3D as was done in 2D to study osteogenic and adipogenic differentiation of MSCs.34,35 In addition, the tunability of microniche size, elasticity and proteolytic degradability can be used to exactly control the time of cellular egress, which is potentially appealing for therapeutic applications where encapsulated cells are ideally protected during initial stages to promote engraftment and viability and later interact with the host’s cells to stimulate repair and regeneration.36 Moreover, our approach allows for encapsulation of heterogeneous cell populations, e.g. heterogeneous tumor cell populations to derive a comprehensive understanding of the fate of individual cells for diagnostic and therapeutic strategies.37 Taken together, due to the presented platform’s versatility and its simple protocol, we expect it to benefit a wide range of applications that require long-term culture of single cells encapsulated in tailor-made microniches.
Supplementary Material
Acknowledgments
We are very grateful to Prof. David A. Weitz (Harvard University, USA) for providing the equipment to perform droplet microfluidics and Thomas Ferrante (Wyss Institute at Harvard University, USA) for support with confocal microscopy. This work has been supported by grants from the Swiss National Science Foundation (SNSF) to P.S.L. (Grant P2ELP3_161850), the German Research Foundation (DFG) to T.R. (GZ: RO 5138/1-1) and the National Institutes of Health (NIH) to D.J.M. (Grant RO1EB014703).
Footnotes
Author contributions
P.S.L., T.R., A.S.M. performed experiments. P.S.L., T.R. and D.J.M. analyzed data and wrote the manuscript. Q.V.M. and M.E. synthesized materials and contributed new reagents. All authors commented on the manuscript.
References
- 1.Velasco D, Tumarkin E, Kumacheva E. Small. 2012;8:1633–1642. doi: 10.1002/smll.201102464. [DOI] [PubMed] [Google Scholar]
- 2.Allazetta S, Lutolf MP. Curr Opin Biotechnol. 2015;35:86–93. doi: 10.1016/j.copbio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Tumarkin E, Kumacheva E. Chem Soc Rev. 2009;38:2161. doi: 10.1039/b809915b. [DOI] [PubMed] [Google Scholar]
- 4.Li CY, Wood DK, Huang JH, Bhatia SN. Lab Chip. 2013;13:1969–78. doi: 10.1039/c3lc41300d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utech S, Prodanovic R, Mao AS, Ostafe R, Mooney DJ, Weitz DA. Adv Healthc Mater. 2015:1628–1633. doi: 10.1002/adhm.201500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Y, Neubauer MP, Thiele J, Fery A, Huck WTS. Biomater Sci. 2014;2:1661–1671. doi: 10.1039/c4bm00104d. [DOI] [PubMed] [Google Scholar]
- 7.Qi M, Strand BL, Mørch Y, Lacík I, Wang Y, Barbaro B, Gangemi A, Kuechle J, Romagnoli T, Hansen MA, Rodriguez LA, Benedetti E, Hunkeler D. Artif Cells Blood Substit Immob Biotechnol. 2013:1–16. doi: 10.1080/10731190802369755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuch BE, Keogh GW, Williams LJ, Wu W, Foster J, Vaithilingam V, Phlips R. Diabetes Care. 2009;32:1887–1889. doi: 10.2337/dc09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott RB, Escobar L, Tan PLJ, Muzina M, Zwain S, Buchanan C. Xenotransplantation. 2007;14:157–161. doi: 10.1111/j.1399-3089.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 10.Köster S, Angilè FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten Ca, Pisignano D, Griffiths AD, Weitz Da. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 11.Yin H, Marshall D. Curr Opin Biotechnol. 2012;23:110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 12.C. Trials, M. S. Cells and A. Update, 2016, 25, 2015–2016.
- 13.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–7. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins DJ, Neild A, deMello A, Liu AQ, Ai Y. Lab Chip. 2015:3439–3459. doi: 10.1039/c5lc00614g. [DOI] [PubMed] [Google Scholar]
- 16.Mao AS, Shin J-W, Utech S, Wang H, Uzun O, Li W, Cooper M, Hu Y, Zhang L, Weitz DA, Mooney DJ. Nat Mater. 2016:1. doi: 10.1038/nmat4781. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gattazzo F, Urciuolo A, Bonaldo P. Biochim Biophys Acta - Gen Subj. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raeber GP, Lutolf MP, Hubbell JA. Acta Biomater. 2007;3:615–629. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Mahoney MJ, Anseth KS. Biomaterials. 2006;27:2265–2274. doi: 10.1016/j.biomaterials.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allazetta S, Kolb L, Zerbib S, Bardy J, Lutolf MP. Small. 2015;11:5647–5656. doi: 10.1002/smll.201501001. [DOI] [PubMed] [Google Scholar]
- 22.Ehrbar M, Rizzi SC, Schoenmakers RG, San Miguel B, Hubbell Ja, Weber FE, Lutoff MP. Biomacromolecules. 2007;8:3000–3007. doi: 10.1021/bm070228f. [DOI] [PubMed] [Google Scholar]
- 23.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Holtze C, Rowat AC, Agresti JJ, Hutchison JB, Angile FE, Schmitz CHJ, Koster S, Duan H, Humphry KJ, Scanga RA, Johnson JS, Pisignano D, Weitz Da, Angilè FE, Schmitz CHJ, Köster S, Duan H, Humphry KJ, Scanga RA, Johnson JS, Pisignano D, Weitz Da. Lab Chip. 2008;8:1632–1639. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
- 25.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. Nat Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lienemann PS, Devaud YR, Reuten R, Simona BR, Karlsson M, Weber W, Koch M, Lutolf MP, Milleret V, Ehrbar M. Integr Biol. 2015;7:101–111. doi: 10.1039/c4ib00152d. [DOI] [PubMed] [Google Scholar]
- 27.Metzger S, Lienemann PS, Ghayor C, Weber W, Martin I, Weber FE, Ehrbar M. Adv Healthc Mater. 2015;4:550–558. doi: 10.1002/adhm.201400547. [DOI] [PubMed] [Google Scholar]
- 28.Caiazzo M, Okawa Y, Ranga A, Piersigilli A, Tabata Y, Lutolf MP. Nat Mater. 2016 doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 29.Ehrbar M, Rizzi SC, Hlushchuk R, Djonov V, Zisch AH, Hubbell JA, Weber FE, Lutolf MP. Biomaterials. 2007;28:3856–3866. doi: 10.1016/j.biomaterials.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Salinas CN, Anseth KS. J Tissue Eng Regen Med. 2008;2:296–304. doi: 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milleret V, Simona BR, Lienemann PS, Vörös J, Ehrbar M. Adv Healthc Mater. 2013;3:508–514. doi: 10.1002/adhm.201300479. [DOI] [PubMed] [Google Scholar]
- 32.Raeber GP, Lutolf MP, Hubbell JA. Biophys J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang J, Peng R, Ding J. Biomaterials. 2010;31:2470–2476. doi: 10.1016/j.biomaterials.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Mao AS, Shin JW, Mooney DJ. Biomaterials. 2016;98:184–191. doi: 10.1016/j.biomaterials.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burdick JA, Mauck RL, Gerecht S. Cell Stem Cell. 2016;18:13–15. doi: 10.1016/j.stem.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Heath JR, Ribas A, Mischel PS. Nat Rev Drug Discov. 2015:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.