Abstract
Tumor-initiating cells (TICs), or cancer stem cells (CSC), possess stem cell-like properties observed in normal adult tissue stem cells. Normal and cancerous stem cells may therefore share regulatory mechanisms for maintaining self-renewing capacity and resisting differentiation elicited by cell-intrinsic or microenvironmental cues. Here, we show that miR-199a promotes stem cell properties in mammary stem cells (MaSCs) and breast CSCs by directly repressing nuclear receptor corepressor LCOR, which primes interferon (IFN) responses. Elevated miR-199a expression in stem cell-enriched populations protects normal and malignant stem-like cells from differentiation and senescence induced by IFNs that are produced by epithelial and immune cells in the mammary gland. Importantly, the miR-199a-LCOR-IFN axis is activated in poorly differentiated ER− breast tumors, functionally promotes tumor initiation and metastasis, and is associated with poor clinical outcome. Our study therefore reveals a common mechanism shared by normal and malignant stem cells to protect them from suppressive immune cytokine signaling.
Keywords: miRNA, mammary gland stem cells, tumor initiation, cancer stem cells, ER− breast cancer, interferon signaling, immune evasion, tumor microenvironment, epithelial-mesenchymal transition
The mammary gland epithelium is a hierarchically organized tissue with multipotent mammary stem cells (MaSCs) capable of generating luminal and basal epithelial cells1. It has been hypothesized that regulators of normal stem cell activity may be exploited by tumor initiating cells (TICs) or cancer stem cells (CSCs)2. Indeed, recent studies have revealed several cell fate regulators as such molecular links between MaSCs and breast TICs that drive their renewal activity in both normal and cancerous mammary gland tissues3–5. Poorly-differentiated tumors, typically basal-like/claudin-low or triple negative breast tumors6, 7, have high TIC activity and are enriched in CD24−/CD44+ breast CSCs8, which resemble some features of normal MaSCs5, 9, 10.
Besides their cell-intrinsic self-renewal ability, normal stem cells need to adopt additional mechanisms to fend off microenvironmental pressure that may deplete the stem cell pool. Although immune cells have been reported to be critical players in mammary gland development11, it is unknown how MaSCs control their interaction with the immune cells to sustain their stem cell activity. Interestingly, stem cells have been shown to downregulate immunogenic factors, such as MHCs, to protect themselves from immune surveillance and assure tissue regeneration12. This mechanism is also used by tumor cells to evade the immune system13, 14. However, the molecular mechanism underlying immune evasion by normal and cancerous stem cells in adult tissues remains poorly understood.
MiRNAs are critical regulators of development and cancer15, 16. MiRNAs have been shown to be expressed in a cell lineage-specific fashion in the mammary gland17, and are functionally involved in mammary gland development5, 18–21. Likewise, miRNAs display distinct expression patterns in different subtypes of breast cancer and are known to promote or suppress tumorigenesis22–24 and regulate breast CSCs25. Despite these progresses, relatively little is known about how miRNAs regulate the interaction of stem cells with immune microenvironment.
In this study, we identified a critical role of miR-199a in promoting MaSC and TIC properties by direct repression of LCOR, a nuclear receptor corepressor that sensitize cells to interferon-induced differentiation and senescence. The miR-199a-LCOR axis represents a conserved molecular pathway in normal and cancer stem cells that mediate their evasion from autocrine and immune microenvirontment suppressive signals.
Results
Systematic screening reveals the MaSC-promoting activity of miR-199a
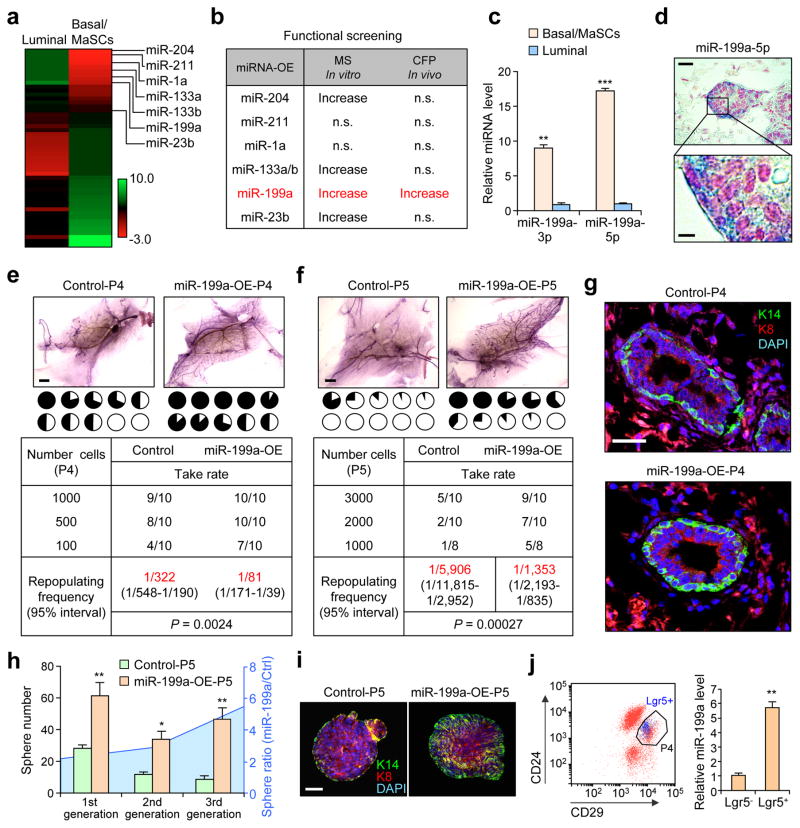
To identify candidate miRNA regulators of MaSCs, we performed miRNA profiling of the Lin−CD24+CD29high MaSC-enriched basal population (hereafter denoted as P4) and the Lin−CD24+CD29low luminal population (P5) of primary mammary epithelial cells (MEC) isolated by fluorescence-activated cell sorting (FACS) from the mouse mammary gland (Supplementary Fig. 1a). Twenty-three miRNAs were significantly up-regulated in P4 versus P5 by more than two fold (Fig. 1a). Based on the degree of differential expression of miRNAs and their predicted mRNA targets of interest, we selected 7 of the 23 up-regulated miRNAs (miR-204, miR-211, miR-1a, miR-133a, miR-133b, miR-199a and miR-23a) for further functional analysis. We used lentiviral vectors to transduce primary MECs to test these candidates in mammosphere assays (MS) in vitro and cleared fat pad (CFP) reconstitution assays in vivo (Fig. 1b and Supplementary Fig. 1a). Interestingly, only miR-199a overexpression (OE) led to a significant increase in both assays (Fig. 1b). We confirmed by qPCR that higher expression of both mature forms (3p and 5p) of miR-199a in P4 versus P5 cells (Fig. 1c). In situ hybridization (ISH) confirmed elevated expression of miR-199a in basal cells compared to luminal cells in the mammary gland (Fig. 1d).
Figure 1. miR-199a is enriched in MaSCs and is functionally critical for MaSC activity.
(a) Heat map representing miRNAs with >2-fold differential expression between P4 and P5 cells. (b) Table of selected miRNAs used for in vitro mammosphere (MS) and in vivo cleared fat pad (CFP) reconstitution analyses. (c) qRT-PCR analysis of the expression levels of the 3′ and 5′ arms (3p and 5p) of miR-199a in P4 compared to P5. n=4 biologically independent samples; data represented mean ± SEM. (d) In situ hybridization analysis (ISH) of miR-199a-5p in the terminal end buds (TEBs). miR-199a is stained blue and nuclei are stained in red. (e) P4 and (f) P5 cells transduced with the indicated constructs are used for limiting dilution cleared fat pad reconstitution assay. Representative images show outgrowth. Each pie chart represents a mammary gland with the blackened area denoting the percentage of mammary gland outgrowth. Tables below represent serial dilution injections with the corresponding take rate. n= number of mammary fat pad injections as indicated in the table. Shown in red are the repopulation frequencies for each condition and P value by Pearson’s Chi-squared test, obtained with the ELDA software. (g) Krt14 (K14-green) and Krt8 (K8-red) staining with reconstituted mammary outgrowths from control and miR-199a-OE P4 cells. (h) Number of P5 mammospheres formed after 3 generations of in vitro passage, and the ratio of sphere number between miR-199-OE group vs. control. 5,000 cells in the indicated conditions were seeded (n=3 biologically independent samples; data represents mean ± SEM). (i) Confocal K14+K8 staining images of mammospheres from control and miR-199-OE P5 cells. (j) Left: Flow cytometry isolation of P4-Lgr5+ and P4-Lgr5− cells from the Lgr5-EGFP knockin reporter mouse glands. Lgr5+ cells are represented in blue dots. Right: qRT-PCR analysis of miR-199a expression in Lgr5+ and Lgr5− P4 cells (n=3 biologically independent samples; data represents mean ± SEM). Scale bars: 20 μm (upper panel) and 5 μm (lower panel) in d, 2 mm in e and f, 25 μm in g, and 40 μm in i. * P<0.05, ** P<0.01, *** P<0.005 by two-tailed Student’s t-test in bar graphs.
We next examined the ability of miR-199a to induce MaSC activity in total MECs (P3), P4 or P5 cells (Fig. 1e, f and Supplementary Fig. 1b, c). Ectopic miR-199a OE enhanced stem cell activity of P4 cells in CFP repopulation assay (Fig. 1e) while the opposite was observed with miR-199a knockdown (KD) (Supplementary Fig. 1d). Importantly, ectopic miR-199a expression in P5 cells also increased the repopulation frequency, indicating that miR-199a can induce a stem cell-like state in luminal cells (Fig. 1f). Characterization of the repopulated mammary gland tissues showed an increase in the basal/MaSC marker Keratin14 (Fig. 1g and Supplementary Fig. 1e, f). Serial passage and transplantation assays further confirmed that an increased and sustained capacity of sphere formation, basal phenotype, and regenerating ability was induced by miR-199a in three successive generations of passage (Fig. 1h, i and Supplementary Fig. 1g). Finally, to investigate the relevance of miR-199a in MaSC populations, we used the Lgr5 knock-in EGFP reporter mouse26 to isolate Lgr5+ and Lgr5− cells. A significant increase of miR-199a expression was observed in Lgr5+ versus Lgr5− P4 cells (Fig. 1j), consistent with reported higher MaSC activity of the Lgr5+ P4 population26. Overall, these results suggest that miR-199a functionally promote MaSC activity.
