Abstract
Obesity is characterized by altered gut homeostasis, including dysbiosis and increased gut permeability closely linked to the development of metabolic disorders. Milk oligosaccharides are complex sugars that selectively enhance the growth of specific beneficial bacteria in the gastrointestinal tract and could be used as prebiotics. The aim of the study was to demonstrate the effects of bovine milk oligosaccharides (BMO) and Bifidobacterium longum ssp. infantis (B. infantis) on restoring diet-induced obesity intestinal microbiota and barrier function defects in mice. Male C57/BL6 mice were fed a Western diet (WD, 40% fat/kcal) or normal chow (C, 14% fat/kcal) for 7 wk. During the final 2 wk of the study, the diet of a subgroup of WD-fed mice was supplemented with BMO (7% wt/wt). Weekly gavage of B. infantis was performed in all mice starting at wk 3, yet B. infantis could not be detected in any luminal contents when mice were killed. Supplementation of the WD with BMO normalized the cecal and colonic microbiota with increased abundance of Lactobacillus compared with both WD and C mice and restoration of Allobaculum and Ruminococcus levels to that of C mice. The BMO supplementation reduced WD-induced increase in paracellular and transcellular flux in the large intestine as well as mRNA levels of the inflammatory marker tumor necrosis factor α. In conclusion, BMO are promising prebiotics to modulate gut microbiota and intestinal barrier function for enhanced health.
Keywords: prebiotic, obesity, gut homeostasis
INTRODUCTION
Impaired intestinal barrier function and microbiota dysbiosis are involved in a wide range of long-term conditions, including digestive and extra-digestive diseases (Sanz et al., 2010; Bischoff et al., 2014). This has stimulated research to find efficient ways to prevent or treat altered gut homeostasis. Among extra-digestive pathologies, obesity and its downstream complications such as metabolic syndrome and cardiovascular risks, constitute a major economic burden to our societies (Finkelstein et al., 2010). In the last decade, research on the role of the gut microbiota in host physiology, behavior, and metabolism has provided new insights into the causal mechanisms in obesity. Intestinal dysbiosis has been observed in obese patients and in animal models of obesity (Ley et al., 2005, 2006; Hamilton et al., 2015). Transfer of the gut microbiota from obese mice into lean germ-free mice recapitulates the obese phenotype (Turnbaugh et al., 2006), suggesting that the gut microbiota may play a role in the development or maintenance of obesity. Changes in gut barrier function are also seen in obesity and have been proposed to connect microbial dysbiosis to obesity (Raybould, 2012). The gut microbiota and barrier function are highly responsive to gut environment (Kashyap et al., 2013) and are able to be targeted by nutritional or pharmacological means. Thus pre-clinical and clinical trials with probiotics or prebiotics have been used to treat obesity and related co-morbidities.
Milk oligosaccharides are complex sugars that selectively enhance the growth of specific beneficial bacteria in the gastrointestinal system (Smilowitz et al., 2014). Bovine milk is a source of oligosaccharides that resemble human milk oligosaccharides (HMO), though with different structures and a much lower concentration than human milk (Chichlowski et al., 2011). The majority of the molecules are more simple in structure compared with those found in human milk (Zivkovic and Barile, 2011). New methods for separation and concentration of bovine milk oligosaccharides (BMO) from bovine milk industrial streams such as whey permeate are now available. Because whey permeate is a byproduct in the production of whey protein concentrate and is readily available, it is an attractive source of oligosaccharides for potential application in human nutrition (Zivkovic and Barile, 2011; Barile and Rastall, 2013). Our group has shown that BMO enhance the growth of Bifidobacterium longum ssp. infantis (B. infantis) in vitro (D. A. Mills, unpublished data). Bifidobacterium longum ssp. infantis is a neonate gut commensal that has been shown to confer both intestinal and extra-intestinal health benefit to the host: it modulates barrier function and protects epithelial cells against cytokine or chemical-induced inflammation (Ganguli et al., 2013; Konieczna et al., 2013; Miyauchi et al., 2013; O’Hara et al., 2006). Previous studies from our laboratories have demonstrated that B. infantis grown on HMO affected intestinal epithelial function in vitro, compared with lactose-grown control culture (Chichlowski et al., 2012; Wickramasinghe et al., 2015).
Our objective was to establish the proof-of-concept that a combination of dietary BMO and B. infantis can reverse the gut microbial dysbiosis and altered gut permeability induced by ingestion of the Western diet (WD). To that aim, we gavaged diet-induced obese mice weekly with fresh culture of B. infantis and supplemented their diet with BMO. The BMO-supplemented mice exhibited improved gut barrier function and microbiota after 2 wk compared with nonsupplemented obese mice; however, we did not detect B. infantis in the luminal contents of the gut 1 wk after the last gavage. This suggests that BMO itself could prove to be beneficial in the treatment of obesity-related gut dysfunction.
MATERIALS AND METHODS
Diets
Bovine milk oligosaccharides were extracted from whey by Hilmar Ingredients (Hilmar, CA). The product was lipid-and protein-free and contained 80% carbohydrates from which lactose represented only 0.02%. Composition of the BMO was obtained by nano-chip HPLC QToF (Figure 1A). The WD+BMO diet was formulated by substituting inulin and part of corn starch by 7% of BMO in a WD (40% fat, #5TJN, TestDiet, St. Louis, MO). The control diet was regular laboratory chow (C, #5001, LabDiet, St. Louis, MO). Composition of diets is presented in Figure 1B and ingredient composition of WD and WD+BMO diets in Supplemental Table S1 (https://doi.org/10.3168/jds.2016-11890).
Figure 1.
