Abstract
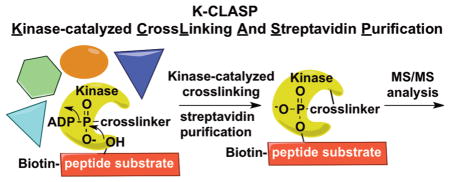
Few methods are available to discover the cellular kinase that phosphorylates a specific amino acid, or phosphosite, on a protein. In addition, identifying the associated proteins bound near a phosphosite during phosphorylation would provide insights into cell biology and signaling. Here, we report K-CLASP (Kinase Catalyzed CrossLinking And Streptavidin Purification) as a method for both phosphosite-specific kinase identification and the discovery of kinase interacting proteins. K-CLASP offers a powerful tool to discover unanticipated protein–protein interactions in phosphorylation-mediated biological events.
Graphical Abstract
Phosphorylation is an important protein post-translational modification that acts as a key chemical switch in signal transduction.1 Aberrant phosphorylation is implicated in many diseases, such as cancer, emphasizing the need to study cellular phosphorylation events.2 Owing to the advancement of powerful mass spectrometric techniques, novel phosphorylated amino acids or phosphosites are being identified at an unprecedented rate.3 The identification of these novel phosphosites poses an important question: which protein kinase catalyzes phosphorylation of a specific phosphosite? With over 500 kinases in the kinome,4 identifying the kinase that phosphorylates a specific phosphosite is a challenge. Although several methods have been established to identify the substrates of a given kinase,5 fewer methods exist to identify the kinase for a specific phosphosite. In silico prediction based on consensus sequence searching represents a useful strategy.6 However, phosphosite consensus sequences have not yet been established for all kinases. Peptide-based phosphosite-specific cross-linking was also developed for kinase-substrate identification, although the method is not widely used.7,8 A second significant challenge is the characterization of proteins interacting with a kinase during phosphorylation. Classic methods, such as immunoprecipitation and yeast two-hybrid analysis, require stable protein–protein interactions, which may miss low abundance or transient partners. To address both of these needs and provide a phosphosite-specific kinase and interactome identification tool, we developed a method based on our previous work with γ-modified adenosine 5′-triphosphate (ATP) analogs.9
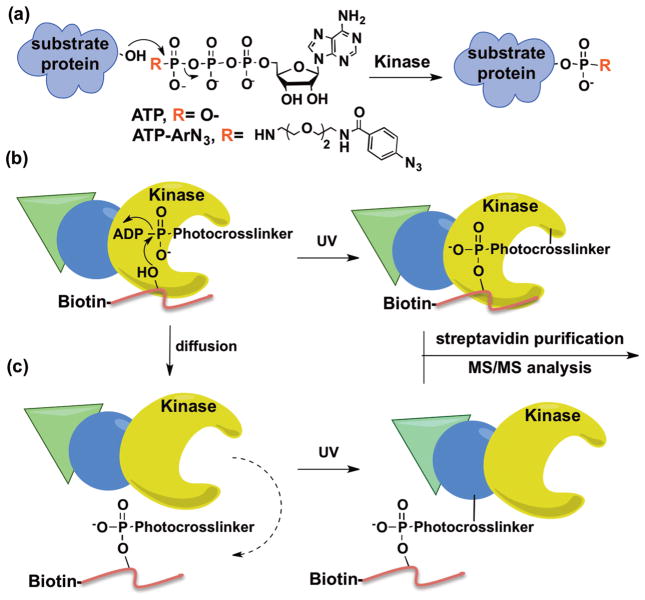
Kinases utilize ATP as a cosubstrate for phosphorylation (Figure 1a). We and others have established that kinases throughout the kinome tree show cosubstrate promiscuity by accepting γ-modified ATP analogs to covalently modify substrate proteins (Figure 1a).9–12 Recently, we used the γ-modified ATP analog ATP-arylazide (ATP-ArN3) to covalently couple kinase-substrate pairs using photo-cross-linking (Figure 1b).13,14 Taking advantage of kinase-catalyzed cross-linking, we describe here a method to identify the kinase that phosphorylates a specific phosphosite, termed K-CLASP (Kinase-catalyzed CrossLinking And Streptavidin Purification). In the K-CLASP method, a biotin-tagged peptide containing the phosphosite of interest is incubated with a cell lysate in the presence of ATP-ArN3 and UV irradiation. The biotin-tagged peptide will covalently cross-link to its respective kinase in the lysate via kinase-catalyzed cross-linking (Figure 1b). The covalently cross-linked peptide–kinase complex can then be streptavidin purified and analyzed by tandem mass spectrometry (MS/MS) to identify the captured kinase. However, kinase–substrate interactions are transient, allowing diffusion of the biotin-tagged peptide away from the kinase after transfer of the photo-cross-linking group (Figure 1c). In this scenario, the biotin-tagged peptide will cross-link to proteins in close proximity of the kinase, including direct and indirect associated proteins (Figure 1c). Therefore, both the target kinase and kinase-associated proteins will be purified and observed in a K-CLASP experiment. Therefore, K-CLASP has the potential not only to discover the kinase that phosphorylates a given phosphosite but to also expose proteins interacting with the kinase under the conditions of the experiment.