miR-199a induces stem cell-like gene signatures and is up-regulated in CSC populations
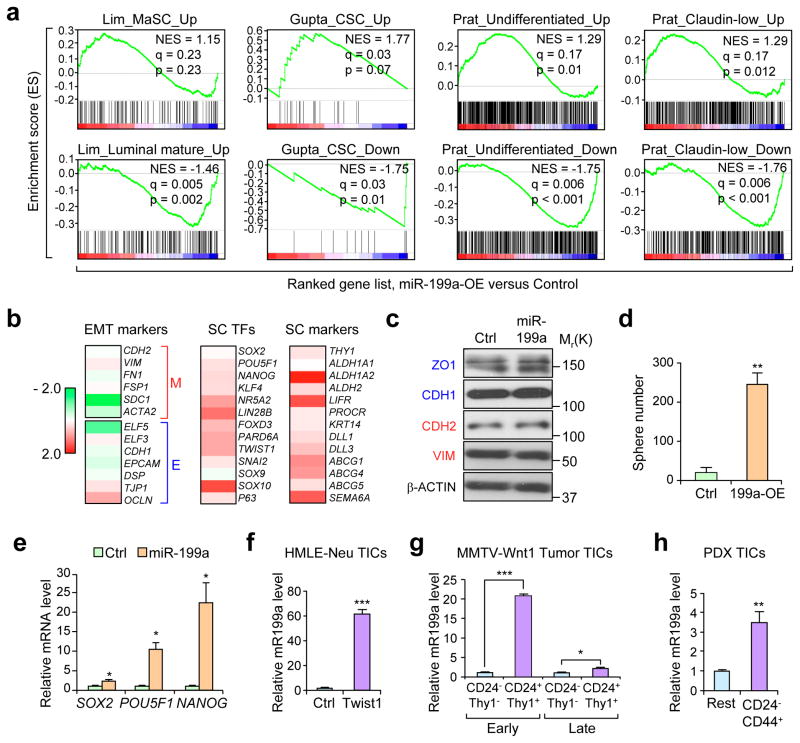
To explore the downstream signaling of miR-199a, we used the immortalized human MEC line HMLE to stably overexpress miR-199a and perform expression profiling. Gene set enrichment analysis (GSEA) revealed that the miR-199a-overexpression resulted in enrichment of gene sets related to MaSC27, CSC28, undifferentiated tumor cell populations and the Claudin-Low (CL) breast cancer signature7 (Fig. 2a). Conversely, negative enrichments of luminal differentiated signatures, CSC downregulated genes, undifferentiated and CL downregulated gene-sets were observed after miR-199a overexpression (Fig. 2a). To further investigate the connection of miR-199a with MaSCs and breast cancer, we next applied a Euclidean centroid-based CL-predictor and stratified the TCGA dataset including the CL subtype, which has been linked to MaSCs based on transcriptomic studies7. Notably, we observed a large overlap between CL tumors and MaSCs at the miRNA level, with miR-199a being one of the most significantly up-regulated common miRNAs among MaSCs and CL tumors (Supplementary Fig. 2a and Supplementary Table 1).
Figure 2. miR-199a induces stem cell-like gene signatures and is enriched in cancer stem cells.
(a) GSEA demonstrating the enrichment of gene sets related to MaSC27, CSC28, undifferentiated tumor cells7, and Claudin-Low tumors7 in the ranked gene list of miR-199a-OE vs. control HMLE cells. (b) Heat map of HMLE-miR-199a-OE microarray data representing fold change expression of EMT markers, stem cell transcription factors (SC-TFs), and stem cell (SC) markers. Fold change is represented as Log2 ratio. (c) Western blot analysis of epithelial (blue) and mesenchymal (red) markers. (d) In vitro quantification of mammospheres formed by 2,000 control or miR-199a-OE HMLE cells seeded. (e) qRT-PCR of mRNA extracted from 5 day HMLE control or miR-199a-OE mammospheres. (f-h) qRT-PCR of miR-199a levels in HMLE-Neu-Twist1-ER-OE tumor initiating cells (TICs) (f), CD24+/Thy1+ TICs isolated from early and late stage spontaneous MMTV-Wnt-1 tumors (g), CD24−/CD44+ TICs isolated from HCI-002 human breast cancer PDX (h) as compared to the non-TIC counterparts (n=3 biologically independent samples; data represents mean ± SEM) in d–h. *P<0.05, **P<0.01, ***P<0.005 by two-tailed Student’s t-test in d–h.
MaSCs resemble cells that undergo EMT based on gene expression signatures, and activated pathways29. Additionally, gene expression profiles of MaSC populations overlap with Claudin-low breast cancers, which harbor EMT-like properties and are enriched for TICs27, 30. Interestingly, although miR-199a is downstream of EMT signaling, being up-regulated by TGF-β and TWIST1 (Supplementary Fig. 2b) as previously reported31, it did not induce EMT in HMLE cells by itself (Fig. 2b, c and Supplementary Fig. 2c). However, miR-199a strongly potentiated sphere formation (Fig. 2d) and increased the expression of several stem cell transcription factors and markers (Fig. 2b, e and Supplementary Fig. 2d), indicating miR-199a as a prominent downstream effector of EMT signaling to induce stemness while having no direct role in influencing epithelial or mesenchymal phenotypes. This is consistent with recent findings suggesting that TWIST1 controls tumor maintenance and stemness independently of its function in regulating EMT32, and the existence of mesenchymal-like10 and epithelial-like CSCs33, 34.
To explore whether miR-199a might also be important in TIC/CSC populations, we isolated different TICs populations in various breast cancer models. We used Twist1-induced TICs in HMLE-Neu cells10, CD24+/Thy1+ cells in the MMTV-Wnt-1 mouse breast tumor35, and the CD24−/CD44+ population8 in HCI-002 patient-derived xenografts (PDX)36. In all of these TIC/CSC populations, miR-199a displayed elevated expression compared to the bulk population (Fig. 2f–h). These data are consistent with a previous study showing upregulation of miR-199a in human breast CSCs5. Overall, these observations suggest miR-199a to be an important regulator of MaSCs and breast CSCs.
LCOR is a direct miR-199a target that suppresses MaSC activity and is downregulated in MaSCs and CSCs
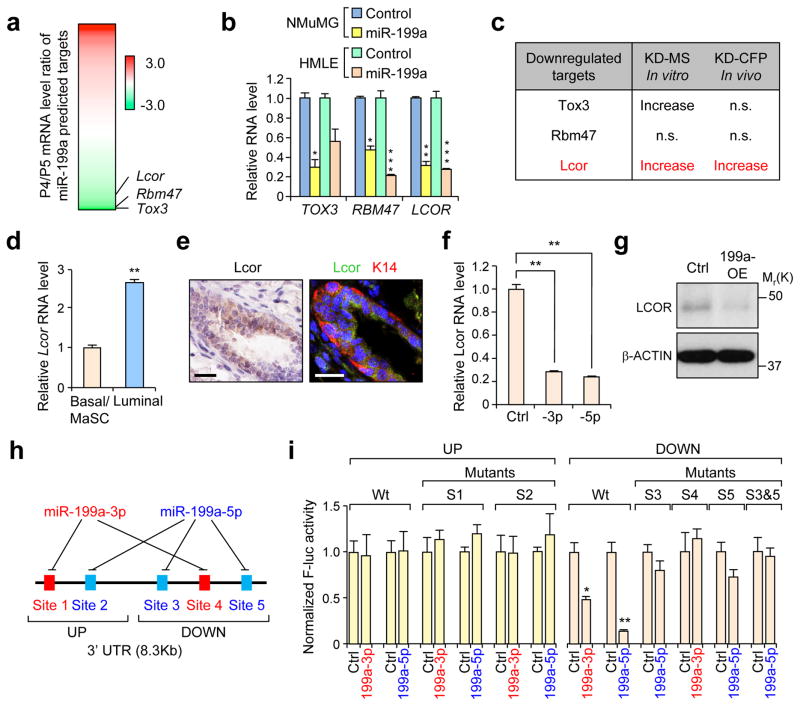
In order to identify potential targets of miR-199a responsible for the observed phenotype, we focused on genes downregulated in MaSCs (P4) that are: 1) predicted to have direct miR-199a binding sites (Fig. 3a and Supplementary Table 2), and 2) downregulated after miR-199a-OE in HMLE and NMuMG cells (Fig. 3b). Based on these criteria, we selected Tox3, Rbm47 and Lcor as candidate functional targets of miR-199a (Fig. 3a). In functional assays for MaSC activity, only Lcor-KD increased both sphere formation in vitro and mammary gland reconstitution in vivo (Fig. 3c). In addition, we validated that Lcor is highly expressed in the luminal compartment (Fig. 3d, e and Supplementary Fig. 3a), and especially in mature luminal cells (P5-CD61−) compared to luminal progenitors (P5-CD61+) (Supplementary Fig. 3b). We next confirmed that transient or stable miR-199a-OE consistently represses LCOR in 10 different normal and malignant mammary cell lines derived from human or mice (Fig. 3f, g and Supplementary Fig. 3c, d). Furthermore, to assess the direct repression of Lcor by miR-199a, we cloned the mouse Lcor 3′UTR into a luciferase reporter plasmid. The Lcor 3′UTR is 8.3Kb long and contains 5 different evolutionarily conserved predicted binding sites for miR-199a: 2 sites for the miR-199a-3p and 3 sites for the miR-199a-5p (Fig. 3h). Due to the length of the 3′UTR, we cloned it as two separate fragments: UP and DOWN (Fig. 3h, i). We confirmed direct targeting of the three binding sites in DOWN 3′UTR that can be blocked by mutations; however, the two sites in UP 3′UTR did not show repression by miR-199a. Overall, this data implicates Lcor as a major candidate downstream effector of miR-199a.
Figure 3. Identification of LCOR as a direct target gene of miR-199a.
(a) Heat map showing the differential expression of predicted miR-199a target genes (TargetScan v7.0) in P4 vs P5 cells. Fold change is represented as Log2 ratio. (b) qRT-PCR analysis of top candidate genes in mouse NMuMG and human HMLE cells after transfection with the indicated miRNA mimics. (c) Summary of the knockdown effect of the selected candidate miR-199a targets in functional assays using mouse MECs. MS: mammosphere formation assay; CFP: cleared fat pad mammary reconsititution assay. (d) qRT-PCR analysis of Lcor expression in different lineages of mouse MECs. (e) Immunohistochemistry (IHC) (left panel) analysis of Lcor and immunofluorescence (right panel) of Lcor and K14 localization in the mammary ducts. (f) qRT-PCR analysis of LCOR after 48h transfection of miR-199a-3p and miR-199a-5p in HMLE cells. (g) Western blot analysis of LCOR in control and miR-199a-OE HMLE cells. (h) Schematic diagram of the miR-199a binding sites on the LCOR 3′UTR. (i) Normalized activity of the luciferase reporter containing the WT mouse Lcor 3′ UTR or various miR-199a seed sequence mutants, after co-transfection with miR-199a-3p or miR-199a-5p, or control miRNAs in Hela cells. The reporters were divided in two groups, UP and DOWN, containing upper and lower halves of the Lcor 3′-UTR, respectively. Scale bars: 20 μm in e. n=3 biologically independent samples; data represents mean ± SEM in b, d, f and i.* P<0.05, ** P<0.01, *** P<0.005 by two-tailed Student’s t-test in bar graphs.