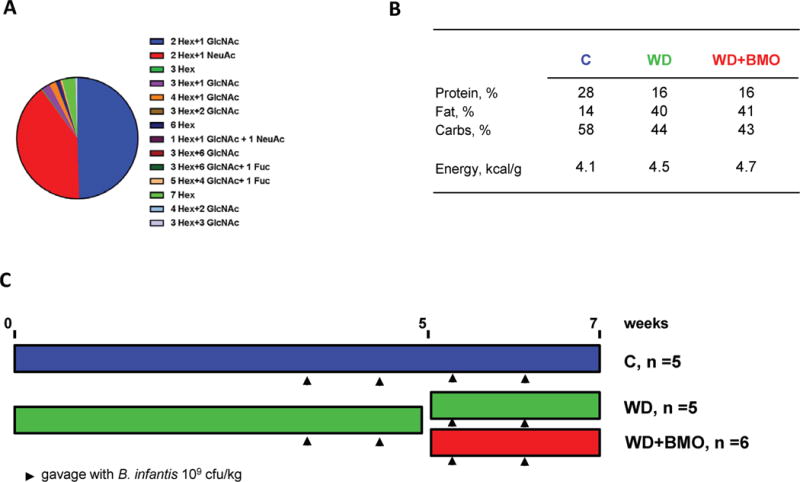
Schematic diagram of the protocol. (A) Composition of the bovine milk oligosaccharides (BMO) extract used in the study, (B) composition of the diets, (C) protocol outline. Hex: glucose or galactose, GlcNAc: N-acetyl glucosamine, NeuAc: sialic acid, C: control diet, WD: Western diet, WD+BMO: WD enriched with BMO. B. infantis = Bifidobacterium longum ssp. infantis. Fuc = fucose. Color version available online.
B. infantis Culture
The B. infantis ATCC 15697 was grown by a foodgrade commercial facility (Culture Systems Inc., Mishawaka, IN) as described previously (Underwood et al., 2013) and stored at −80°C. Purity and number of viable bacteria per gram were confirmed by the investigators every 6 mo by culture. Cultures from stocks frozen at −80°C were grown anaerobically at 37°C in a semisynthetic de Man, Rogosa, Sharpe broth (Becton Dickinson, Franklin Lakes, NJ) supplemented with 1% (wt/vol) l-cysteine hydrochloride. After centrifugation, bacteria were re-suspended in PBS before gavage.
Animals
Animals were maintained and handled in accordance with protocols approved by the Institutional Animal Care and Use Committee (University of California, Davis) under #16076. Male C57/BL6 mice (18.6 ± 0.3 g, n = 18, Harlan, San Diego, CA) were acclimatized to the animal facility and fed the C diet ad libitum. All animals were housed individually at 22°C with a 12:12 h light-dark cycle. After 1 wk of acclimation, 12 mice were switched to the WD diet for a total of 5 wk to establish obesity. At that time, 6 mice continued on the WD, whereas 6 mice switched to the WD+BMO diet for another 2 wk. All animals were gavaged once a week with 0.2 mL of 109 cfu/mL of B. infantis solution, starting at wk 3. Food and water were provided ad libitum. Body weight and food intake were measured weekly. The protocol is summarized on Figure 1C.
At experiment completion, mice were euthanized by cardiac puncture under deep anesthesia induced with isoflurane. Blood was collected via cardiac puncture in heparinized tubes. Plasma was obtained after centrifugation (4°C; 1,500 × g, 10 min) and stored at −20°C. Luminal contents of the cecum and proximal colon were flash frozen in liquid nitrogen and stored at −80°C. Segments of cecum and proximal colon were collected and stored in Krebs buffer for immediate measure of intestinal permeability in Ussing chambers. Sections of proximal colon were flash frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Ussing Chamber Analysis
Gut tissue was opened along the mesenteric border and mounted in Ussing chambers (Physiologic Instruments, San Diego, CA), exposing 0.5 cm2 of tissue surface area to 2.5 mL of oxygenated Krebs-glucose (10 mM) and Krebs-mannitol (10 mM) at 37°C on the serosal and luminal sides, respectively. Tissue conductance was measured using a voltage/current clamp (VCC MC8, Physiologic Instruments). Flux of FITC-4000 (FD-4, Sigma Aldrich, St. Louis, MO) and horseradish peroxidase (HRP Type II, Sigma Aldrich) were measured by addition of FD-4 (0.4 mg/mL) and HRP (0.2 mg/mL) to the mucosal chamber, respectively. Samples were collected from the serosal chamber after 30 and 60 min. Concentration of FD-4 was measured via fluorescence at excitation 485 nm, emission 538 nm. Oxidation of O-dianisidine (Sigma Aldrich) by HRP and evaluation of absorbance of the formed product at 450 nm were used to measure HRP concentrations.
Microbiota Analysis
The DNA was extracted from cecum and colonic luminal contents using the Zymo Research fecal DNA mini prep kit according to the manufacturer’s protocol (catalog D6010, Zymo Research, Irvine, CA). Quantification of DNA was performed on a Quibit Fluorometer (Life Technologies, Carlsbad, CA). The PCR amplification was performed on the V4 region of the 16S rRNA gene primers F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACHVGGGTWTCTAAT-3′). Barcode and primer linker sequences were added to allow for mulitplexing. Sequencing was performed with Illumina MiSEQ paired-end 250-bp runs.
Bioinformatic Analysis
Quantitative Insights Into Microbial Ecology (QIIME, version 1.9; Caporaso et al., 2010) software was used for demultiplexing and quality filtering. Operational taxonomic unit picking was performed using an open reference workflow script with Greengenes 97 (version 13_9) for taxonomy assignment at 97% similarity (McDonald et al., 2012). Chimera sequences were identified via Chimera Slyer (Haas et al., 2011) and removed. Additional filtering of lowread operational taxonomic units was also performed (Bokulich et al., 2013). Principal coordinate analysis taxa plots using weighted and unweighted UNIFRAC and α rarefaction plots were created through QIIME. Multivariable statistical analysis was performed in the LEfSe package (Segata et al., 2011) with default parameters.