Figure 1.
Kinase-catalyzed labeling and the K-CLASP method. (a) Kinases accept ATP and γ-modified ATP analogs, such as ATP-ArN3, to phosphorylate substrate proteins. (b) An N-biotinylated peptide (red) carrying a phosphosite will bind to its phosphorylating kinase (yellow) and ATP-ArN3 in lysates. The kinase catalyzes transfer of the photo-cross-linker group onto the peptide, while UV irradiation leads to covalent attachment of biotinylated peptide and kinase. The biotin-linked kinase can then be streptavidin purified and identified by MS/MS analysis. (c) Once the photo-cross-linker is transferred, the N-biotinylated peptide can diffuse away from the kinase to cross-link with nearby proteins, including direct (blue) and indirect (green) associated proteins of the kinase.
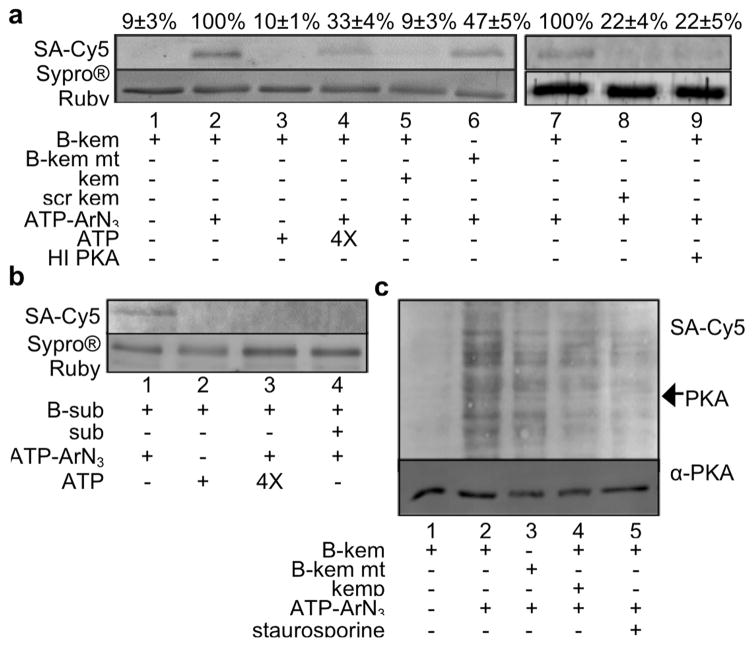
To establish K-CLASP for kinase identification, we chose a well-known kinase–substrate peptide pair. Protein kinase A (PKA) is widely reported to phosphorylate the peptide substrate kemptide (LRRASLG).15 As a first step to show that ATP-ArN3 cross-links kemptide with PKA, a recombinant PKAα catalytic subunit was incubated with ATP-ArN3 in the presence of N-biotinylated kemptide and UV irradiation. We used a modified kemptide sequence containing four N-terminal glycine residues (biotin-GGGGLRRASLG) to separate the biotin tag from the phosphosite and avoid steric issues. After cross-linking, samples were separated by SDS-PAGE, transferred onto a PVDF membrane, and then stained with a streptavidin–Cy5 conjugate to detect biotin-linked PKA kinase. Biotinylated PKA was observed in the presence of ATP-ArN3 (Figure 2a, lane 2) but not ATP (Figure 2a, lane 3), demonstrating dependence on photo-cross-linking. As controls, the biotin signal was reduced when excess ATP (Figure 2a, lane 4) or nonbiotinylated kemptide (Figure 2a, lane 5) were included. In addition, biotinylation was also reduced when a scrambled biotin peptide (Figure 2a, lane 8) or heat inactivated PKA was used (Figure 2a, lane 9).