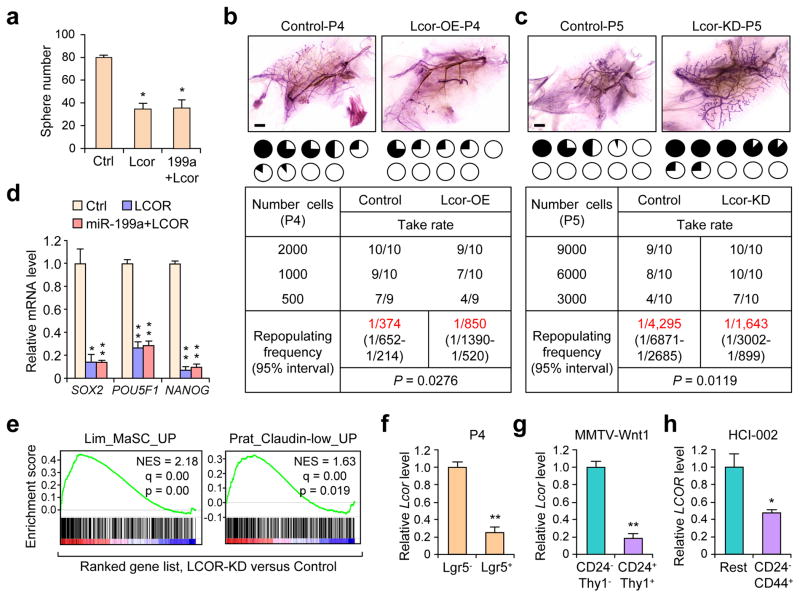
We next examined the functional role of Lcor in the mammary gland. Ectopic overexpression of Lcor in P4 cells (Supplementary Fig. 4a) decreased MaSC activity as measured in sphere-forming (Fig. 4a) and reconstitution assays (Fig. 4b). Conversely, Lcor-KD in P5 cells increased MaSC activity and phenocopied miR-199a overexpression (Fig. 4c and Supplementary Fig. 4b). Moreover, Lcor rescue in miR-199a-OE P4 cells completely nullified the MaSC-promoting effect of miR-199a, and suppressed the induction of stem cell transcription factors by miR-199a in HMLE cells (Fig. 4d) (Fig. 4a and Supplementary Fig. 4c). Likewise, LCOR-KD in HMLE cells increased sphere formation (Supplementary Fig. 4d, e), and LCOR-OE reduced it (Supplementary Fig. 4f). GSEA showed enrichment of the MaSC signature and the CL predictor gene set in HMLE-LCOR-KD cells versus control (Fig. 4e), consistent with the miR-199a-OE transcriptional changes. Moreover, LCOR is strongly downregulated in the Lgr5+ MaSC population (Fig. 4f). Consistent with its inverse expression with miR-199a in normal mammary gland, we also observed that Lcor is downregulated in various TIC populations (Fig. 4g, h).
Figure 4. LCOR suppresses MaSC function and is downregulated in stem cell populations.
(a) Quantification of mammosphere formation of 20,000 P4 cells in the indicated conditions (n=3 biologically independent samples; data represents mean ± SEM). (b–c) Limited dilution cleared fat pad reconstitution assay of P4 (b) and (c) P5 cells after transduction with the indicated constructs. Representative images show the outgrowth. Each pie chart represents a mammary gland with blackened area showing the percentage of mammary gland outgrowth. Tables below represent serial dilution injections with the corresponding take rate. n= number of mammary fat pad injections as indicated in the table. Shown in red are the repopulation frequencies for each condition and P value by Pearson’s Chi-squared test, obtained with the ELDA software. (d) qRT-PCR of mRNA extracted from mammospheres formed by HMLE cells after transduction with the indicated constructs (n=3 biologically independent samples; data represents mean ± SEM). (e) GSEA demonstrating the enrichment of gene sets related to MaSC27 and Claudin-low tumors7 in the ranked gene list of LCOR-KD vs control HMLE cells. (f–g) qRT-PCR analysis of Lcor expression in Lgr5+ MaSC-enriched P4 cells (f), CD24+/Thy1+ MMTV-Wnt-1 TICs (g) and CD24−/CD44+ TICs isolated from HCI-002 PDX (h) as compared to their non-stem cell counterparts (n=3 biologically independent samples; data represents mean ± SEM). Scale bars: 2 mm in b and c. * P<0.05, ** P<0.01 by two-tailed Student’s t-test in bar graphs.
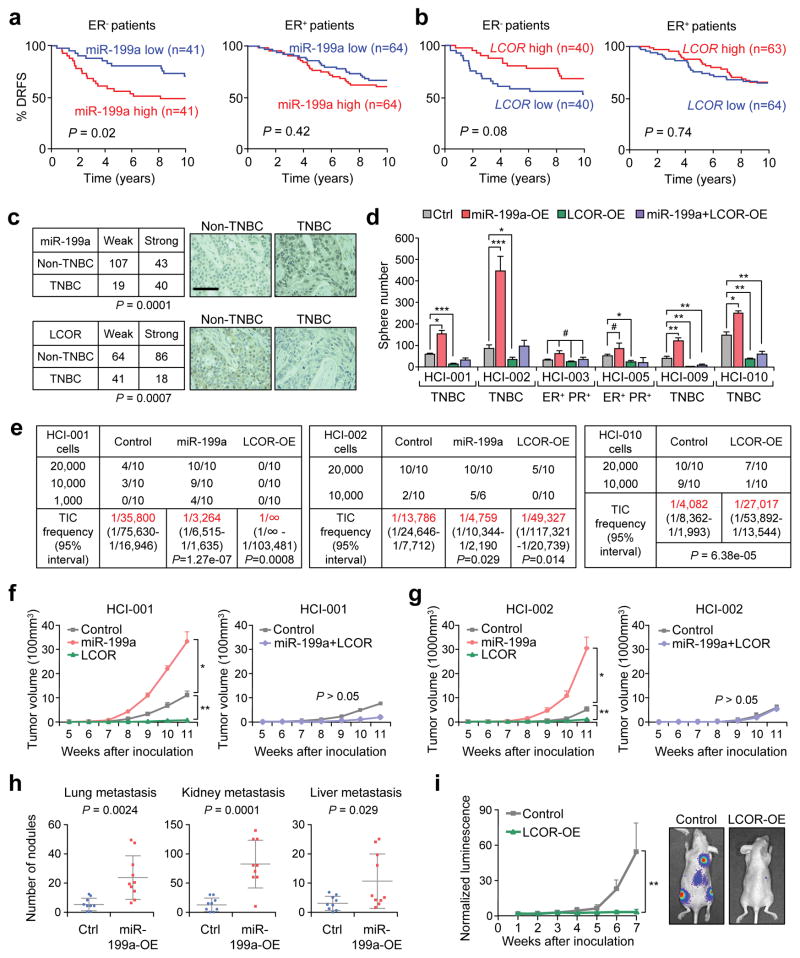
The miR-199a-LCOR axis promotes TIC activities in ER− breast cancer
We evaluated the clinical relevance of the miR-199a-LCOR axis using the Buffa dataset37. Interestingly, miR-199a displayed a poor prognosis value and LCOR a good prognosis in the ER− patients but not in ER+ patients (Fig. 5a, b). We confirmed miR-199a and LCOR as independent prognostic markers in a triple negative breast cancer (TNBC)-specific dataset38 (Supplementary Fig. 5a, b). Additionally, LCOR mRNA is downregulated in TNBC compared to non-TNBC tumors (Supplementary Fig. 5c), and the analysis of 209 human breast tumor tissues showed high miR-199a and low LCOR levels in TNBC by ISH and IHC, respectively (Fig. 5c), with inverse correlation to each other (Supplementary Fig. 5d). In the NKI295 dataset, LCOR also shows good prognosis for distant metastasis-free survival (Supplementary Fig. 5e). Overall, these analyses indicate that the miR-199a-LCOR axis is clinically relevant in ER− breast cancer.
Figure 5. miR-199a and LCOR functionally influence the initiation of ER− breast tumors in vivo.
(a–b) Kaplan-Meier distant relapse-free survival (DRFS) curve of breast cancer patients with higher or lower than median RNA expression levels of (a) miR-199a and (b) LCOR in their tumors37. (c) miR-199a and LCOR protein expression levels in TNBC (n=59 patient samples) and non-TNBC tumors (n=150 patient samples). Each sample was scored as weak (low expression) or strong (high expression) according to staining intensities of miR-199a by ISH and LCOR by IHC. (d) Quantification of tumorspheres formed by 10,000 cells from multiple human breast cancer PDXs in different tumor subtypes with the indicated conditions (n=3 biologically independent samples; data represents mean ± SEM). (e) Tumor take rate of HCI-001, HCI-002 and HCI-010 upon MFP injection of indicated cells. n=number of MFP injections as indicated in the table. Tumor initiating cell (TIC) frequency calculated by the ELDA software is indicated in red. (f–g) Tumor growth of HCI-001 (f) and HCI-002 (g) upon MFP injection of 20,000 cells in the indicated conditions (n=10 mouse mammary glands). (h) Metastatic nodule counts in the indicated organs 10 days after intracardiac (I.C.) injection of 100,000 4TO7 cells in Balb/c mice (n=10 mice). Each dot represents a value and the lines represent the mean and SD. (i) Bioluminescence imaging (BLI) quantification of the metastatic growth of the control and LCOR-KD MDA-MB-231 cells after intracardiac (I.C.) injection of 100,000 cells in Ncr-nu/nu mice (n=10 mice). Scale bar: 100 μm in c. P-value by log-rank test in a and b, Fisher’s exact test in c. P value by Pearson’s Chi-squared test in e. * P<0.05, ** P<0.01, *** P<0.005 by two-tailed Student’s t-test in d, f, g, h and i. # P>0.05 in d.
In order to investigate the functional relevance of the miR-199a-LCOR axis in ER− breast cancer, we next used PDX36 in tumorsphere forming assays in vitro and mammary fat pad (MFP) tumorigenesis assay in vivo using NSG mice (Supplementary Fig. 5f). Intriguingly, ectopic miR-199a expression significantly increased tumorsphere formation of the TNBC PDXs (HCI-001; HCI-002; HCI-009; and HCI-010) but not in the ER+PR+ HCI-003 and HCI-005 (Fig. 5d). Likewise, LCOR OE reduced tumorsphere formation in the TNBC PDXs, and nullified the tumorsphere-promoting effect of miR-199a (Fig. 5d). Consistent with the tumorsphere assays, miR-199a expression strongly increased tumor initiation of ER− PDXs (Fig. 5e–g and Supplementary Fig. 5g–h); but not in the ER+ HCI-003 (Supplementary Fig. 5g). Conversely, LCOR-OE reduced tumor initiation in HCI-001, HCI-002 and HCI-010 (Fig. 5e–g). In addition, miR-199a OE stimulated the development of systemic metastasis of the weakly metastatic 4TO7 mouse tumor cells. Conversely, LCOR overexpression strongly suppressed metastasis of MDA-MB-231 breast cancer cells (Fig. 5h, i). Collectively, these data demonstrate that the miR-199a-LCOR axis, besides its function in regulating normal MaSCs, is also a critical modulator of CSCs/TICs in ER− breast cancer, which are known to be enriched in TIC/CSCs.