Quantity of Bifidobacterium longum Via Real-Time PCR
Real-time quantitative PCR was performed to quantify the presence of B. longum (species that contains subspecies infantis). Uniquely designed TaqMan primers and probe used were as follows: forward primer BiLONF (CAGTTGATCGCATGGTCTT), reverse primer BiLONR (TACCCGTCGAAGCCAC) and BifSpP probe [(6-FAM)TGGGATGGGGTCGCG TCCTATCAG(TAMRA)]. Data were analyzed against a standard curve of known concentrations of B. infantis (Malinen et al., 2005).
Quantitative PCR
The quantitative PCR for the pro-inflammatory cytokine tumor necrosis factor (TNF)-α was performed on colonic tissues as already described (Vegezzi et al., 2014) using a TaqMan Gene Expression assay (Mm 99999068_M1, Applied Biosystems, Foster City, CA). β-Actin was used as housekeeping gene (Mm00607939_ s1). The TNF-α expression was analyzed according to the 2−ΔΔCT method.
Biochemical Analyses
Leptin and LPS-binding protein (LBP) levels were detected in plasma samples via dedicated ELISA kits (Alpco Diagnostics, Salem, NH; and Biometec, Greifswald, Germany, respectively) according to the manufacturer recommendations.
Statistical Analysis
Data are presented as means ± SEM. Data were analyzed by one-way ANOVA using Prism (GraphPad Software Inc., La Jolla, CA). Post hoc analysis with Student test and Tukey correction was performed to compare the 3 means.
RESULTS
Obesity and Metabolic Markers
Ingestion of the WD for 4 wk significantly increased BW compared with C mice (Figure 2A). Addition of BMO to the WD had no effect on BW in wk 5 to 6 compared with WD alone (Figure 2A). At wk 7, plasma leptin was significantly greater in both WD and WD+BMO mice compared with C mice (Figure 2D), however, there was no change in fasting plasma levels of glucose between the 3 dietary groups (Figure 2E). Plasma levels of LBP tended to be increased in WD fed mice compared with C mice (P = 0.09, Figure 2F).
Figure 2.
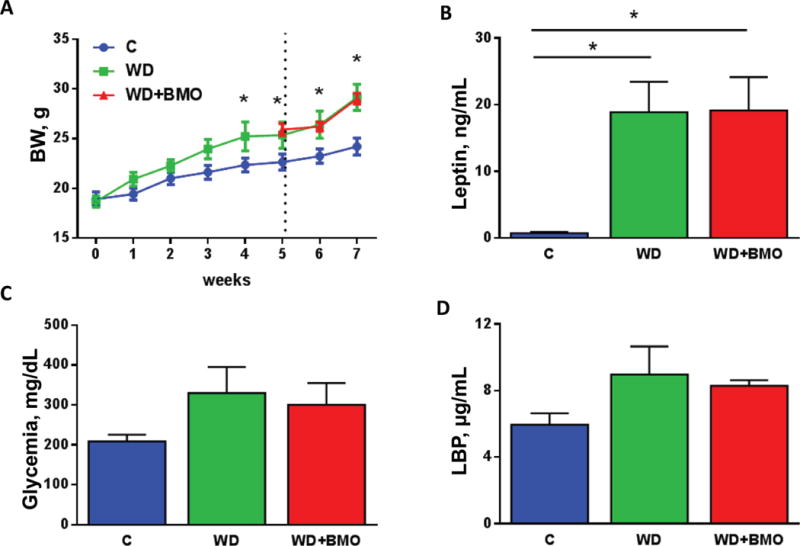
Obesity and metabolic parameters. Body weight (A), plasma leptin levels (B), fasted glycemia (C), and plasma LPS-binding protein (LBP) levels (D) were measured in mice fed a control chow (C) diet or Western diet (WD) for 7 wk or WD for 5 wk followed by WD supplemented with bovine milk oligosaccharides (WD+BMO) for 2 wk. Dotted vertical line in (A) indicates switch from WD to WD+BMO diet for half of the WD-fed mice. Data are means ± SEM; n = 5 to 6 per group. * denotes a significant difference (P < 0.05) compared with control. Color version available online.
Cecal and Colonic Microbiota
Richness and Diversity
The effects of diet upon cecal and colonic microbiota were similar. Therefore, microbiota analysis was performed on all data, taking into account the region as a subclass. Alpha rarefaction plot indicates that cecal and colonic microbiota richness was lower in WD and WD+BMO fed mice compared with C mice (Figure 3A). Simpson diversity index in the cecum and the colon was reduced by the WD diet consumption but restored (cecum) or partially restored (colon) by addition of BMO to the WD (Table 1). The Shannon diversity index showed the same pattern although addition of BMO did not significantly restore the index to the control level (Table 1). Unweighted unifrac analysis clearly distinguishes the 3 dietary groups, with the C samples separating from WD and WD+BMO samples along the first axis, whereas WD and WD+BMO separated along the second axis (Figure 3B). Weighted unifrac analysis, which considers relative abundance, showed a clear clustering of WD mice compared with C and WD+BMO samples along the first axis (Figure 3C). Samples from C and WD+BMO mice were less separated, suggesting more similar microbiota composition.
Figure 3.
