Abstract
Abstract
In the 1H NMR-guided fractionation of extracts from the edible mushroom Lactarius deliciosus, two new azulene-type sesquiterpenoids, 7-isopropenyl-4-methyl-azulene-1-carboxylic acid (1) and 15-hydroxy-3,6-dihydrolactarazulene (2), together with seven known compounds were characterized. Their structures were determined on basis of spectroscopic evidence, as well as by comparing with literature data. Amongst the known metabolites, the 13C NMR assignment of 15-hydroxy-6,7-dihydrolactarazulene (3) is reported here for the first time. Moreover, 7-acetyl-4-methylazulene-1-carbaldehyde (5) displayed a moderate antibacterial activity against Staphylococcus aureus.
Graphical Abstract
*Digital image of L. deliciosus. Retrieved March 17, 2017 from https://upload.wikimedia.org/wikipedia/commons/e/e3/Lactarius_deliciosus_1_(1).jpg
 .
.
Electronic supplementary material
The online version of this article (doi:10.1007/s13659-017-0130-1) contains supplementary material, which is available to authorized users.
Keywords: Lactarius deliciosus, Fungal pigments, Azulene sesquiterpenoids, Antibacterial activity
Introduction
The genus Lactarius belongs to the family Russulaceae (class Basidiomycetes) and is widely distributed allover the world. Many Lactarius species are edible; chemically, these mushrooms are appreciated for their metabolites, including sesquiterpenes, steroids, nitrogen-containing compounds and other secondary metabolites with e.g. antitumor or antiviral activities [1, 2]. Lactarius deliciosus, L. aureus, L. hatsudake, and others are well known for their colorful pigments, some of which are formed as response to injury of the fruiting bodies [3–5]. Previous chemical and biological investigation of this genus showed that these pigments belong to the group of azulene-type sesquiterpenoids, some of them possessing potent biological activities [6–10]. This class of metabolites is also considered as chemotaxonomic marker of the genus Lactarius [11].
As part of our search for fungal metabolites, we are reporting herein the structures and complete 1H and 13C NMR assignments of two new pigments 1 and 2 from the edible mushroom L. deliciosus collected in the forests near Göttingen (Germany). In addition to these two new compounds, four other pigments (3–6) and three fatty acids were also isolated. Although 1H NMR data of the unstable dihydroazulene alcohol 3 have been reported [3, 12], to the best of our knowledge, no 13C NMR data were published for this compound. We also evaluated the antimicrobial activities of the pigments 1 and 4–6.
Results and Discussion
After extraction of the freshly collected mushrooms with methanol and repetitive chromatography on Sephadex LH-20, compound 1 was obtained as a purple amorphous solid, with violet color in solution. Electrospray high resolution mass spectrometry (ESI HRMS) displayed an [M + H]+ ion peak at m/z 227.1075, suggesting C15H14O2 as molecular formula. The 1H and 13C NMR data of 1 (Table 1) were very similar to those reported for lactaroviolin (4) [13], a further Lactarius constituent also isolated in the present investigation. The 1H–1H COSY spectrum of 1 exhibited three spin systems of H-2/H-3, H-5/H-6/H-8/H-14 and H-12/H2-13, indicating that 1 and 4 might have the same azulene substitution pattern. Interestingly, the 1H NMR spectrum (Table 1) of 1 displayed a proton at δ H 10.0 (d, J = 2.1 Hz), which was attached to a carbon at δ C 136.8 (HSQC), excluding thereby an aldehyde. This finding corroborated with the 13C NMR spectrum (Table 1) that did not show resonances at lower field of an aldehyde group, but exhibited the characteristic signal of a conjugated carboxyl group at δ C 170.0. Further comparison of 1H and 13C NMR data of 1 with those of lactaroviolin (4) and 7-acetyl-4-methylazulene-1carboxylic acid [13] were in good agreement with the presence of the carboxy group at C-1. This was supported in the HMBC experiment by long-range couplings between the aromatic proton at δ H 8.43 (H-2) and the carboxy group (Fig. 2). The HMBC spectrum also exhibited cross-peaks of the olefinic methylene protons at δ H 5.46 and 5.33 (H2-12) with the carbon atoms at δ C 23.3 (C-13), 146.9 (C-11) and 141.2 (C-7), and of the methyl protons at δ H 2.96 (H-14) with the carbons at δ C 143.3 (C-10), 148.0 (C-4) and 129.9 (C-5). From the above data, the structure of 1 was established as 7-isopropenyl-4-methyl-azulene-1-carboxylic acid (Fig. 1). Related azulene-1-carboxylic acids have been characterized previously from L. deliciosus [13] and L. hadsudake [8].