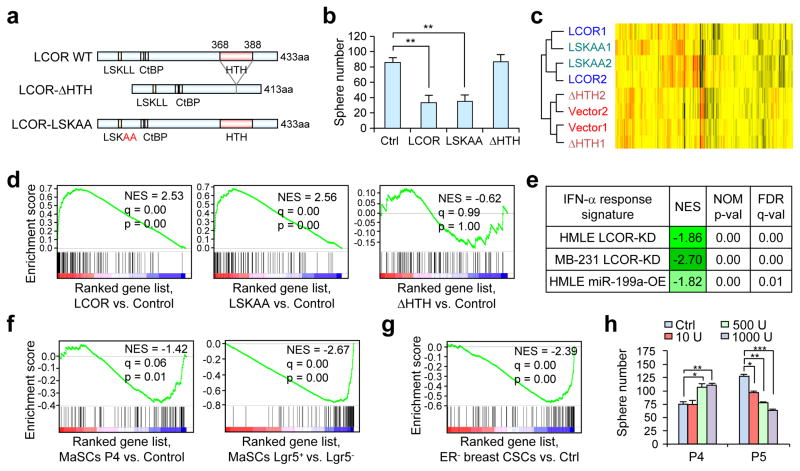
miR-199a-LCOR modulates the IFN-α response of normal and cancerous stem cells
LCOR is a co-repressor of agonist-bound nuclear receptors (NR), but can also directly bind to DNA through its HTH domain39. We generated a double-point mutation of the NR box (LSKLL to LSKAA) to abolish NR binding40, and a HTH domain deletion mutant defective in direct DNA binding (Fig. 6a). Co-immunoprecipitation revealed near complete loss of interaction of LSKAA with the estrogen receptor (ER) (Supplementary Fig. 6a), and immunofluorescence analysis confirmed that the ΔHTH mutant maintains nuclear localization (Supplementary Fig. 6b). Strikingly, disruption of the NR interaction did not diminish the ability of LCOR to suppress sphere formation of HMLE cells while ΔHTH lost such function (Fig. 6b). These results suggest that LCOR acts through DNA binding to suppress stem cell properties.
Figure 6. LCOR primes the IFN-α response.
(a) Schematic representation of LCOR mutants. (b) Quantification of mammospheres formed by 5,000 HMLE cells with ectopic expression of the indicated LCOR constructs (n=3 biologically independent samples; data represents mean ± SEM). (c) Unsupervised hierarchical clustering of the HMLE expressing various LCOR constructs based on transcriptomic profiles. (d) GSEA of the IFN-α response gene-set (M5911) in the ranked gene list of LCOR, LSKAA and ΔHTH vs. control HMLE cells. (e) GSEA of the IFN-α response gene set in the ranked gene list of the LCOR-KD or miR-199a-OE vs. control HMLE and MDA-MB-231 cells. (f) GSEA of the IFN-α response gene set in the indicated gene list from the current study. (g) GSEA of the IFN-α response gene set in the gene list of CD24−/CD44+ CSC vs. non-CSCs in ER− breast cancer9. (h) Quantification of mammospheres formed by P4 (20,000) and P5 (10,000) cells treated with different doses of IFN-α, from 10 to 1000 U/ml in 8 days (n=3 biologically independent samples; data represented mean ± SEM). * P<0.05, ** P<0.01 by two-tailed Student’s t-test in b and h.
Global transcriptomic profiling clustered LCOR-OE cells together with LCOR-LSKAA, and LCOR-ΔHTH with control cells (Fig. 6c). Using GSEA, we observed that the most enriched gene sets in LCOR-OE cells were related to the IFN-α and IFN-γ responses (Supplementary Table 3). Importantly, the ΔHTH mutant, but not LSKAA, completely lost this enrichment of the IFN-α signature (Fig. 6d and Supplementary Table 3). Consistently, LCOR-KD HMLE and MDA-MB-231 cells showed a strong negative enrichment of the IFN-α response gene set (Fig. 6e). Such negative enrichment is also observed in HMLE-miR-199a-OE cells, suggesting that miR-199a, through LCOR repression, reprograms the transcriptome to suppress the IFN-α response. We further performed GSEA of the same gene sets in stem cell enriched populations versus non-stem cell populations. Strikingly, the IFN-α signature showed negative enrichment in P4-MaSC, and this negative enrichment bias was even more accentuated in the Lgr5+ MaSCs (Fig. 6f). Similar negative enrichment of the IFN-α signature was also observed in ER− CD24−/CD44+ breast CSCs9 (Fig. 6g). These findings further suggest a muted interferon response in MaSCs and breast CSCs.
To directly evaluate the functional importance of interferon signaling in stem cell regulation, we performed mammosphere assays using P4 and P5 (luminal cells have high sphere formation capacity due to progenitor proliferation41) treated with IFN-α and IFN-γ. The P5 cells, which have higher LCOR levels, responded to the IFN-α treatment with reduced sphere formation ability (Fig. 6h and Supplementary Fig. 6c). In contrast, P4 cells showed an increase in sphere formation (Fig. 6h and Supplementary Fig. 6c), suggesting that MaSCs and luminal cells respond differently to IFN-α.
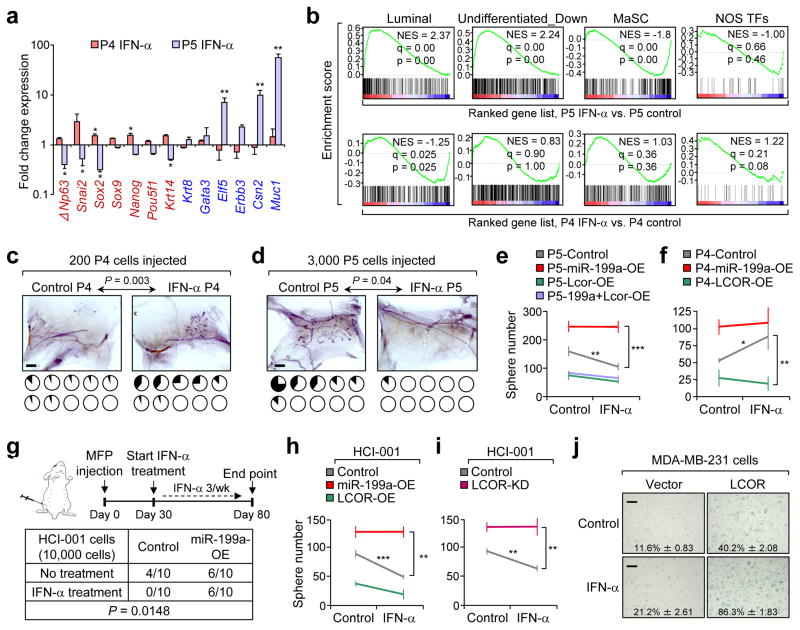
Interestingly, stem cell-related genes (in red) were up-regulated in P4 spheres and downregulated in P5 spheres upon IFN-α treatment (Fig. 7a). Moreover, luminal differentiation genes (in blue) were up-regulated in P5, and not in P4, upon IFN-α treatment (Fig. 7a). GSEA revealed enrichment of luminal and differentiated gene sets in P5 cells after IFN-α treatment, while MaSC and stemness gene sets were enriched in P4 treated cells (Fig. 7b). Consistent with the genomic profiling result, IFN-α induced the basal/MaSC marker K14 in P4 spheres, and the luminal marker K8 in Lcor-OE P4 spheres (Supplementary Fig. 7a).To further investigate the IFN-α effects in vivo, we performed CFP injections of P4 and P5, followed by IFN-α administration for 3 weeks. P4 cell reconstitution was slightly increased by the subcutaneous IFN-α treatment while P5 cell reconstitution was severely suppressed (Fig. 7c, d), again highlighting the differential response of P4 and P5 cells to IFN-α in vivo.
Figure 7. Stem cells and differentiated cells respond differently to the IFN-α signaling.
(a) qRT-PCR result showing the fold change of stem cell-related genes (red) and luminal differentiation-related genes (blue) in mammospheres formed by P4 and P5 with or without IFN-α 1000 U/ml treatment for 6 days. (n=3 biologically independent samples; data represented mean ± SEM of fold change in IFN-α vs Ctrl). (b) GSEA of the MaSC and luminal upregulated gene sets generated in this study, as well as Nanog-Oct4-Sox2 transcriptional factors (NOS-TFs) targets gene set6 and undifferentiated downregulated genes7 in the ranked gene list of IFN-α-treated P5 and P4 cells cells vs. control. (c–d) Cleared fat pad reconstitution assay of 200 P4 (c) and 3,000 P5 (d) cells after trice a week treatment of 100,000 U IFN-α for 3 weeks. Representative images show the outgrowth. Each pie chart represents a mammary gland with blackened area showing the percentage of mammary gland outgrowth (n=10 mammary glands injected); P value by two-tailed Student’s t–test). (e–f) Quantification of mammospheres formed by 10,000 P5 (e) and P4 (f) cells with or without IFN-α treatment after transduction of the indicated constructs (n=3 biologically independent samples; data represents mean ± SEM). Plots represent the enhanced difference of treated conditions vs. the control conditions (without treatment). (g) Tumor take rate upon MFP injection of 10,000 control and miR-199a-OE HCI-001 cells, with or without treatment with 100,000 U IFN-α as indicated in the schematics. P value calculated by one-way ANOVA of the tumor incidence. (h–i) Quantification of PDX cell tumorspheres formed by 10,000 HCI-001 cells, with the indicated conditions (n=3 biologically independent samples; data represents mean ± SEM). (j) Senescence-associated β-galactosidase (SA-β gal) assay of MDA-MB-231 cells comparing control and LCOR-OE cells, and with or without IFN-α treatment for 72 hours (n=3 biologically independent samples; data represents mean ± SEM; source of data in Supplementary Table 4). Scale bars: 2 mm in c, d, and 100 μm in j. * P<0.05, ** P<0.01, *** P<0.005 by two-tailed Student’s t-test in a, e, f, h and i.
We next determined how miR-199a and ectopic LCOR expression affects these responses to IFN-α. Ectopic expression miR-199a in P5 prevented mammosphere reduction upon IFN-α treatment (Fig. 7e), while Lcor expression in P4 and HMLE cells sensitized them to the suppressive effect of IFN-α (Fig. 7f and Supplementary Fig. 7b). Similarly, IFN-α treatment in the HCI-010, a PDX highly enriched in TICs (Fig. 5e), did not reduce sphere formation; however, LCOR-OE sensitized HCI-010 to IFN-α-induced reduction of tumorspheres (Supplementary Fig. 7c, d). In contrast, IFN-α treatment in miR-199a-OE or LCOR-KD HCI-010 cells increased sphere formation (Supplementary Fig. 7c, d), suggesting that the TIC-enriched population resists the interferon blockage on sphere formation. Another PDX line (HCI-001), which has relatively lower TIC activity, is sensitive to IFN-α treatment in vivo and in vitro (Fig. 7g–i), but was rendered insensitive to IFN-α after miR-199a-OE (Fig. 7m, n) or LCOR-KD (Fig. 7i).
Overexpression of WT but not the ΔHTH mutant of LCOR partially induced senescence in MDA-MB-231 and HMLE cells, and strongly sensitizes them to interferon-mediated senescence (Fig. 7j and Supplementary Fig. 7e). Consistent with these findings, GSEA revealed a significant enrichment of the senescence gene set in LCOR-OE but not in ΔHTH-OE HMLE cells, and a negative enrichment in miR-199a-OE HMLE cells (Supplementary Fig. 7f).Overall, these findings indicate that the miR-199a-LCOR axis modulates the sensitivity of normal and cancerous stem cells to the differentiation and senescence effects of IFN-α.