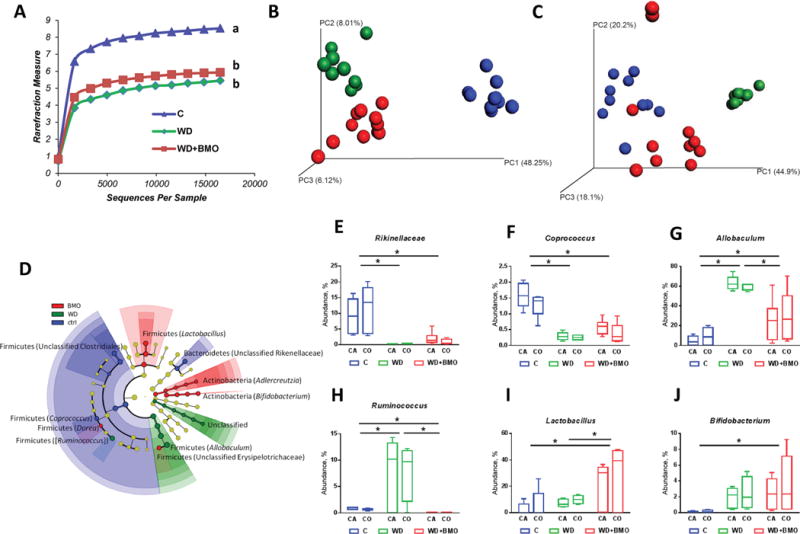
Cecal and colonic microbiota. Rarefaction curves (A), unweighted (B) and weighted (C) UniFrac analysis, significant changes in relative genus abundance (D) and average relative abundance of Rikinellaceae (E), Coprococcus (F), Allobaculum (G), Ruminococcus (H), Lactobacillus (I), and Bifidobacterium (J) in cecal (CA) and colonic (CO) microbiota of mice fed a control chow (C; blue) or Western diet (WD) for 7 wk (green) or WD for 5 wk followed by WD supplemented with bovine milk oligosaccharides (WD+BMO) for 2 wk (red). Whisker plots: minimum to maximum with median. Curves with different letters are significantly different. * denotes a significant difference (P < 0.05).
Table 1.
Simpson and Shannon diversity indexes1
| Simpson index
|
Shannon index
|
|||||
|---|---|---|---|---|---|---|
| Item | C | WD | WD+BMO | C | WD | WD+BMO |
| Cecum | 0.95 ± 0.04a | 0.60 ± 0.04b | 0.82 ± 0.04a | 0.93 ± 0.01a | 0.64 ± 0.02b | 0.74 ± 0.06b |
| Colon | 5.40 ± 0.04a | 2.52 ± 0.15b | 3.91 ± 0.28c | 5.06 ± 0.22a | 2.61 ± 0.11b | 3.28 ± 0.30b |
Means with different letters differ significantly (P < 0.05).
Data are means ± SEM; n = 5 to 6 per group. C: mice fed a control chow diet, WD: mice fed Western diet for 7 wk, WD+BMO: mice fed WD for 5 wk followed by WD supplemented with bovine milk oligosaccharides for 2 wk.
Relative Abundances
The gut microbiota of C mice was characterized by high abundance of bacteria within the Clostridia class (Figure 3D), in particular in Coprococcus (Figure 3D and 3F) and Clostridiaceae family (Figure 3D). Bacteria belonging to the Rikenellaceae family (Figure 3D and 3E) were also a strong component of the microbiota of C mice. A significant increase occurred in the abundance of Allobaculum and Ruminococcus in mice fed WD; this was reduced by addition of BMO to the WD diet (Figure 3D, 3G, and 3H). Addition of BMO to the WD increased the abundance of Lactobacillus (Figure 3D and 3I).
Bifidobacterium and B. longum Detection
The abundance of Bifidobacterium was greater in cecal and colonic contents of WD+BMO than C mice, with no difference between WD and WD+BMO groups (Figure 3J). Quantitative PCR did not reveal the presence of Bifidobacterium longum in the cecal and colonic luminal contents, suggesting that B. infantis did not persist in the gut of any of the mice. Further investigations indicated that B. infantis did not persist longer than 2 d, regardless of the diet (data not shown).
Cecal and Colonic Permeability
A trend was observed for an increase in the flux of FD-4 across the cecal mucosa of WD mice compared with C mice (Figure 4A); however, there was no significant effect of WD on either conductance or HRP flux (Figure 4B and 4C). In the colon, flux of FD-4 and HRP significantly increased (Figure 4D and 4E) and conductance increased (Figure 4F) in mice fed the WD compared with C mice. These impairments of colonic barrier function were reversed by the addition of BMO to the WD (Figure 4D–4F). There was a significant increase in levels of tnf-α mRNA in WD versus C mice in the colonic mucosa; addition of BMO to the WD normalized the levels of this inflammatory marker (Figure 4G).
Figure 4.
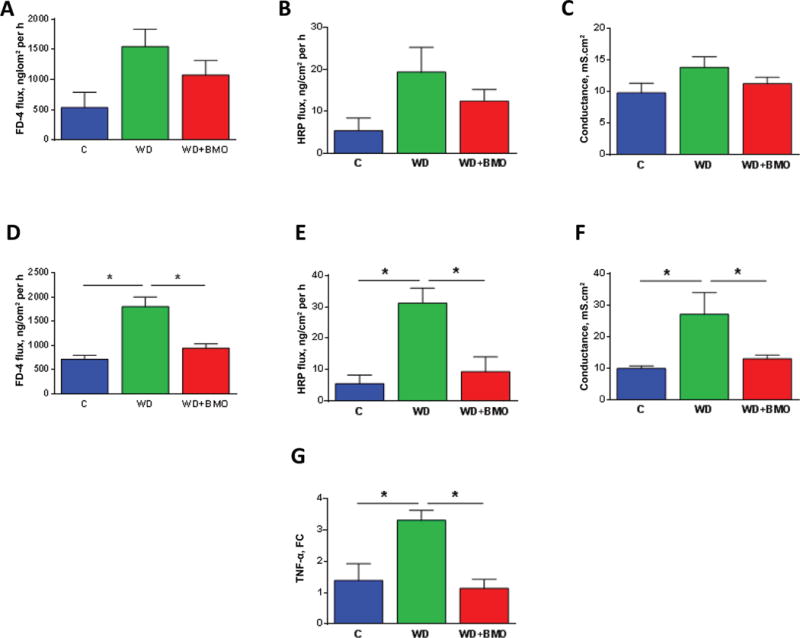
Cecal and colonic barrier function. Cecal ex vivo permeability to FITC-4000 (FD-4, Sigma Aldrich, St. Louis, MO; A) and horseradish peroxidase (HRP, Sigma Aldrich; B), cecal conductance (C), colonic ex vivo permeability to FD-4 (D) and HRP (E), colonic conductance (F), and colonic tumor necrosis factor (TNF)-α mRNA levels (G) were evaluated in mice fed a control chow (C) diet or Western diet (WD) for 7 wk, or WD for 5 wk followed by WD supplemented with bovine milk oligosaccharides (WD+BMO) for 2 wk. Data are means ± SEM; n = 5 to 6 per group. * denotes a significant difference (P < 0.05). FC: fold-change compared with control. Color version available online.