Figure 2.
N-biotin kemptide cross-linking with recombinant PKA and lysates. (a) Cross-linking reactions with recombinant PKA, biotinylated kemptide (B-kem), and ATP-ArN3. Control reactions were carried out with N-biotin mutant kemptide (B-kem mt) lacking the phosphorylated Ser, 4× excess ATP, 2× excess nonbiotinylated kemptide (kem), an N-biotinylated scrambled kemptide (scr kem), or heat inactivated PKA (HI PKA). The biotin signal was quantified from independent trials with mean and standard error shown above each lane (Figure S1). (b) Cross-linking reactions with recombinant CK2, biotinylated CK2 substrate peptide (B-sub), and ATP-ArN3. Control reactions were carried out with ATP, 4× excess ATP, and 2× excess nonbiotinylated CK2 peptide (sub). Reproducible trials are shown in Figure S4. (c) Biotin kemptide (B-kem) cross-linking with ATP-ArN3 in HeLa lysates. Optimization of reaction conditions is shown in Figure S5. Reproducible trials are shown in Figure S6. For all parts, proteins were separated by SDS-PAGE and visualized with Sypro Ruby total proteins stain, streptavidin-Cy5 (SA-Cy5) biotin stain, or PKA antibody (α-PKA).
As a further control, we tested an N-biotinylated kemptide mutant where Ala replaced the phosphorylation site Ser (biotin-GGGGLRRAALG). PKA was biotinylated in the presence of N-biotin mutant kemptide, although at a reduced level compared to wild type kemptide (47 ± 5%, Figure 2a, lane 6 and Figure S1). We speculate that cross-linking of PKA with mutant kemptide is due to nonselective reactivity of the arylazide photo-cross-linker with biotin–kemptide before PKA autophosphorylation (Figure S2). In agreement, when N-biotinylated kemptide was preincubated with ATP-ArN3 before the addition of PKA, biotinylation was observed (Figure S3, lanes 2 and 3). Interestingly, the N-biotinylated scrambled peptide generated minimal PKA biotinylation (Figure S3, lane 4), suggesting that kemptide–PKA interaction was necessary. The results highlight the fact that a negative control reaction will be critical to account for and remove background cross-linking due to the highly reactive arylazide in the K-CLASP method. To be rigorous, reactions with mutant kemptide were included as critical negative controls in future experiments.
To establish that K-CLASP is applicable with other kinases, we also carried out cross-linking reactions with recombinant casein kinase II (CK2) and its N-biotinylated peptide substrate (biotin-RRREEETEEE). As observed with PKA, CK2 biotinylation was observed with ATP-ArN3 (Figure 2b, lane 1) but not with ATP (Figure 2b, lane 2). Similarly, CK2 labeling was reduced when excess ATP (Figure 2b, lane 3) or a nonbiotinylated CK2 peptide (Figure 2b, lane 4) was present. The results with CK2 demonstrate that kinase–peptide cross-linking with ATP-ArN3 is compatible with other kinases.
With the ultimate goal of identifying endogenous, cellular kinases of kemptide, we carried out cross-linking in lysates. N-biotin kemptide and ATP-ArN3 were incubated with HeLa lysates in the presence of UV light before SDS-PAGE analysis. A biotinylated band at the size of the PKA catalytic subunit (~41 kDa) was observed (Figure 2c, lane 2, see arrow). In addition to PKA, many other proteins were also biotinylated (Figure 2c, lane 2), consistent with possible diffusion of N-biotin kemptide away from the kinase after photo-cross-linker transfer (Figure 1c). The biotin signal, including that for PKA, was lost in the absence of ATP-ArN3 (Figure 2c, lane 1). Reduced biotinylation was also observed with N-biotin mutant kemptide (Figure 2c, lane 3) or in the presence of excess nonbiotinylated kemptide (Figure 2c, lane 4) or the kinase inhibitor staurosporine (Figure 2c, lane 5). These results suggest that peptide cross-linking is kinase-dependent and phosphosite-specific in complex mixtures, such as lysates.