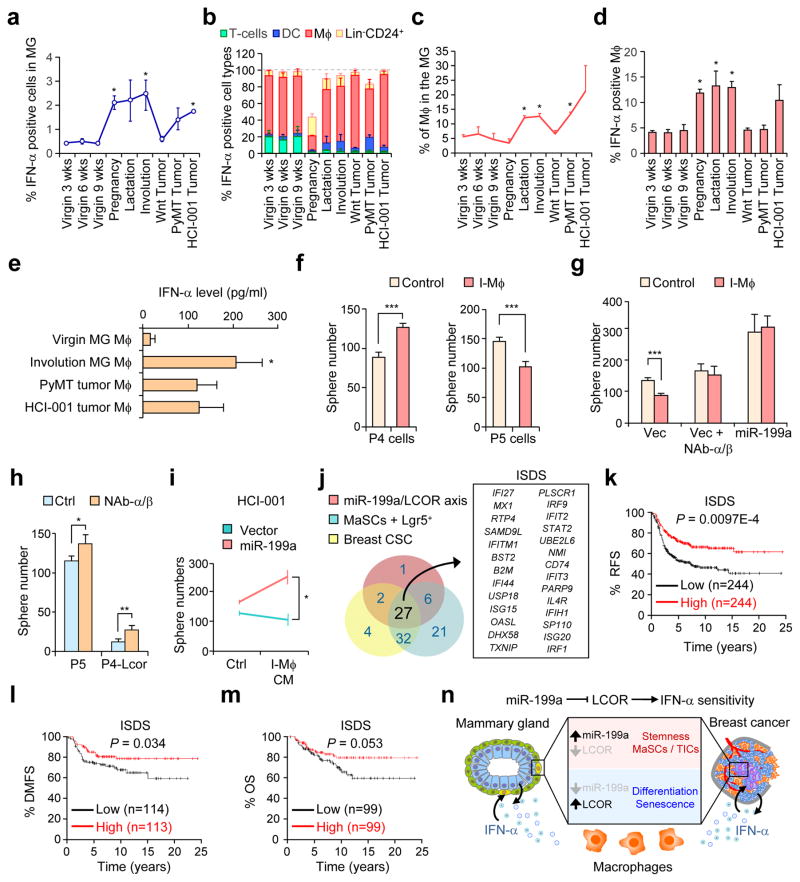
MaSCs and breast CSCs are protected from suppressive effects of immune and autocrine IFN-α
To investigate the expression levels and sources of IFN-α in the mammary gland during different physiological and malignant states, we performed flow cytometry after co-staining intracellular IFN-α with various lineage markers of immune cells. Importantly, virgin mice already had presence of IFN-α expressing cells in the mammary gland and a substantial increase was observed during pregnancy, lactation and involution (Fig. 8a and Supplementary Fig. 8a). Interestingly, most of the IFN-α-expressing cells were macrophages across all the different stages of the mammary gland, while T-cells and dendritic cells were a relatively insignificant source of IFN-α (Fig. 8b). Importantly, high levels of infiltration (Fig. 8c) and activation (Fig. 8d) of macrophages in the mammary gland were observed during lactation and involution. In addition, the macrophages from the virgin mammary glands already expressed more interferons than peritoneal macrophages (Supplementary Fig. 8b). Beside immune cells, a small source of IFN-α-expressing cells corresponded to the Lin−CD24+ epithelial cells (Fig. 8b). This IFN-α-positive population increases significantly during pregnancy, consistent with the major expansion of the epithelial tissue at this stage. In breast tumors, different tumor types also showed elevated IFN-α levels and increased infiltration of IFN-α-positive macrophages (Fig. 8a, c, e and Supplementary Fig. 8a).
Figure 8. Immune and autocrine IFN related effects on mammary gland and tumor cells.
(a) Percentage of total IFN-α-expressing cells from digested mammary glands and tumors at the indicated stages, analyzed by flow cytometry after intracellular IFN-α staining of single cell suspensions. (b) Relative percentage of IFN-α positive cell types of the mammary gland, analyzed by flow cytometry after co-staining intracellular IFN-α with CD3e (T-cells), CD11c (dendritic cells), F4/80 (macrophages) and Lin−CD24+ (epithelial cells). (c) Flow cytometry analysis showing the percentage of the F4/80 (macrophages) positive cells in the mammary gland at different stages. (d) Flow cytometry analysis of the percentage of IFN-α positive cells within the total mcarophage population. In a–d n=4 biologically independent samples; data represents mean ± SEM. (e) Quantification of IFN-α levels in the CM of the indicated cells, detected by ELISA (n=3 biologically independent samples; data represents mean ± SEM). (f) Quantification of mammospheres formed by P4 (20,000 cells), P5 (10,000 cells) and (g) P5-miR-199a-OE (10,000 cells) cells treated 1:3 with CM from involution macrophages and neutralizing antibodies (NAb) against IFN-α/β (2.5 μg/ml) (n=3 biologically independent samples; data represents mean ± SEM). (h) Quantification of mammospheres formed by 10,000 P5 and P4-Lcor cells with or without treatment with NAb against IFN-α/β (2.5 μg/ml) (n=5 biologically independent samples; data represents mean ± SEM). (i) Quantification of PDX cell tumorspheres formed by 10,000 HCI-001 treated 1:3 with CM from involution macrophages and conditions indicated (n=3 biologically independent samples; data represents mean ± SEM). (j) Schematic diagram showing the compilation of the 27-gene ISDS. See Methods for details. (k–m) Kaplan-Meier relapse-free survival (RFS) (k), distant metastasis-free survival (DMFS) (l), and overall survival (OS) (m) analysis of the ISDS gene signature in ER− breast cancer using the KM plotter42. (n) Schematic model for the conserved function of the miR-199a-LCOR axis in allowing the evasion of normal mammary gland and breast cancer cells from macrophage-derived and autocrine IFN-α. * P<0.05 by Student’s t-test in a, c and d respect to the virgin 9 week condition. * P<0.05, ** P<0.01, *** P<0.005 by Student’s t-test in e–i. P-value by log-rank tests in k–m.
We next isolated F4/80+ macrophages from the mammary gland at different states to generate conditioned media (CM) and validated IFN-α secretion by ELISA (Fig. 8e). Interestingly, CM of mammary gland macrophages (MG-Mϕ) significantly increased P4 and decreased P5 sphere formation, but such an effect was lost after miR-199a OE in P5 cells (Fig. 8f, g). The effect of CM from MG-Mϕ on P5 spheres was abolished after treatment with neutralizing antibodies (NAbs) against IFN-α/β (Fig. 8g), proving that these effects were mediated by IFNs. Moreover, NAbs against IFN-α/β consistently increased P5 and P4-Lcor-OE sphere formation even without the presence of immune cells (Fig. 8h), indicating that P5 or P4-Lcor cells are also constrained by autocrine IFN-α/β signaling. Similarly, MG-Mϕ CM also reduced the HCI-001 tumorsphere formation, and this is avoided by miR-199a-OE (Fig. 8i). These results demonstrate that, by virtue of elevated miR-199a expression and reduced LCOR levels, both normal and cancerous stem cells are protected from immune or autocrine/paracrine interferon-mediated suppressive effects.
To further explore the clinical significance of the miR-199a-LCOR-IFN-related regulatory pathway in breast cancer, we generated an IFN-Stem Cell-Down signature (ISDS) from the Interferon-α response gene set (M5911) to represent Interferon-α responsive genes that are regulated by the miR-199a-LCOR axis and also contribute to negative enrichment of the M5911 in normal and cancerous stem cells. The combined 27-genes constitute the ISDS (Fig. 8j) and have a good prognosis value for relapse-free survival (RFS), overall survival (OS) and distant metastasis-free survival (DMFS) in ER− breast cancer patients from the KM plotter dataset42 (Fig. 8k–m). The individual analysis of the ISDS genes in ER− breast cancer showed a good prognosis value for all of them, with 21 out of 27 being significant (Supplementary Fig. 8c), highlighting the clinical relevance of the miR-199a-LCOR-IFN axis in ER− breast cancer.
Discussion
In this study, we identified a miRNA-199a-mediated pathway shared by both MaSCs and breast CSCs to maintain their self-renewal competence and avoid differentiation or senescence induced by suppressive immune cytokines such as IFN-α (Fig. 8n). MiR-199a have been reported to have either tumor suppressive functions43, 44 or tumor promoting activities45 across different cancer types, including breast cancer46, 47. Here, we provide evidence for an important functional role of miR-199a in promoting MaSC activity by directly repressing LCOR, a nuclear receptor corepressor. LCOR has been proposed as a tumor suppressor in prostate cancer48; however, there is no previous report of LCOR function in mammary gland development or breast cancer. We identified LCOR as a direct functional target of miR-199a in regulating MaSC and breast CSC activities. Our experimental and clinical data show that the miR-199a-LCOR axis mainly influences tumorigenesis of ER− breast cancer, suggesting LCOR is not acting through ER binding. This is also consistent with our results showing the maintenance of LCOR function in the LSKAA mutant. Therefore, the LCOR action on stem cells and breast cancer is independent of the ER, which is consistent with its function in MaSCs (which are ER−) and ER− breast cancer.
Exploring the downstream effects of miR-199a-LCOR, we showed that LCOR negatively regulates stem cells by sensitizing them to interferon responses. Interferon signaling is known to be critical in anticancer immunesurveillance of primary tumors and metastasis49, 50. Interferons can also induce tumor cell-intrinsic inhibitory effects, including differentiation, growth arrest, and cell death50, 51. However, less is known about the effects of interferon on stem cells. Some studies have found different effects of IFN-α on hematopoietic stem cells, as it can activate dormant hematopoietic stem cells (HSCs) but inhibit active HSCs52, or drive exhaustion of quiescent HSCs53. Therefore, IFNs can play opposite roles in stem cell fate in the hematopoetic system, depending on target cell status and on acute or chronic signaling52. Whether this is also the case in other adult stem cell systems and in cancer stem cells was unknown. Here we show that IFN can have different effects on MaSCs or differentiated cells, depending on the status of the miR-199a-LCOR axis. Remarkably, we found that IFN-α response is attenuated in normal and malignant stem cell populations based on GSEA, indicating that low IFN sensitivity is a critical and general mechanism to maintain the stem cell phenotype.
Taken together, our study reveals a miR-199a-LCOR-IFN-dependent mechanism that is commonly used by MaSCs and CSCs to escape from differentiation and senescence induced by IFN signaling, which is particularly relevant during mammary gland lactation and involution54, and in immune cell-rich Claudin-low and TNBC tumors7, 55. Moreover, normal stem cells may use this mechanism to acquire immune privilege properties, as they do by downregulating MHC to assure tissue homeostasis. Since MHCs are regulated by IFN12, stem cells may downregulate MHCs by suppressing IFN signaling. Accordingly, CSCs are less responsive to IFN and can escape immune surveillance, which is a critical ability during tumor and metastasis initiation events56, 57. However, CSCs may have a defective anti-viral interferon-mediated response, which may explain why oncolytic viruses specifically target CSC populations58, 59. In fact, the interferon response is frequently defective in multiple cancer types by genetic or epigenetic alteration of related genes60, 61, suggesting that defective IFN responses are advantageous for tumors. As IFNs have been widely used as adjuvant therapy in multiple cancer types, such treatments may become more effective if the IFN-resistant CSCs can be rendered sensitive by targeting the miR-199a-LCOR axis.