DISCUSSION
We demonstrated that whey-extracted BMO ameliorates WD-induced gut microbiota dysbiosis and intestinal barrier function in mice. Obese mice fed the BMO-supplemented diet for 2 wk had completely restored colonic permeability and tnf-α gene expression levels similar to chow-fed mice. Large intestinal microbiota of obese mice fed BMO had a unique bacterial signature compared with both chow-fed and nonsupplemented obese mice. In particular, BMO increased the relative abundance of Lactobacillus and reduced the overabundance of Allobaculum and Ruminococcus, which had bloomed with the WD diet.
The main effect on gut microbiota induced by addition of BMO to the WD diet in our model was the increased abundance of Lactobacillus, from an average 6–10% to 30–39%. Lactobacillus is not known to be the best bacteria to metabolize milk oligosaccharides, as opposed to Bifidobacterium species, which harbor distinctive gene clusters that are specifically induced during growth on HMO (Barile and Rastall, 2013). Yet, several studies reported weak growth of a few species of Lactobacillus on HMO in vitro (Ward et al., 2006; Sela et al., 2008; Marcobal et al., 2010; Schwab and Gänzle, 2011). Hydrolysis activity of milk oligosaccharides by Lactobacillus seems limited to tri-or tetrasaccharides (Schwab and Ganzle, 2011), presumably because larger oligosaccharides are not transported into the cytoplasm Andersen et al. (2011) recently demonstrated that the lacS permease of Lactobacillus acidophilus was required to allow growth on galactooligosaccharides with a degree of polymerization from 2 to 6. Our BMO extract consisted primarily of trimers (i.e., degree of polymerization of 3), suggesting transport and intracellular metabolism by Lactobacillus is possible. Extracellular hydrolysis of BMO by other microorganisms in the intestine may provide another route for growth of Lactobacillus by cross-feeding on liberated monosaccharides. Several reports indicated that Lactobacillus spp. could use milk oligosaccharide monomers as substrate (Schwab and Gänzle, 2011; Bidart et al., 2014; Young et al., 2015).
Bifidobacterium longum ssp. infantis is a human isolate highly adapted to growth on HMO as a carbon source (Ward et al., 2006) and we had hypothesized that BMO with their close structure to HMO would help the probiotic colonization. However, B. infantis was not detected in any of the luminal contents from chow-, WD-, or WD+BMO-fed mice. The lack of B. infantis persistence in our study suggests that consumption of BMO does not override the colonization resistance inherent to the mouse intestinal microbiota, that the BMO resource is consumed by a wider array of mouse intestinal bacteria, or both. The human origin of B. infantis may be another factor. Recently, Oh et al. (2010) showed that human-derived Lactobacillus strains could not colonize mice in comparison to rodent-derived Lactobacillus strains.
The increased abundance of Lactobacillus was parallel to decreased abundances of Ruminococcus (from an average of 10 to 0.01–0.03%) and Allobaculum (from an average of 62 to 25–27%). A similar pattern of changes was observed in rats when soy milk and fiber were added to a cholesterol-enriched diet, resulting in restoration of Allobaculum to control levels while increasing Lactobacillus abundance (Lee et al., 2015). Kaplan et al. (2001) reported that consumption of the probiotic Lactobacillus acidophilus inhibited the growth of a Ruminococcus flavifaciens strain in rats. Similarly, several studies reported increased proportion of Lactobacillus and decreased proportions of Ruminococcus in human feces in different conditions (i.e., aging or probiotic consumption; Mäkivuokko et al., 2010; Ferrario et al., 2014). In these later studies, other shifts in bacterial populations were also observed; establishing a direct link between Lactobacillus and Ruminococcus abundances is therefore difficult. Finally, a relative rise in Lactobacillus species was accompanied by a corresponding reduction in the genera Escherichia/Shigella, Ruminococcus, and Eubacterium in piglet cecal microbiota cultivated in vitro with sialic acid (Young et al., 2015). These later data suggest a possible antagonism between Lactobacillus and Ruminococcus in the presence of sialic acid found in BMO. The bacterial family Ruminococcus was also decreased in mice fed 6’sialyllactose (Tarr et al., 2015). Further research is needed to understand the ecological interactions between those different genera in the presence of BMO.