To perform a full K-CLASP experiment, cross-linked complexes in lysates were purified using streptavidin resin (Figure S7) and analyzed by tandem mass spectrometry. Cross-linked samples from N-biotin mutant kemptide were used as the negative control to remove nonspecific background cross-linking. Compared to the mutant samples, 324 proteins were enriched in N-biotin kemptide cross-linked samples (Table S1). Out of the 324 proteins, only three were protein kinases (Table S2). Importantly, the known kemptide kinase PKA (alpha catalytic subunit PRKACA) was observed. In addition to PKA, two other kinases, CHEK1 and MAP2K7, and many nonkinase proteins were also observed by K-CLASP. To rule out the possibility that K-CLASP is preferentially pulling down high abundant proteins, we analyzed the abundance of the K-CLASP hits using previously published values.16 A range of abundances were observed among the K-CLASP hits (Figure S8), indicating that K-CLASP is not biased toward highly expressed proteins.
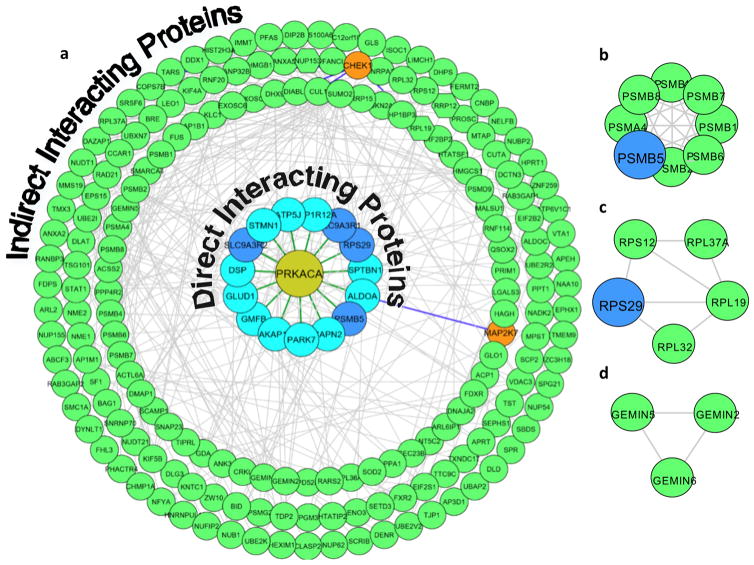
Given that K-CLASP can also identify proteins associated with the kinase during a cross-linking event, the many nonkinase proteins and the two other kinases observed could be either direct or indirect interactors of PKA. In this scenario, diffusion of N-biotin kemptide after PKA-catalyzed photo-cross-linker labeling (Figure 1c) may result in the biotinylation of the nearby proteins of PKA. To validate that CHEK1, MAP2K7, and the other proteins were biotinylated due to their proximity to PKA, we performed an interactome analysis. For the interactome analysis, protein–protein interactions among the 324 K-CLASP enriched proteins were determined using the GeneMANIA17 application in Cytoscape. In addition, PKA, CHEK1, and MAP2K7 substrates among the enriched proteins were identified using Phosphosite Plus and KinaseNET.18 The known substrates and interacting proteins in the K-CLASP hit list were then graphically depicted as an interactome map using Cytoscape (Figure 3a).19
Figure 3.
Kinase hits and interactome analysis of the PKA K-CLASP study. (a) The 324 proteins enriched in N-biotin kemptide but not N-biotin mutant kemptide samples (Supporting Information Table S1) were analyzed to identify known PKA, CHEK1, and MAP2K7 substrates using Phospho site Plus and KinaseNET. Known physical protein–protein interactions were analyzed using GeneMANIA in Cytoscape. All interacting proteins and substrates of PKA (60% of K-CLASP hits) were mapped using Cytoscape. The blue inner circles represent known PKA substrates (light) or PKA binding proteins (dark). Orange circles represent the other two kinases observed. Green circles represent indirect interacting proteins of PKA. Green hexagons represent indirect proteins of PKA that are also substrates of CHEK1. Green lines show direct interactions with PKA. Gray lines indicate interactions among the enriched proteins. Blue lines show the interactions of CHEK1 and MAP2K7. Line thickness and length were adjusted arbitrarily to improve clarity. An enlarged version of this figure is available as Figure S9. The Cytoscape file is also available as Supporting Information, ZIP file. (b) Proteins of the proteasome core complex identified by K-CLASP. (c) Proteins of the ribosome complex identified by K-CLASP. (d) Proteins of the SMN complex identified by K-CLASP.