METHODS
Animal studies
Animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Princeton University. For mammary stem cell isolation, 8 to 9 week-old female FVB virgin mice were used. The Lgr5-EGFP-IRES-creERT2 (C57/B6 background) model was used for MaSCs Lgr5+ isolation (Lin−CD24+CD29highGFP+) as previously described26. For cleared fat pad injections, 3 to 4 week-old female FVB mice were anesthetized and minimal incisions were made to expose the mammary gland. Randomization among litters was performed before the injection time, and animals were similar age and female sex (apply to the rest of the experiments using mice). No statistical method was used to predetermined the sample size (apply to the rest of the experiments using mice). Following standard protocol, the mammary gland clearing was done above the central lymph node in inguinal gland #4, and different quantities of cells (as indicated in the figure or figure legend) were injected into the cleared fat pad as previously described62. The investigators were not blinded to allocation during experiments and outcome assessment. For the IFN-α treatment experiment, 100,000 U of recombinant mouse IFN-α2 was subcutaneously administered 48 hours after surgery, three times a week for 3 weeks. A MMTV-Wnt1 mouse model of spontaneous breast cancer was used to isolate mouse tumor-initiating cells. MMTV-PyMT and MMTV-Wnt1 mouse model of spontaneous breast cancer were used to measure IFN-α level in various immune cell populations in tumors and isolate macrophages. For orthotopic mammary tumor experiments of human patient-derived xenografts (PDX; kindly provided by Dr. Alana Welm) including HCI-001, HCI-002, HCI-003, HCI-005, HCI-009 and HCI-010, immunocompromised NOD Scid Gamma (NSG) mice were used. Note that NSG mice lack mature T-cells, B-cells and NK cells, but they have dendritic cells and macrophages, although with reduced activity63, 64. We followed the standard protocol for PDX transplantation, maintenance and digestion of the tumors36. The lentiviral transduction and orthotopic injections of PDX single cell suspension were optimized for primary tumor initiation experiments. 6 to 10 mice or glands were used for each experimental group and the primary tumors were monitored weekly by palpation. For the IFN-α treatment experiment, subcutaneously injection of 100,000 U of recombinant human IFN-α2A was administered three times a week for 50 days, started at day 30 after tumor inoculation (prior to the formation of palpable tumors) and ended at the endpoint of the experiment (Fig. 6m). Tumor monitoring and measurement were performed by trained technicians in a blinded fashion. Tumors were measured by calipers for calculation of tumor volumes (π x length x width2/6). For systemic metastasis experiments using 4TO7 and the MDA-MB-231 cell line, intracardiac injection of 100,000 cells in the left ventricle was performed in anesthetized female athymic Ncr-nu/nu mice. Development of metastases was monitored by blinded investigators and measuring photon flux of metastatic lesions based on bioluminescence imaging (BLI) as previously described65, and nodule counts were obtained after dissection of the different organs (investigators were not blinded to outcome assessment).
Cell lines, culture conditions and treatments
All cell lines used in the study, including mammary epithelial cell lines (human HMLE, MCF10A and mouse NMuMG cell lines), breast cancer cell lines (human MDA-MB-231, HMLE-Neu, T47D, MCF7, BT474, and mouse 4TO7 cell lines) and other cell lines (HEK293T, H29 and HeLa), were cultured using the standard conditions according the American Type Culture Collection (ATCC) instructions. HMLE and HMLE-Neu cells were obtained from Dr. Robert Weinberg at MIT. iMMEC cells were obtained from Dr. Vassiliki Karantza at CINJ. Primary isolated mammary epithelial cells (MECs) were cultured with MEGM (Lonza) and immortalized murine MECs (iMMECs) were cultured as described66. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell lines were not authenticated. Mycoplasma contamination was routinely checked (monthly) in the lab by PCR analysis; all cell lines used in the study were confirmed to be mycoplasma negative. TGF-β1 (R&D systems) and Z-4-hydroxytamoxifen (4-OHT) (Sigma-Aldrich) cell culture treatments in vitro were done for 12 days, at 100 pM and 20 nM, respectively.
Limiting dilution assays
For mammary gland reconstitution assays, we prepared single cell suspensions of MECs and sorted for P4/P5 cells by flow cytometry. Lentivirally transduced cells were injected in serial dilution numbers into cleared mammary fat pads. Cells were injected in 50% matrigel and outgrowth was analyzed after 6 weeks. ELDA (Extreme limiting dilution analysis) software was used to calculate the frequency of MaSCs with 95% of confidence. The same quantitative method was used in limiting dilution mammary fat pad injections of PDX cells to calculate TIC frequency. The PDX transduced single cell suspensions were also injected in 50% matrigel into the mammary fat pad of NSG mice. Tumors were considered established when they became palpable for 2 consecutive weeks.
Viral production and infection of cell lines and primary cells
Lentiviral plasmids were transfected into HEK293T cells together with the envelope plasmid (VSVG) and gag-pol plasmid (pCMV-dR8.91) following the standard lentiviral packaging protocol to generate lentiviruses. For retrovirus, the pWZL-ER-Blast retroviral vectors were transfected into the H29 packaging cell line and viruses were collected 48 and 72 hours after transfection. Primary cells were spin infected in conical tubes for 2 hours at 1000g at 4°C with concentrated viruses in media containing 8 μg/ml polybrene. After spin infection, cells were counted and used for in vitro or in vivo experiments. For established cell lines, cells were transduced in culture and selected with the corresponding antibiotic resistance.
Immune cell isolation
F4/80+ macrophages were obtained by FACS from from 3 day involuting mammary glands of 9–10 week FVB mice. Primary cells were cultured in mammosphere media (MSM) media for 48 hours to generate the CM, which was then used to treat mammospheres.
Senescence assays
Cell lines were cultured and senescence was evaluated after 2–3 days of IFN-α treatment (1000U/ml human IFN-α2A) using the β-Galactosidase staining kit (Cell Signaling Technology) following manufacturer instructions. Images were taken with 10x objective in bright field.
Molecular cloning and Plasmids
Multiple miRNA (mouse miR199a-2, miR-204, miR-211, miR-1a, miR-133a, miR-133b, miR-23b) and gene (mouse Lcor and human LCOR-HA, LCOR-LSKAA-HA and LCOR-ΔHTH-HA) expression constructs were generated using the pLEX-MCS-Puro lentiviral vector. cDNA was introduced into pLEX using the SpeI and AgeI cutting sites. For LCOR-HA, LCOR cDNA was first introduced into pRVPTO-HA using EcoRI and NotI. The resultant LCOR-HA was then subcloned into the pLEX-MCS using SpeI and AgeI. The LCOR mutants (LCOR-LSKAA-HA and LCOR-HTH-HA) were generated from the pLEX-LCOR-HA construct by two separate PCR amplifications with the mutation region joining the two PCR fragments. The 5′ fragment was amplified using a forward primer paired with specifically designed mutation-containing reverse primers, which end on the intended mutation region or right before it. The 3′ fragment was amplified using the mutation-containing forward primers paired with a reverse primer (see sequence in the table below). To enable blunt end ligation and cloning, the mutant harboring primers were phosphorylated using T4 kinase before the PCR reactions. The amplified 5′ and 3′ fragments were digested with SpeI and AgeI respectively and ligated into the pLEX-MCS. Retroviral tamoxifen inducible overexpression plasmids pWZL-ER-Blast and pWZL-Twist1-ER-Blast were obtained from Addgene. For miR-199a knock down, miRZIP technology from System Biosciences (Moutain View, CA) was used to generate pGreenPuro (scrambled hairpin control), miR-ZIP-199a-5p, and miR-ZIP-199a-3p. For gene knock-down studies, shRNA lentiviral vectors were purchased from Sigma-Aldrich for targeting mouse genes Lcor (TRCN0000085107), Tox3 (TRCN0000413123), and Rbm47 (TRCN0000123514), and human LCOR (TRCN0000016306 and TRCN0000436034). The pMIR-REPORT vector (Ambion) was used to generate luciferase reporters for miRNA targeting activity. Wild type and mutant mouse Lcor 3′UTRs were cloned into pMIR-REPORT using SpeI and HindIII sites. The 3′UTR-Lcor sequence were divided into UP and DOWN segments, as the full length 3′UTR exceeded 8Kb and could not be cloned in its entirety. Single and double mutants were generated for the 5 different predicted seed sequences. Mutated 3′UTR seed sequences were generated using the QuikChange multi site-directed mutagenesis kit (Stratagene). All primer sequences used for cloning are listed in the Supplementary Table 5 (restriction sites are in green and mutated nucleotides are in red and bolded).
Flow cytometry analysis and cell sorting
For mammary gland lineage cell isolation, we followed the standard protocol used previously62. Briefly, mammary glands from 8–9 week-old female mice were digested to form single cell suspensions of primary MECs. These cells were sorted using Lin−CD24+CD29high markers to obtain P4 (MaSCs/basal) cells, Lin−CD24+CD29low markers to obtain P5 (luminal) cells, and total Lin− for P3 (total MECs). We used CD61+ to isolate luminal progenitors and CD61− for mature luminal cells within the P5 population. For Lgr5+ MaSC isolation, the Lgr5-EGFP-IRES-creERT2 mice were used and Lgr5+ MaSCs were isolated by FACS using the Lin−CD24+CD29highGFP+ marker combination. F4/80+ primary macrophages were isolated by FACS from single cell suspensions of digested mammary glands from FVB mice. Mouse TICs were obtained from MMTV-Wnt1 tumors after sorting for the CD45−CD24+Thy1+ population. Digestion of PDX tumors and preparation of single cell suspension were performed using the standard protocol36 and human primary TICs were isolated by sorting the Lin−CD24−CD44+ population.
Intracellular IFN-α flow cytometry analysis
Mammary glands and tumors were digested and brought to single cell suspension as described above and with the presence of Golgi-plug as indicated by the manufacturer (BD Biosciences). After fixation with 2% paraformaldehyde 30 minutes on ice, cells were permeabilized and blocked with 0.2% saponin, 5 mM EDTA, 2 mM NaN3, 5% NGS and 4 μg/ml anti-Fcγ receptor (Clone 2.4 G2; BD Biosciences). IFN-α was stained using FITC-conjugated anti-mouse IFN-α (Clone RMMA-1; PBL)67. T-cells were stained with PE anti-mouse CD3ε (Clone 145-2C11; BioLegend), macrophages with APC/Cy7 anti-mouse F4/80 (Clone BM8; BioLegend), and dendritic cells with APC anti-mouse CD11c (Clone HL3; BD Pharmingen). FITC-conjugated rat IgG1 was used as negative isotype control (Clone RTK2071; BioLegend). Note: pregnancy, lactation and involution cell samples display small levels of auto-fluorescence for reasons unknown. We thoroughly excluded this population of our analysis and applied the same gating for all the all samples.
Histological analysis, immunohistochemistry (IHC); immunofluorescence (IF) and in situ hybridization (ISH)
Histology, IHC, and IF analysis of mouse mammary and tumor tissue samples was done as previously described68 using the following antibody and dilution ratio (Supplementary Table 6).