Addition of BMO to the WD greatly improved large intestinal barrier function, which was dramatically altered in WD-fed mice as already described (Cani et al., 2009; Teixeira et al., 2012; Hamilton et al., 2015). This intestinal barrier improvement parallels the BMO-induced increase in Lactobacillus abundance. Lam et al. (2012) observed a positive correlation between Lactobacillus abundance and proximal colon trans-epithelial resistance, suggesting inverse relationship between this genus abundance and barrier function properties, although the mechanisms are not understood. The BMO-induced increase in Lactobacillus abundance observed in the present study could be at the origin of the improvement of gut barrier function. However, we cannot exclude a direct effect of BMO or its individual components on intestinal epithelial cell barrier function as it was reported recently that HMO inhibit the growth of intestinal epithelial cells in vitro by suppressing cell cycle progression through induction of differentiation, by influencing apoptosis, or both (Kuntz et al., 2008; Holscher et al., 2014). Another possibility is that BMO itself or its individual components downregulate gut inflammation, as illustrated by the decreased colonic tnf-α expression in the present study. Such direct effect of milk oligosaccharides on inflammation has been recently demonstrated for HMO in in vitro models of intestinal epithelial cells (He et al., 2016). Reduction of local inflammation will in turn restore barrier function (Watson and Hughes, 2012).
Our diet-induced obesity model was characterized by increased BW and greater plasma leptin, indicative of greater adiposity in WD compared with C mice. Yet metabolic endotoxemia and metabolic disorders were not obvious as plasma LBP and fasting glycemia were not significantly different from control mice. On the other hand, at the gut level, the WD diet had a profound effect on gut barrier function and gut microbiota as already described (Cani et al., 2009; Teixeira et al., 2012; Hamilton et al., 2015). The main characteristics of the WD microbiota were a bloom of Allobaculum and Ruminococcus relative abundance together with a dramatic decrease in Rikinellaceae and Coprococcus relative abundances. Dysbiosis in diet-induced obesity is well established (Ojeda et al., 2016). However, due to differences in animal strain and environment among studies, the specific community of microbes that are altered by a high fat diet is still not clearly established (Walker et al., 2014). Moreover, dietary components and timing of intervention alter gut microbiota differently (Clarke et al., 2013; Hamilton et al., 2015). The alteration of cecal and colonic microbiota observed in our WD-fed mice resembles some of the alterations already observed. Indeed, relative abundance of Ruminococcus has been positively correlated with serum triglycerides in several reports (Lahti et al., 2013; Chen et al., 2014). Increased relative abundance of Ruminococcus gnavus has also been observed in obese Zucker rats (Petriz et al., 2014). Similarly Allobaculum relative abundance was increased in rats fed high fat (Zhang et al., 2012), although this association is controversial because Allobaculum relative abundance decreased in mice fed high fat in 2 other reports (Ravussin et al., 2012; Everard et al., 2014; Qiao et al., 2014; Baldwin et al., 2016).
In conclusion, our data strongly suggest that BMO act as prebiotics to restore gut health in obesity. Yet, metabolic parameters were not affected by BMO supplementation, probably due to the short time of the intervention. This study needs to be taken as a proof-of-concept of BMO prebiotics properties. Further studies are now needed to define a minimal effective dose and time of treatment as well as to design specific BMO-enriched human food to be tested in clinical trials. Moreover, whether this effect is also pertinent to other diseases where gut health is compromised needs further investigation. Given that BMO can be extracted from byproducts in the production of cheeses worldwide, this opens avenues for translation of original protective mechanisms from milk into wide-reaching nutritional strategies aimed at gut health.
Acknowledgments
G.B. was funded by a FP7-PEOPLE-2011-IOF Grant (Symobigut # 298697). The work was funded by NIH AT006642 (M.C.), DK41004 (H.E.R.), AT006180 (H.E.R., D.A.M.), and AT008759 (H.E.R., D.A.M.) and AT007079 (D.A.M.) and the Peter J. Shields Endowed Chair in Dairy Food Science (D.A.M.). The authors thank Kanika Verma (Interlake High School, Bellevue, WA) for technical help and Catia Sternini (University of California–Los Angeles School of Medicine) for assistance with measurement of tnf-α expression. Author contribution: G.B., M.C., D.A.M., and H.E.R. conceived and designed research; G.B. and S.W. performed experiments; G.B., M.K.H., K.M.K., and H.E.R. analyzed and interpreted data; G.B., M.K.H., and H.E.R. drafted the manuscript; and G.B., K.H.M., M.C., SW, DB, K.M.K., D.A.M., and H.E.R. edited and revised the manuscript and approved the final version of the manuscript. The authors have no conflict of interest to declare. D.B. and D.A.M. are co-founders of Evolve Biosystems Inc., a company focused on diet-based manipulation of the gut microbiota.