Table 1.
1H NMR and 13C NMR data of compounds 1–3 (δ in ppm, J in MHz)
| No. | 1 a | 2 b | 3 b | |||
|---|---|---|---|---|---|---|
| δ H | δ C | δ H | δ C | δ H | δ C | |
| 1 | – | 116.5 | – | 147.4 | – | 139.1 |
| 2 | 8.43 (d, 4.3) | 140.6 | 6.23 (br s) | 125.6 | 6.24 (brs) | 127.3 |
| 3 | 7.30 (d, 4.3) | 115.3 | 3.20 (br s) | 40.4 | 6.33 (brs) | 129.0 |
| 4 | – | 148.0 | – | 132.7 | – | 132.1 |
| 5 | 7.44 (d, 10.8) | 129.9 | 5.13 (td, 7.1, 1.5) | 115.8 | 5.50 (br t, 7.1) | 121.8 |
| 6 | 7.88 (dd, 10, 2.1) | 136.2 | 2.48 (m) | 28.5 | 2.46 (m) | 30.8 |
| 2.26 (ddd, 15.4, 7.1, 3.1) | ||||||
| 7 | – | 141.2 | – | 130.8 | 3.23 (m) | 44.8 |
| 8 | 10.00 (d, 2.1) | 136.8 | 6.49 (br s) | 117.4 | 6.77 (d, 4.5) | 143.0 |
| 9 | – | 140.2 | – | 141.8 | – | 142.4 |
| 10 | – | 143.3 | – | 145.7 | – | 135.2 |
| 11 | – | 146.9 | – | 141.7 | – | 146.4 |
| 12 | 5.46 (s) | 116.3 | 5.42 (s) | 114.0 | 4.81 (m) | 111.5 |
| 5.33 (s) | 5.08 (s) | 4.75 (m) | ||||
| 13 | 2.36 (s) | 23.3 | 1.96 (s) | 21.1 | 1.77 (s) | 20.9 |
| 14 | 2.96 (s) | 25.0 | 1.93 (s) | 20.5 | 1.88 (s) | 21.9 |
| 15 | – | 170.0c | 4.33 (br s) | 58.1 | 4.31 (br s) | 56.6 |
a In CDCl3
b In DMSO
c From HMBC data
Fig. 2.
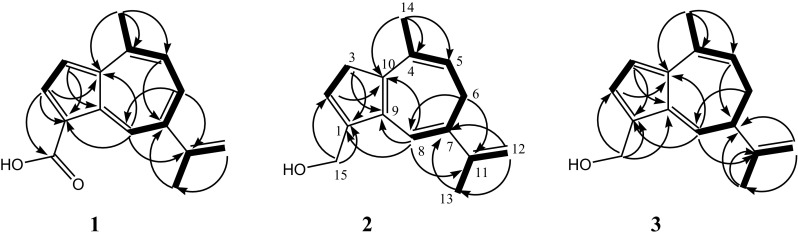
Key HMBC (arrows) and COSY (bold bonds) correlations for compounds 1–3
Fig. 1.