For IHC analysis of clinical specimens, paraffin slides of 4 μm thickness or tissue microarrays were baked overnight at 60°C. Tissue slides were washed with PBS after deparaffinization and hydration and then boiled in citrate buffer at 100°C for 40min. After treated with 3% H2O2 for 30 min to block endogenous peroxidase, slides were incubated at 4°C overnight with rabbit anti-human LCOR antibody (Sigma). Following washes with PBS, slides were then incubated with HRP-conjugated goat anti-rabbit secondary antibody (Genetech) for 30 min at RT. Sections were stained by DAB and then counterstained with Gill hematoxylin.
IF analysis of cell culture was performed in HMLE cells using anti-HA to determine the localization of ectopically expressed LCOR-HA. Sterile coverslips placed at the bottom of 24-well plates were seeded, washed with PBS, and fixed for 1 hour with methanol at −20°C. After fixation, samples were washed with acetone, then 5 times with PBS, blocked for 30 minutes with blocking buffer (5% normal goat serum, 0.5% Triton X-100 in PBS) and incubated with anti-HA for 2 hours at room temperature (RT). This was followed by PBS washes, and 1 hour incubation with secondary antibody conjugated with Alexa Fluor 488. Images were taken using a Nikon A1 confocal microscope and Zeiss fluorescence microscope. For mammosphere staining, spheres were collected with a 5 min spin at 450g, fixed with PFA 4% for 15 min at 4°C and washed with PBS, followed by centrifuge colony precipitation. Blocking was performed using PBS-Tween 0.3% and M.O.M. kit blocking reagent (Vector Laboratories), followed by 1 hour co-staining with anti-Keratin-14 and anti-Keratin-8, and then by 1 hour incubation with the respective species-specific secondary antibodies.
For mammary gland in situ hybridization (ISH) experiments, we used the miRCURY LNA™ microRNA ISH Optimization Kit from Exiqon. LNA probes were double DIG labeled to specifically detect miR-199-5p and U6 snRNA as a positive control (Exiqon). Manufacturer’s protocol was strictly followed to perform the ISH in mouse and human samples. For clinical breast cancer ISH, paraffin slides (4-μm thick) of paraformaldehyde-fixed tissues were baked overnight at 60°C, deparaffinized and hydrated, then washed with phosphate-buffered saline (PBS). To block endogenous peroxidase activity, slides were treated with 3% H2O2 for 10 min at RT. After pepsin digestion for 30 min at 37°C, slides were incubated with pre-hybridization buffer (all reagents from sensitivity enhanced in situ hybridization kits, Boster, Wuhan, China) for 3 hr at 37°C. Slides were hybridized with double digoxigenin-labeled probes (30 nM for miR-199a, 30 nM for the internal control U6, Exiqon, Vedbæk, Denmark) in hybridization buffer at 65°C overnight, then washed sequentially with 2×SSC, 1×SSC and 0.2×SSC buffers. Slides were incubated sequentially with the following reagents: blocking buffer, biotinylated digoxin, streptavidin-biotin complex, and peroxidase. Sections were stained by 3,3-diaminobenzidine (DAB) and then counterstained with Gill hematoxylin.
Mammosphere and Tumorsphere assays
For mammosphere and tumorsphere assays, single cells were plated in ultra-low attachment plates (Corning) with the standard mammosphere media69. The number of cells plated is indicated for each specific experiment in the figures. The mammospheres were counted 5–12 days later depending on the experiment, which is indicated in figure legends. Tumorspheres were counted after 5–8 days. For multiple generation sphere formation experiments, colonies were collected by 2 min centrifugation at 200g and dissociated with trypsin for 5 min at 37°C. Single dissociated cells were then centrifuged and counted to allow seeding of equal numbers of cells for the next round of sphere formation assays.
Interferon treatment (1000 U/ml) was started 24 hours after cell seeding. Recombinant mouse IFN-α2 and IFN-γ were purchased from Novoprotein. Neutralizing antibodies (NAb) against mouse IFN-α (Clone RMMA-1, PBL) and IFN-β (Clone RMMB-1, PBL) were used at 2.5 μg/ml. Conditioned media (CM) was generated by culturing primary immune cells in mammosphere media (MSM) during 48 hours, using 1 ml of MSM for every 600,000 primary cells seeded. Conditioned media (CM) treatments were done as indicated in Fig. 7a.
Tissue microarrays and other tumor samples
A total of 200 stage I to III primary breast cancer samples from females with invasive ductal carcinoma were randomly collected at the Fudan University Shanghai Cancer Center (FUSCC, Shanghai, P.R. China) between March 2003 and January 2008. Tissue microarrays were constructed using paraffin-embedded blocks of these samples, consisting of duplicate cores from different areas of the same tumor to compare staining patterns. We also included nine cases of primary metaplastic carcinoma diagnosed in the same time period. Paraffin-embedded sections were used for in situ hybridization and immunohistochemical staining. We used the staining index to interpret the staining of miR-199a and LCOR. Briefly, the staining score was determined by 3 blinded independent researchers to the tumor information. Each sample was scored as weak (1) or strong (2) according to staining intensities and the average of the resultant scores was computed. Our study was approved by the independent ethics committee/institutional review board of FUSCC (Shanghai Cancer Center Ethics Committee). All patients gave their written informed consent before inclusion.
Murine IFN-α ELISA
Specific cell populations (500,000 cells) were plated for 48 hours with subsequent collection of conditioned media for cytokine quantification. IFN-α levels were quantified from the CM for all cell cultures using the mouse IFN-α Platinum ELISA kit (Fisher/eBioscience; San Diego, CA) following the manufacturer’s instructions.
Luciferase reporter assays
Wildtype and mutant pMIR-LCOR-3′UTR repoters were transfected into HeLa cells together with the Renilla-luciferase control plasmid (Ambion). 200ng of reporter plasmid was co-transfected with the renilla-luciferase control plasmid and 10pM of miRNA mimics (Applied Biosystems; Life Technologies). Lipofectamine 2000 was used as the transfection reagent. Cells were lysed 24 hours after transfection and analyzed for luciferase activity using the Glomax 96 Luminometer (Promega).
qRT-PCR analyses
Total mRNA and miRNAs were isolated using the mirVana miRNA isolation Kit (Ambion). mRNA reverse transcription was done using Superscript III kit (Invitrogen) and real-time quantitative PCR performed using the Power SYBR green PCR master mix (Applied Biosystems). miRNAs were reverse transcribed using the TaqMan Reverse Transcription Kit (Applied Biosystems) and followed by real-time qPCR using TaqMan miRNA assays (Applied Biosystems). All analyses were performed using an ABI 7900HT PCR machine. mRNA expression was normalized by the expression of GAPDH, and miRNA expression by RNU6B in each sample. qRT-PCR primers used are listed in the Supplementary Table 5.
Immunoprecipitation and western blot analysis
For immunoprecipitation (IP) experiments, cells were lysed in lP lysis buffer (20 mM Tris pH7.4, 0.15 M NaCl, 1 mM EDTA, 1mM EGTA, 1% Tx-100) with complete protease inhibitor cocktail (Roche). Cell lysates were incubated on ice for 20 minutes, centrifuged, and incubated with 5μg of anti-HA (Abcam, Ab9110) at 4°C over-night. Protein G magnetic beads (Life Technologies) were pre-cleared and 20 μl were added to the sample for 2 hours at 4°C. Beads were washed 5 times and boiled with SDS laemmli buffer to elute bound protein for western blotting (WB). For WB analysis of cultured cells, proteins were extracted using RIPA buffer and SDS laemmli buffer. Gel preparation and immunoblotting were performed according to standard protocols. Antibodies and dilutions for WB are listed in the Supplementary Table 6.
Microarray analysis
The P4 and P5 subpopulations of mammary epithelial cells (MECs) were isolated from the mammary glands (4–5 mammary glands from each group) of 8–9 week old virgin FVB or Lgr5-EGFP-IRES-creERT2 mice. MECs were isolated using FACS as previously described62. Total RNA was prepared from MECs and various human cell lines including HMLE-miR-199a, -LCOR variants and -LCOR-KDs or MDA-MB-231 LCOR-KD cells, using the mirVana kit as described above. P4 and P5 with IFN-α treatment (1000 U/ml) were maintained in mammosphere conditions during 8 days of mammosphere culture. The expression of miRNA in MEC subpopulations was determined using Agilent mouse miRNA_V19 array (G4872A). The RNA samples were labeled and hybridized using a miRNA complete labeling and hybridization kit (Agilent, 5190-0456). The expression of mRNA in MECs was determined with Agilent Mouse GEv1 8x60K Microarray (G4852A). The mRNA expression in human cell lines was determined with Agilent human GEv2 8x60k microarray (G4851B). The mRNA microarray analyses were performed using a two-color system. Briefly, the RNA samples and universal mouse reference RNA (Agilent 740100) were labeled with CTP-cy5 and CTP-cy3, respectively, using the Agilent Quick Amp Labeling Kit. Labeled testing and reference RNA samples were mixed in equal proportions, and hybridized to the arrays as described above. After hybridization, the miRNA and mRNA arrays were scanned with an Agilent G2565BA scanner and raw data was extracted using Agilent Feature Extraction software (v10.7). Data were analyzed using the GeneSpring GX software (Agilent). In brief, for one color miRNA array data, the intensity values of multiple probes for the same miRNA are first summed up and then log2 transformed. The values were then normalized to 90 percentiles of each sample and finally baseline transformed with median across all samples. For two color mRNA array data, for each probe, the Log2(Cy5/Cy3) ratio is computed and used as expression value.
Gene set enrichment analysis (GSEA)
Normalized microarray Log2 ratio expression data was rank-ordered by differential expression between cell populations using a fold change metric. Multiple probes for the same gene were collapsed into one value by the highest probe reading when there were fewer than 3 probe matches, and median when there were 3 or more probe matches. Interrogated signatures include HALLMARK gene sets from the MSigDB database v5.1 release; MaSCs (GSE19446) with 489_UP and 428_DOWN genes qualifying for >1.5-FC (fold change) and FDR<0.05; and Senescence dataset (M9143) with 77_UP genes. Other interrogated datasets include the CSC dataset (GSE17215) with 25_UP and 14_DOWN genes qualifying for >3-FC; Undifferentiated tumor cell dataset (GSE18229) with 558_UP and 490_DOWN and Claudin-low dataset (GSE18229) with 437_UP and 370_DOWN genes qualifying for FDR<0.05. The NOS_TFs gene set with 37 genes were derived from published gene list6, and the MaSCs 230_UP genes and Luminal 230_UP genes were derived from the current study (GSE85808). In addition, the published breast cancer stem cell expression dataset9 was extracted from Gene Expression Omnibus (GEO) GSE52262 and analyzed using GeneSpring GX software. Using this dataset, we generated an ER− BCSC dataset by a median compilation of the 4 different ER− breast cancer cell lines and xenografts (HCC1954, MC1, SUM149, and SUM159) isolated by CD24−/CD44+9. Gene signatures were tested using default enrichment GSEA statistics and compared to enrichment results from 1000 random permutations to obtain p-value, q-value, and normalized enrichment score (NES).