References
- Andersen JM, Barrangou R, Hachem M Abou, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc Natl Acad Sci USA. 2011;108:17785–17790. doi: 10.1073/pnas.1114152108. https://doi.org/10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J, Collins B, Wolf PG, Martinez K, Shen W, Chuang CC, Zhong W, Cooney P, Cockrell C, Chang E, Gaskins HR, McIntosh MK. Table grape consumption reduces adiposity and markers of hepatic lipogenesis and alters gut microbiota in butter fat-fed mice. J Nutr Biochem. 2016;27:23–135. doi: 10.1016/j.jnutbio.2015.08.027. https://doi.org/10.1016/j.jnutbio.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Rastall RA. Human milk and related oligosaccharides as prebiotics. Curr Opin Biotechnol. 2013;24:14–219. doi: 10.1016/j.copbio.2013.01.008. https://doi.org/10.1016/j.copbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Bidart GN, Rodríguez-Díaz J, Monedero V, Yebra MJ. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol Microbiol. 2014;93:21–538. doi: 10.1111/mmi.12678. https://doi.org/10.1111/mmi.12678. [DOI] [PubMed] [Google Scholar]
- Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A, Wells JM. Intestinal permeability–A new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:89. doi: 10.1186/s12876-014-0189-7. https://doi.org/10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Qualityfiltering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:7–59. doi: 10.1038/nmeth.2276. https://doi.org/10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, Muccioli GG, Delzenne NM. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:091–1103. doi: 10.1136/gut.2008.165886. https://doi.org/10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:35–336. doi: 10.1038/nmeth.f.303. https://doi.org/10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Yang Z, Chen X, Huang Y, Yin B, Guo F, Zhao H, Zhao T, Qu H, Huang J, Wu Y, Gu R. The effect of Lactobacillus rhamnosus hsryfm 1301 on the intestinal microbiota of a hyperlipidemic rat model. BMC Complement Altern Med. 2014;14:86. doi: 10.1186/1472-6882-14-386. https://doi.org/10.1186/1472-6882-14-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr. 2012;55:21–327. doi: 10.1097/MPG.0b013e31824fb899. https://doi.org/10.1097/MPG.0b013e31824fb899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M, German JB, Lebrilla CB, Mills DA. The influence of milk oligosaccharides on microbiota of infants: Opportunities for formulas. Annu Rev Food Sci Technol. 2011;2:31–351. doi: 10.1146/annurev-food-022510-133743. https://doi.org/10.1146/annurev-food-022510-133743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, O’Sullivan O, Ross RP, O’Toole PW, Shanahan F, Cotter PD. Targeting the microbiota to address diet-induced obesity: A time dependent challenge. PLoS One. 2013;8:e65790. doi: 10.1371/journal.pone.0065790. https://doi.org/10.1371/journal.pone.0065790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Gaïa N, Johansson M, Ståhlman M, Backhed F, Delzenne NM, Schrenzel J, François P, Cani PD. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:116–2130. doi: 10.1038/ismej.2014.45. https://doi.org/10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, Noni I De, Stuknyte M, Chouaia B, Riso P, Guglielmetti S. Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr. 2014;144:787–1796. doi: 10.3945/jn.114.197723. https://doi.org/10.3945/jn.114.197723. [DOI] [PubMed] [Google Scholar]
- Finkelstein EA, Brown DS, Wrage LA, Allaire BT, Hoerger TJ. Individual and aggregate years-of-life-lost associated with overweight and obesity. Obesity (Silver Spring) 2010;18:33–339. doi: 10.1038/oby.2009.253. https://doi.org/10.1038/oby.2009.253. [DOI] [PubMed] [Google Scholar]
- Ganguli K, Meng D, Rautava S, Lu L, Walker WA, Nanthakumar N. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G132–G141. doi: 10.1152/ajpgi.00142.2012. https://doi.org/10.1152/ajpgi.00142.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:94–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MK, Boudry G, Lemay DG, Raybould HE. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am J Physiol Gastrointest Liver Physiol. 2015;308:G840–G851. doi: 10.1152/ajpgi.00029.2015. https://doi.org/10.1152/ajpgi.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Liu S, Kling DE, Leone S, Lawlor NT, Huang Y, Feinberg SB, Hill DR, Newburg DS. The human milk oligosaccharide 2′-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation. Gut. 2016 doi: 10.1136/gutjnl-2014-307544. https://doi.org/10.1136/gutjnl-2014-307544. [DOI] [PubMed]
- Holscher HD, Davis SR, Tappenden KA. Human milk oligosaccharides influence maturation of human intestinal Caco-2Bbe and HT-29 cell lines. J Nutr. 2014;144:86–591. doi: 10.3945/jn.113.189704. https://doi.org/10.3945/jn.113.189704. [DOI] [PubMed] [Google Scholar]
- Kaplan CW, Astaire JC, Sanders ME, Reddy BS, Kitts CL. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2001;67:935–1939. doi: 10.1128/AEM.67.4.1935-1939.2001. https://doi.org/10.1128/AEM.67.4.1935-1939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA. 2013;110:7059–17064. doi: 10.1073/pnas.1306070110. https://doi.org/10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczna P, Ferstl R, Ziegler M, Frei R, Nehrbass D, Lauener RP, Akdis CA, O’Mahony L. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS One. 2013;8:e62617. doi: 10.1371/journal.pone.0062617. https://doi.org/10.1371/journal.pone.0062617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz S, Rudloff S, Kunz C. Oligosaccharides from human milk influence growth-related characteristics of intestinally transformed and non-transformed intestinal cells. Br J Nutr. 2008;99:62–471. doi: 10.1017/S0007114507824068. https://doi.org/10.1017/S0007114507824068. [DOI] [PubMed] [Google Scholar]
- Lahti L, Salonen A, Kekkonen RA, Salojärvi J, Jalanka-Tuovinen J, Palva A, Orešič M, Vos WM de. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. Peer J. 2013;1:e32. doi: 10.7717/peerj.32. https://doi.org/10.7717/peerj.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YY, Ha CWY, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, Cook DI, Hunt NH, Caterson ID, Holmes AJ, Storlien LH. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7:e34233. doi: 10.1371/journal.pone.0034233. https://doi.org/10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Han HW, Yim SY. Beneficial effects of soy milk and fiber on high cholesterol diet-induced alteration of gut microbiota and inflammatory gene expression in rats. Food Funct. 2015;6:92–500. doi: 10.1039/c4fo00731j. https://doi.org/10.1039/c4fo00731j. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:1070–11075. doi: 10.1073/pnas.0504978102. https://doi.org/10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:022–1023. doi: 10.1038/4441022a. https://doi.org/10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Mäkivuokko H, Tiihonen K, Tynkkynen S, Paulin L, Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br J Nutr. 2010;103:27–234. doi: 10.1017/S0007114509991553. https://doi.org/10.1017/S0007114509991553. [DOI] [PubMed] [Google Scholar]
- Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:73–382. doi: 10.1111/j.1572-0241.2005.40312.x. https://doi.org/10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:334–5340. doi: 10.1021/jf9044205. https://doi.org/10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, De-Santis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:10–618. doi: 10.1038/ismej.2011.139. https://doi.org/10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi E, Ogita T, Miyamoto J, Kawamoto S, Morita H, Ohno H, Suzuki T, Tanabe S. Bifidobacterium longum alleviates dextran sulfate sodium-induced colitis by suppressing IL-17A response: involvement of intestinal epithelial costimulatory molecules. PLoS One. 2013;8:e79735. doi: 10.1371/journal.pone.0079735. https://doi.org/10.1371/journal.pone.0079735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, O’Regan P, Fanning A, O’Mahony C, Macsharry J, Lyons A, Bienenstock J, O’Mahony L, Shanahan F. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology. 2006;118:02–215. doi: 10.1111/j.1365-2567.2006.02358.x. https://doi.org/10.1111/j.1365-2567.2006.02358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:77–387. doi: 10.1038/ismej.2009.123. https://doi.org/10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- Ojeda P, Bobe A, Dolan K, Leone V, Martinez K. Nutritional modulation of gut microbiota—The impact on metabolic disease pathophysiology. J Nutr Biochem. 2016 doi: 10.1016/j.jnutbio.2015.08.013. https://doi.org/10.1016/j.jnutbio.2015.08.013. [DOI] [PMC free article] [PubMed]
- Petriz BA, Castro AP, Almeida JA, Gomes CP, Fernandes GR, Kruger RH, Pereira RW, Franco OL. Exercise induction of gut microbiota modifications in obese, nonobese and hypertensive rats. BMC Genomics. 2014;15:11. doi: 10.1186/1471-2164-15-511. https://doi.org/10.1186/1471-2164-15-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Sun J, Xie Z, Shi Y, Le G. Propensity to highfat diet-induced obesity in mice is associated with the indigenous opportunistic bacteria on the interior of Peyer’s patches. J Clin Biochem Nutr. 2014;55:20–128. doi: 10.3164/jcbn.14-38. https://doi.org/10.3164/jcbn.14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20:38–747. doi: 10.1038/oby.2011.111. https://doi.org/10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J Physiol. 2012;590:41–446. doi: 10.1113/jphysiol.2011.222133. https://doi.org/10.1113/jphysiol.2011.222133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz Y, Santacruz A, Gauffin P. Gut microbiota in obesity and metabolic disorders. Proc Nutr Soc. 2010;69:34–441. doi: 10.1017/S0029665110001813. https://doi.org/10.1017/S0029665110001813. [DOI] [PubMed] [Google Scholar]
- Schwab C, Gänzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol Lett. 2011;315:41–148. doi: 10.1111/j.1574-6968.2010.02185.x. https://doi.org/10.1111/j.1574-6968.2010.02185.x. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. https://doi.org/10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum ssp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:8964–18969. doi: 10.1073/pnas.0809584105. https://doi.org/10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:43–169. doi: 10.1146/annurev-nutr-071813-105721. https://doi.org/10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr AJ, Galley JD, Fisher SE, Chichlowski M, Berg BM, Bailey MT. The prebiotics 3′Sialyllactose and 6′Sialyllactose diminish stressor-induced anxiety-like behavior and colonic microbiota alterations: Evidence for effects on the gut–brain axis. Brain Behav Immun. 2015;50:66–177. doi: 10.1016/j.bbi.2015.06.025. https://doi.org/10.1016/j.bbi.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira TFS, Collado MC, Ferreira CLLF, Bressan J, do M, Peluzio C. Potential mechanisms for the emerging link between obesity and increased intestinal permeability. Nutr Res. 2012;32:37–647. doi: 10.1016/j.nutres.2012.07.003. https://doi.org/10.1016/j.nutres.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:027–1031. doi: 10.1038/nature05414. https://doi.org/10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Underwood MA, Kalanetra KM, Bokulich NA, Lewis ZT, Mirmiran M, Tancredi DJ, Mills DA. A comparison of two probiotic strains of bifidobacteria in premature infants. J Pediatr. 2013;163:585–1591. doi: 10.1016/j.jpeds.2013.07.017. https://doi.org/10.1016/j.jpeds.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegezzi G, Anselmi L, Huynh J, Barocelli E, Rozengurt E, Raybould H, Sternini C. Diet-induced regulation of bitter taste receptor subtypes in the mouse gastrointestinal tract. PLoS One. 2014;9:e107732. doi: 10.1371/journal.pone.0107732. https://doi.org/10.1371/journal.pone.0107732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A, Pfitzner B, Neschen S, Kahle M, Harir M, Lucio M, Moritz F, Tziotis D, Witting M, Rothballer M, Engel M, Schmid M, Endesfelder D, Klingenspor M, Rattei T, Castell WZ, de Angelis MH, Hartmann A, Schmitt-Kopplin P. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J. 2014;8:380–2396. doi: 10.1038/ismej.2014.79. https://doi.org/10.1038/ismej.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:497–4499. doi: 10.1128/AEM.02515-05. https://doi.org/10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AJM, Hughes KR. TNF-α-induced intestinal epithelial cell shedding: Implications for intestinal barrier function: Cell shedding and intestinal barrier function. Ann N Y Acad Sci. 2012;1258:1–8. doi: 10.1111/j.1749-6632.2012.06523.x. https://doi.org/10.1111/j.1749-6632.2012.06523.x. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe S, Pacheco AR, Lemay DG, Mills DA. Bifidobacteria grown on human milk oligosaccharides downregulate the expression of inflammation-related genes in Caco-2 cells. BMC Microbiol. 2015;15 doi: 10.1186/s12866-015-0508-3. https://doi.org/10.1186/s12866-015-0508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young W, Egert M, Bassett SA, Bibiloni R. Detection of sialic acid-utilising bacteria in a caecal community batch culture using RNA-based stable isotope probing. Nutrients. 2015;7:109–2124. doi: 10.3390/nu7042109. https://doi.org/10.3390/nu7042109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, Li X, Ning G, Zhao L. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat dietfed rats. PLoS One. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. https://doi.org/10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr. 2011;2:84–289. doi: 10.3945/an.111.000455. https://doi.org/10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]