The interactome analysis confirmed that known interacting proteins (Figure 3a, dark blue circles) and substrates of PKA (Figure 3a, light blue circles) were among the 324 K-CLASP hits. Beyond direct interacting proteins and substrates, many proteins that interact indirectly with PKA through complexes containing substrates or binding partners were observed (Figure 3a, green and orange circles). When all direct and indirect interactors were considered, 60% of the K-CLASP hits were accounted for in the PKA interactome. The presence of direct and indirect PKA interacting proteins among the K-CLASP enriched hits is consistent with diffusion of N-biotin kemptide away from PKA after labeling (Figure 1c), resulting in the cross-linking and biotinylation of proteins located in close proximity to PKA.
Importantly, the interactome analysis revealed that CHEK1 and MAP2K7 (Figure 3a, orange circles) interact indirectly with PKA through common interacting partners (Figure 3a, see blue lines). Therefore, CHEK1 and MAP2K7 biotinylation was likely due to their proximity to PKA, rather than by direct kinase-catalyzed cross-linking. In agreement, fewer direct interacting proteins or substrates were identified for CHEK1 and MAP2K7 compared to PKA (Figure S10). The small number of associated proteins or substrates of CHEK1 and MAP2K7 among K-CLASP hits is consistent with biotinylation due to peptide diffusion rather than a direct kinase labeling. By considering the direct interacting proteins of each kinase, the interactome analysis revealed false positive kinases.
Beyond PKA, kemptide is phosphorylated by cyclic GMP-dependent protein kinase15 and RPS6KA120 in vitro and predicted to be phosphorylated by AKT and protein kinase C based on consensus sequence.21 However, none of these kinases were identified by K-CLASP. Thus, K-CLASP identified PKA as the main kinase that phosphorylates kemptide in lysates. Given the wide use of kemptide in PKA studies,22–24 these results reinforce that PKA is the predominant kinase for kemptide.
In addition to eliminating false positive hits, the interactome analysis revealed cellular complexes associated with PKA under the conditions of the experiment. For example, PKA is known to phosphorylate several proteasome subunits.25 In agreement, K-CLASP identified multiple proteins belonging to the proteasome core complex (Figure 3b). Additionally, many members of the ribosome complex (Figure 3c) were also identified, consistent with reports that PKA regulates translation in response to starvation.26 Finally, PKA is known to regulate the stability of the SMN complex, which plays a role in the assembly of snRNPs (small nuclear Ribo Nucleo Proteins),27 and K-CLASP identified several members of the SMN complex (Figure 3d). While K-CLASP is primarily a tool for phosphosite-specific identification of kinases, these interactome results demonstrate that cellular complexes containing the kinase can also be uncovered.
In summary, K-CLASP was developed as a discovery tool to identify the cellular kinase of a given phosphosite. Using a well-known kinase–substrate pair, K-CLASP identified PKA as the predominant cellular kinase of kemptide from a complex mixture of other proteins and kinases. In addition, interactome analysis of K-CLASP hits identified the cellular complexes containing PKA and ruled out false positives. K-CLASP represents a simple, yet powerful, method to probe the function of a specific phosphosite. Importantly, K-CLASP only requires a biotinylated peptide containing a known phosphosite and cell lysate where phosphorylation of that specific site is observed. Studies are currently ongoing to use K-CLASP to identify the unknown kinase of a specific phosphosite. Given the importance of studying the many phosphorylation events in a cell, K-CLASP provides an enabling technology to understand the intricate regulatory networks governing cellular events.
METHODS
Detailed experimental procedures are available in the Supporting Information.
Supplementary Material
Acknowledgments
Funding
National Institutes of Health (R01 GM079529, P30 ES020957, P30 CA022453, and S10 OD010700) and Wayne State University.