The IFN-Stem Cell-Down signature (ISDS) was generated to represent genes that are regulated by the miR-199a-LCOR axis and also contribute to negative enrichment of the Interferon-α response gene set (M5911) in normal and cancerous stem cells. We first identify three subsets of genes from the M5911 Interferon-α response gene set that represent: 1) common genes downregulated by miR-199a-OE and LCOR-KD, and upregulated by LCOR-OE in HMLE cells; 2) genes downregulated in MaSCs or in Lgr5+-MaSCs; and 3) genes downregulated in ER− breast CSCs (HCC1954, MC1, SUM149, and SUM159). The 27-gene ISDS represents the common overlap among these three set of genes.
Clinical data set analysis
The Cancer Genome Atlas (TCGA) data portal was used to access the TCGA breast cancer expression data. The RNA-Seq by Expectation Maximization (RSEM) expression data were median centered and all samples were standardized to zero-mean and unit variance before subtype classification, as recommended by previous authors to remove platform biases7. The TCGA breast cancer population analyzed contains 794 ER+ (77%) and 233 ER− (23%) samples. Prior to any subtype classification, ER populations were balanced using 233 ER− samples and 233 randomly sampled ER+ samples to calculate a median gene expression, then normalized by subtracting this median gene expression. TCGA miRNA expression data were normalized with the R voom package from the limma library70. Comparisons between subtype miRNA expression levels was performed using a Wilcoxon Unpaired Two-Sample Test. Intrinsic subtype classification of breast cancer samples was performed using the previously described PAM50 centroid-based classifier. The Claudin-Low (CL) classifier was constructed according to the work listed by Prat et. al7. Briefly, for each sample we calculated the Euclidean distance to the nice-cell CL predictor “CL” and “others” centroids. The samples were classified based on their proximity to the nearest centroid.
To evaluate the prognosis value of matched miRNA-target samples in breast cancer, the Buffa dataset37 was used, with 210 patients (82 ER−) and 10 year follow-up. The patients were stratified by median expression of miR-199a and LCOR. Other mRNA datasets were also used: The Jiang et al. mRNA dataset with a total of 168 triple negative breast cancer (TNBC) and 5 year follow-up, samples were collected at the Fudan University Shanghai Cancer Center (FDUSCC, Shanghai, P.R. China) as previously described38. We used 65 frozen sample from the Jiang dataset to extract miRNAs with Trizol and perform qRT-PCR analysis of miR-199a using TaqMan miRNA assays (Applied Biosystems). The NKI295 dataset gene expression was also used to determine distant metastasis-free survival prognosis of a total of 295 patients. Patients were stratified by median expression of LCOR (C10orf12). To evaluate the prognosis of gene signatures in ER− breast cancer, the KM plotter with a total of 807 ER− patients was used42. Patients were stratified by the median score.
Statistics and reproducibility
Results were represented as indicated in figure legends, generally as mean ± s.e. (standard error). For experiments with two groups, a small sample size (less than 30), and normally distributed data, the significance was evaluated using a two-tailed unpaired Student’s t-test under the assumption of unequal variance. Asterisks will denote p-value significance: p<0.05*; p<0.01**; p<0.005***. For multiple independent groups, one-way ANOVA was evaluated. Non-parametric data sets were evaluated using the Mann-Whitney-Wilcoxon U test. Stem cell and TIC frequency was calculated with the ELDA software by Pearson’s χ2 test. For free-survival analysis, Kaplan-Meier plots and significance with P log-rank test were used. For correlation analysis of clinical samples, Chi-square was used to assess significance. For the clinical multivariate analysis, Cox proportional hazards modeling was used to assess independent prognosis value. All statistics were calculated with the commercial software: GraphPad Prism6 and Microsoft Excel, except for the stem cell and TIC frequency using the online ELDA software. All of the experiments with images (BLI, FACS, IF, IHC and senescence) were repeated >3 times and representative images are shown. If apply, data corresponding to representative images have been included in Supplementary Table 4.
Data availability
All microarray data generated in this study have been deposited as a superseries at the NCBI Gene Expression Omnibus with the accession code GSE85808. Previously published microarray data that were reanalyzed or used for GSEA are available under the origin accession codes: GSE76250 (Jiang dataset), GSE22220 (Buffa dataset), GSE19446, GSE17215, GSE18229, GSE18229, and GSE52262 at the Gene Expression Omnibus. Other gene sets used for GSEA analysis are found in the MSigDB database v5.1 release under the code: M5911, M9143 and the Hallmark gene set collection. Other analyzed prognosis sources: KM plotter breast cancer (http://kmplot.com/analysis/). Previously published RNA-seq data reanalyzed are available in the TCGA Genomic Data Commons (https://gdc-portal.nci.nih.gov/projects/TCGA-BRCA): TCGA-BRCA (Breast Invasive Carcinoma) containing mRNA and miRNA data. Source data for Supplementary Fig. 1b, Fig. 7j and Supplementary Fig. 7e have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank A. Welm for providing the PDX and A. Prat for technical advice for CL subtype classification. This work was supported by a Susan G. Komen Fellowship to T. C-T (PDF15332075), and grants from the Brewster Foundation, the Breast Cancer Research Foundation, Department of Defense (BC123187), and the National Institutes of Health (R01CA141062) to Y.K. This research was also supported by the Genomic Editing and Flow Cytometry Shared Resources of the Cancer Institute of New Jersey (P30CA072720).
References
- 1.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti R, et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol. 2014;16:1004–1015. 1001–1013. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast cancer research : BCR. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gyorki DE, Asselin-Labat ML, van Rooijen N, Lindeman GJ, Visvader JE. Resident macrophages influence stem cell activity in the mammary gland. Breast Cancer Res. 2009;11:R62. doi: 10.1186/bcr2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drukker M, Benvenisty N. The immunogenicity of human embryonic stem-derived cells. Trends Biotechnol. 2004;22:136–141. doi: 10.1016/j.tibtech.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Cordon-Cardo C, et al. Expression of HLA-A,B,C antigens on primary and metastatic tumor cell populations of human carcinomas. Cancer research. 1991;51:6372–6380. [PubMed] [Google Scholar]
- 14.Bruttel VS, Wischhusen J. Cancer stem cell immunology: key to understanding tumorigenesis and tumor immune escape? Frontiers in immunology. 2014;5:360. doi: 10.3389/fimmu.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal B, et al. Integration of microRNA signatures of distinct mammary epithelial cell types with their gene expression and epigenetic portraits. Breast cancer research : BCR. 2015;17:85. doi: 10.1186/s13058-015-0585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123:606–618. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llobet-Navas D, et al. The miR-424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev. 2014;28:765–782. doi: 10.1101/gad.237404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ucar A, et al. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat Genet. 2010;42:1101–1108. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- 22.Bockmeyer CL, et al. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat. 2011;130:735–745. doi: 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 23.Dvinge H, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 24.Zhu M, et al. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol. 2011;12:R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Clouthier SG, Wicha MS. Role of microRNAs in the regulation of breast cancer stem cells. J Mammary Gland Biol Neoplasia. 2012;17:15–21. doi: 10.1007/s10911-012-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plaks V, et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast cancer research : BCR. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YB, et al. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck B, et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Celia-Terrassa T, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest. 2012;122:1849–1868. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ocana OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Cho RW, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 36.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buffa FM, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer research. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 38.Jiang YZ, et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 39.Calderon MR, et al. Ligand-dependent corepressor (LCoR) recruitment by Kruppel-like factor 6 (KLF6) regulates expression of the cyclin-dependent kinase inhibitor CDKN1A gene. J Biol Chem. 2012;287:8662–8674. doi: 10.1074/jbc.M111.311605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandes I, et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 41.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 42.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, et al. miR-199a-3p targets stemness-related and mitogenic signaling pathways to suppress the expansion and tumorigenic capabilities of prostate cancer stem cells. Oncotarget. 2016 doi: 10.18632/oncotarget.10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin G, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29:3545–3553. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alemdehy MF, et al. ICL-induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood. 2015;125:3937–3948. doi: 10.1182/blood-2014-11-612507. [DOI] [PubMed] [Google Scholar]
- 46.Cuiffo BG, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. 2014;15:762–774. doi: 10.1016/j.stem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, et al. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16:887. doi: 10.1186/s12885-016-2916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asim M, et al. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J Biol Chem. 2011;286:37108–37117. doi: 10.1074/jbc.M111.292771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bidwell BN, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18:1224–1231. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- 50.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nature reviews. Immunology. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 51.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nature reviews. Immunology. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 53.Sato T, et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 54.Schedin P. Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer. 2006;6:281–291. doi: 10.1038/nrc1839. [DOI] [PubMed] [Google Scholar]
- 55.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Celia-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30:892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malladi S, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schatton T, Frank NY, Frank MH. Identification and targeting of cancer stem cells. Bioessays. 2009;31:1038–1049. doi: 10.1002/bies.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eriksson M, et al. Oncolytic adenoviruses kill breast cancer initiating CD44+CD24−/low cells. Mol Ther. 2007;15:2088–2093. doi: 10.1038/sj.mt.6300300. [DOI] [PubMed] [Google Scholar]
- 60.James CD, et al. Chromosome 9 deletion mapping reveals interferon alpha and interferon beta-1 gene deletions in human glial tumors. Cancer research. 1991;51:1684–1688. [PubMed] [Google Scholar]
- 61.Lu R, Au WC, Yeow WS, Hageman N, Pitha PM. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon snd silencing by hypermethylation. J Biol Chem. 2000;275:31805–31812. doi: 10.1074/jbc.M005288200. [DOI] [PubMed] [Google Scholar]
- 62.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 63.Behan JW, et al. Activation of adipose tissue macrophages in obese mice does not require lymphocytes. Obesity (Silver Spring) 2013;21:1380–1388. doi: 10.1002/oby.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shultz LD, et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- 65.Chakrabarti R, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karantza-Wadsworth V, White E. A mouse mammary epithelial cell model to identify molecular mechanisms regulating breast cancer progression. Methods Enzymol. 2008;446:61–76. doi: 10.1016/S0076-6879(08)01604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonnefoy F, et al. Plasmacytoid dendritic cells play a major role in apoptotic leukocyte-induced immune modulation. J Immunol. 2011;186:5696–5705. doi: 10.4049/jimmunol.1001523. [DOI] [PubMed] [Google Scholar]
- 68.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 2009;329:227–241. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 69.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microarray data generated in this study have been deposited as a superseries at the NCBI Gene Expression Omnibus with the accession code GSE85808. Previously published microarray data that were reanalyzed or used for GSEA are available under the origin accession codes: GSE76250 (Jiang dataset), GSE22220 (Buffa dataset), GSE19446, GSE17215, GSE18229, GSE18229, and GSE52262 at the Gene Expression Omnibus. Other gene sets used for GSEA analysis are found in the MSigDB database v5.1 release under the code: M5911, M9143 and the Hallmark gene set collection. Other analyzed prognosis sources: KM plotter breast cancer (http://kmplot.com/analysis/). Previously published RNA-seq data reanalyzed are available in the TCGA Genomic Data Commons (https://gdc-portal.nci.nih.gov/projects/TCGA-BRCA): TCGA-BRCA (Breast Invasive Carcinoma) containing mRNA and miRNA data. Source data for Supplementary Fig. 1b, Fig. 7j and Supplementary Fig. 7e have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author upon request.