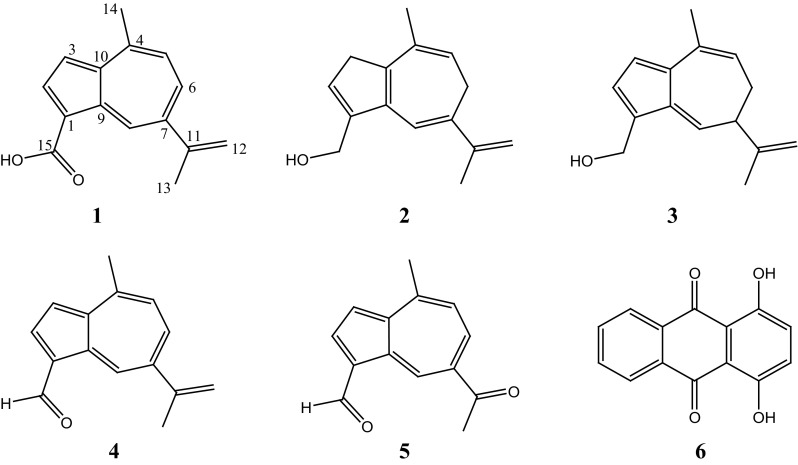
Structures of pigments 1–6 isolated from the fruiting bodies of Lactarius deliciosus
Further compounds were isolated as inseparable orange pigments, which decomposed rapidly. ESI MS exhibited pseudo-molecular ion peaks at m/z 237 [M + Na]+, 253 [M + K]+, and 467 [2M + K]+, consistent with the molecular formula C15H18O. The 1H and 13C NMR spectra indicated, however, that it was a mixture of two isomeric compounds 2 and 3 in a ratio of 1:3. The 1H and 13C NMR spectra of compounds 2 and 3 showed significant differences to those of 1 and 4, notably the signals of two aliphatic methylene groups in 2 and one methylene and one methine group in 3. Carbonyl signals were absent, and the methyl signal of C-14 at the azulene system was shifted upfield. The 1H NMR spectra of both compounds indicated two additional oxymethylene protons, which were connected with sp 3 carbons at δ C 58.1 and 56.6, respectively, indicating the presence of the dihydroazulene alcohols 2 and 3; the latter is known to be very unstable [3, 6, 12].
We were unable to separate both compounds, however, succeeded to assign all NMR signals in the mixture and elucidated the structures of 2 and 3 unambiguously; the 1H–1H COSY spectrum was clearly resolved and indicated three spin systems for each component (see Fig. 2). The different signal intensities in the 1:3 mixture of 2 and 3 allowed us to assign the HSQC and HMBC signals without doubt. For the less intensive peaks (compound 2), the methylene protons at δ H 4.33 (H2-15) correlated with the carbons at δ C 147.4 (C-1) and 125.6 (C-2); the expected correlation with C-9 was observed for compound 3, but not for 2 (Fig. 2). These two carbons in turn correlated with the methylene protons at δ H 3.20 (H2-3). HMBC correlation was also observed between C-1 and the olefinic proton at δ H 6.49 (H-8), which showed further correlations with the carbons at δ C 28.5 (C-6), 141.7 (C-11), and 145.7 (C-10). A long-range correlation was seen from the methyl at δ H 1.93 (H-14) to C-10, and to the carbons at δ C 132.7 (C-4) and 115.8 (C-5). Thereby, the minor component was finally characterized as 15-hydroxy-3,6-dihydrolactarazulene (2).
In a similar way, the main component was identified as 15-hydroxy-6,7-dihydrolactarazulene (3). Its structure was further supported by comparison of the 1H NMR data with those published by Sterner et al. [3]. To the best of our knowledge, the 13C NMR data (Table 1) for this compound are reported here for the first time.
Further components of the extract were identified on basis of their NMR data as lactaroviolin (4) [13, 14], 7-acetyl-4-methylazulene-1-carbaldehyde (5) [13], quinizarin (6) [15], stearic acid, and a mixture of oleic and linoleic acid. Among them, quinizarin (6) is reported here from higher fungi for the first time.