We thank the National Institutes of Health (GM079529) and Wayne State University for funding; the Wayne State University and Karmanos Cancer Center Proteomics Core, which is supported by NIH Grants P30 ES020957, P30 CA022453, and S10 OD010700; T. Faner, J. Caruso, and N. Carruthers for technical support; and A. Fouda, D. M. Embogama, and N. Acharige for comments on the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions
P.M.D.-A. performed all experiments and assisted in experimental design and interpretation. M.K.H.P. conceived of the project and assisted in experimental design and interpretation. Both authors wrote the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio. 6b00289.
Table S1 (XLSX)
Methods, Figures S1–S10, Table S2 (PDF)
Cytoscape file (ZIP)
References
- 1.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsatsanis C, Spandidos DA. The role of oncogenic kinases in human cancer (Review) Int J Mol Med. 2000;5:583–590. doi: 10.3892/ijmm.5.6.583. [DOI] [PubMed] [Google Scholar]
- 3.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Xue L, Tao WA. Current technologies to identify protein kinase substrates in high throughput. Front Biol (Beijing, China) 2013;8:216–227. doi: 10.1007/s11515-013-1257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maly DJ, Allen JA, Shokat KM. A Mechanism-Based Cross-Linker for the Identification of Kinase-Substrate Pairs. J Am Chem Soc. 2004;126:9160–9161. doi: 10.1021/ja048659i. [DOI] [PubMed] [Google Scholar]
- 8.Statsuk AV, Shokat KM. Covalent cross-linking of kinases with their corresponding peptide substrates. Methods Mol Biol. 2012;795:179–190. doi: 10.1007/978-1-61779-337-0_12. [DOI] [PubMed] [Google Scholar]
- 9.Green KD, Pflum MK. Kinase-catalyzed biotinylation for phosphoprotein detection. J Am Chem Soc. 2007;129:10–11. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]
- 10.Senevirathne C, Embogama DM, Anthony TA, Fouda AE, Pflum MK. The generality of kinase-catalyzed biotinylation. Bioorg Med Chem. 2016;24:12–19. doi: 10.1016/j.bmc.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martić S, Gabriel M, Turowec JP, Litchfield DW, Kraatz HB. Versatile Strategy for Biochemical, Electrochemical and Immunoarray Detection of Protein Phosphorylations. J Am Chem Soc. 2012;134:17036–17045. doi: 10.1021/ja302586q. [DOI] [PubMed] [Google Scholar]
- 12.Wang N, She Z, Lin YC, Martic S, Mann DJ, Kraatz HB. Clickable 5′-gamma-ferrocenyl adenosine triphosphate bioconjugates in kinase-catalyzed phosphorylations. Chem - Eur J. 2015;21:4988–4999. doi: 10.1002/chem.201405510. [DOI] [PubMed] [Google Scholar]
- 13.Suwal S, Pflum MK. Phosphorylation-dependent kinase-substrate cross-linking. Angew Chem, Int Ed. 2010;49:1627–1630. doi: 10.1002/anie.200905244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garre S, Senevirathne C, Pflum MK. A comparative study of ATP analogs for phosphorylation-dependent kinase-substrate crosslinking. Bioorg Med Chem. 2014;22:1620–1625. doi: 10.1016/j.bmc.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maller JL, Kemp BE, Krebs EG. In vivo phosphorylation of a synthetic peptide substrate of cyclic AMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1978;75:248–251. doi: 10.1073/pnas.75.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11:M111 014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjorbaek C, Zhao Y, Moller DE. Divergent functional roles for p90rsk kinase domains. J Biol Chem. 1995;270:18848–18852. doi: 10.1074/jbc.270.32.18848. [DOI] [PubMed] [Google Scholar]
- 21.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, McKnight GS. Hypothalamic PKA regulates leptin sensitivity and adiposity. Nat Commun. 2015;6:8237. doi: 10.1038/ncomms9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangoda L, Doerflinger M, Lee YY, Rahimi A, Etemadi N, Chau D, Milla L, O’Connor L, Puthalakath H. Cre transgene results in global attenuation of the cAMP/PKA pathway. Cell Death Dis. 2012;3:e365. doi: 10.1038/cddis.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22:46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.