Compounds 1 and 4–6 were screened for their antimicrobial activities against Bacillus subtilis, Staphylococcus aureus, Escherichia coli, Candida albicans and Mucor miehei. Only 7-acetyl-4-methylazulene-1-carbaldehyde (5) displayed a moderate antibacterial activity against S. aureus in the agar diffusion test, with an inhibition diameter of 15 mm at a concentration of 50 μg/disc.
Experimental Section
General Experimental Procedures
The NMR spectra were recorded on a Varian Inova-500 spectrometer at 599.737 MHz (1H) or 150.818 MHz (13C), respectively. The chemical shifts are given in δ values with TMS as internal reference, and coupling constants are given in [Hz]. The ESI and ESI HR mass spectra were recorded on a Bruker micrOTOF mass spectrometer. Open column chromatography was done on silica gel 60 (0.063–0.20 mm), and PTLC was performed on silica gel P/UV254 (both obtained from Macherey–Nagel, Düren, Germany). Size exclusion chromatography was performed on Sephadex LH-20 (Lipophilic Sephadex; Amersham Biosciences, Ltd., purchased from Sigma-Aldrich Chemie, Steinheim, Germany). Pre-coated silica gel plates (Polygram SIL G/UV254, Macherey–Nagel & Co.) were used for TLC. Spots were visualized at 254 or 365 nm, and sprayed with an anisaldehyde/sulfuric acid reagent followed by heating.
Extraction and Isolation
The fruiting bodies of L. deliciosus were collected in forests near Göttingen (Germany) in September 2015. The fresh material (7.8 kg) was grinded and macerated with 2 × 4 L of MeOH at ~20 °C. The solution was evaporated under reduced pressure to afford a dark brown crude extract (110 g) that was suspended in water and further extracted with ethyl acetate (EtOAc). The evaporation residue (38 g) of the EtOAc fraction was chromatographed on Sephadex LH-20 (column 7 × 60 cm) with MeOH to give four main fractions A–D. According to their 1H NMR profiles, the colorless fractions A (8.5 g) and B (6.3 g) contained fatty acids and glycerol derivatives, while the orange and violet fractions C (11.7 g) and D (10.5 g), respectively, contained azulene-type sesquiterpenoids. Fraction C was again chromatographed on Sephadex LH-20 (column 4 × 90 cm) and further on silica gel (column, CH2Cl2) to afford a mixture of the unstable compounds 2 and 3 (35 mg). Fraction D crystallized from MeOH to yield stearic acid (200 mg). The mother liquor from D was purified again on Sephadex LH-20 (column, CH2Cl2/MeOH 1:1) to afford compound 1 (5 mg), quinizarine (6; 2 mg, R f = 0.90 CH2Cl2/MeOH 95:5) and two subfractions D-1 (violet) and D-2 (red). By PTLC (CH2Cl2/MeOH 99:1), D-1 gave lactaroviolin (4; 70 mg, R f = 0.78 CH2Cl2/MeOH 95:5). Purification of D-2 on a silica gel (column, CH2Cl2/MeOH 98:2) yielded 7-acetyl-4-methylazulene-1-carbaldehyde (5; 7 mg, R f = 0.72 CH2Cl2/MeOH 95:5) and a mixture of stearic, oleic and linoleic acid (150 mg).
7-Isopropenyl-4-methyl-azulene-1-carboxylic acid (1)
Purple amorphous solid, R f = 0.41 (CH2Cl2/MeOH 95:5); UV (MeOH) λ max (log ε) 238 (4.10), 300 (4.10), 380 (3.59); IR (film) νmax 3300-2500 (br, OH), 2920, 1652 (C=O), 1496, 1415, 1252, 890 cm−1; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3,125 MHz) data, see Table 1; (+)-ESI MS m/z 227 ([M + H]+), 249 ([M + Na]+), 475 ([2M + Na]+); (+)-ESI HRMS; m/z 227.1075 (calcd for C15H15O2 [M + H]+, 227.1067).
15-Hydroxy-3,6-dihydrolactarazulene (2) and 15-hydroxy-6,7-dihydrolactarazulene (3)
Orange gum, R f = 0.30 (CH2Cl2/MeOH 95:5); 1H NMR (DMSO-d 6, 300 MHz) and 13C NMR (DMSO-d 6, 125 MHz) data, see Table 1; (+)-ESI MS: m/z = 237 ([M + Na]+), 253 ([M + K]+), 475 ([2M + K]+); (+)-ESI HRMS; m/z 237.1250 (calcd for C15H18ONa [M + Na]+, 237.1255).
Antimicrobial Test
The antimicrobial test was performed according to a previously described procedure [16].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. H. Frauendorf and Dr. M. John for MS and NMR measurements, respectively. JQ thanks the Natural Science Foundation for a Chinese Government Scholarship Fund for Study Abroad (31470414, 20140101126JC).
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13659-017-0130-1) contains supplementary material, which is available to authorized users.
References
- 1.Yang Y, Bao NM, Shao HJ, Wang YL, Zhu L, Duan YF. Nat. Prod. Res. Dev. 2013;25:274–279. [Google Scholar]
- 2.Dong D, Li G. Nat. Prod. Res. Dev. 1991;4:66–80. [Google Scholar]
- 3.Bergendorff O, Sterner O. Phytochemistry. 1988;27:97–100. doi: 10.1016/0031-9422(88)80597-1. [DOI] [Google Scholar]
- 4.Spiteller P. Chem. Eur. J. 2008;14:9100–9110. doi: 10.1002/chem.200800292. [DOI] [PubMed] [Google Scholar]
- 5.Velíšek J, Cejpek K. J. Food Sci. 2011;29:87–102. [Google Scholar]
- 6.Bertelli DJ, Crabtree JH. Tetrahedron. 1968;24:2019–2089. doi: 10.1016/S0040-4020(01)82507-2. [DOI] [PubMed] [Google Scholar]
- 7.Harmon AD, Weisgraber KH, Weiss U. Experientia. 1979;36:54–56. doi: 10.1007/BF02003967. [DOI] [Google Scholar]
- 8.Fang LZ, Shao HJ, Yang WQ, Liu JK. Helv. Chim. Acta. 2006;89:1463–1466. doi: 10.1002/hlca.200690147. [DOI] [Google Scholar]
- 9.Fang LZ, Dong ZJ, Shao HJ, Liu JK. Acta Bot. Yunn. 2007;29:122–124. [Google Scholar]
- 10.Xu GH, Kim JW, Ryoo IJ, Choo SJ, Kim YH, Seok SJ, Ahn JS, Yoo ID. J. Antibiot. 2010;63:335–337. doi: 10.1038/ja.2010.43. [DOI] [PubMed] [Google Scholar]
- 11.Vidari G, Vita-Finzi P. Studies in natural products chemistry. In: Atta-ur-Rahman, editor. Stucture and Chemistry (Part D) Amsterdam: Elsevier; 1995. p. 153. [Google Scholar]
- 12.Vokáč K, Samek Z, Herout V, Šorm F. Collect. Czech. Chem. Commun. 1970;35:1296–1301. doi: 10.1135/cccc19701296. [DOI] [Google Scholar]
- 13.Yang XL, Luo DQ, Liu JK. Z. Naturforsch. 2006;61b:1180–1182. [Google Scholar]
- 14.Yang XL, Luo DQ, Dong ZJ, Liu JK. Helv. Chem. Acta. 2006;89:988–990. doi: 10.1002/hlca.200690103. [DOI] [Google Scholar]
- 15.Lee HS. J. Microbiol. Biotechnol. 2003;13:529–536. [Google Scholar]
- 16.Sajid I, Fotso Fondja Yao CB, Shaaban KA, Hasnain S, Laatsch H. World J. Microbiol. Biotechnol. 2009;25:601–610. doi: 10.1007/s11274-008-9928-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


