Abstract
Staphylococcus aureus is a common colonizer in pigs, with methicillin-resistant S. aureus (MRSA) in particular being a potential health risk to humans. To reduce the exposure to humans, the colonization in pigs should be reduced. The aim of this study was to quantitatively compare the susceptibility of pig lineages for S. aureus colonization, and if the absence of S. aureus could be associated with the presence or absence of other staphylococcal species. Nasal samples (n = 129) were obtained from seven different pig lineages in the Netherlands, France, and Germany. S. aureus and other staphylococci were enumerated from these samples by real-time (RT)-PCR and culture. Associations were explored between the presence of S. aureus and other staphylococci. S. aureus was detected by RT-PCR on all farms and in samples from pigs of all lineages. Twenty-five percent of the pigs from lineage F (from two farms) were colonized with S. aureus, while in all other lineages it was more than 50% (p < 0.01). Moreover, in S. aureus-positive samples from pigs of lineage F smaller amounts of S. aureus were found than in other lineages. Staphylococcus sciuri, Staphylococcus cohnii, and Staphylococcus saprophyticus were usually not found in combination with S. aureus in these samples. In conclusion: (i) pigs from different genetic lineages have different susceptibilities for colonization with S. aureus. These pigs might contain a genetic factor influencing nasal colonization. (ii) Colonization of S. aureus is also associated with the absence of S. sciuri, S. cohnii, or S. saprophyticus. (iii) The farm environment seems to influence the presence of S. aureus in pigs.
Keywords: Staphylococcus aureus, pigs, methicillin-resistant S. aureus, colonization, staphylococci
Introduction
Staphylococcus aureus is one of the most common opportunistic pathogens. Its methicillin-resistant variant—methicillin-resistant S. aureus (MRSA)—is often multiresistant and can pose a therapeutic challenge when it causes an infection. MRSA can be distinguished in community-associated, hospital-associated (HA-MRSA), and livestock-associated (LA-MRSA). LA-MRSA—in Western countries belonging to clonal complex 398—is commonly found in pigs and calves and is a poor colonizer in humans (1). However, people with occupational exposure to LA-MRSA-colonized livestock (e.g., farmers) are at increased risk of becoming a carrier (2). Despite the large proportion of LA-MRSA-positive individuals, individuals showing clinical signs due to LA-MRSA are limited. This may be attributed to the fact that the largest fraction of LA-MRSA-positive individuals are healthy and are less likely to develop clinical disease. Furthermore, the capacity of LA-MRSA to spread in health-care institutions is limited compared to HA-MRSA (3, 4). Nonetheless, the incidence of infections with LA-MRSA in Denmark has in recent years increased among people without livestock contact (5, 6).
To lower the risk for acquiring LA-MRSA, the exposure needs to be limited either by reduction of shedding of LA-MRSA by livestock or physical protection of people who are exposed (e.g., by using a nose-and-mouth mask). Because of the increasing problem of antimicrobial resistance, approaches to reduce LA-MRSA colonization need to be explored and assessed for their potential and efficacy. In general, reduction of colonization of animals can be studied by different approaches: (1) the colonizing MRSA (and its interaction with the microbiota) and (2) the host, e.g., epithelial adhesion.
Interactions of MRSA with its environment provide possibilities for interference with colonization. In a study from Japan, S. aureus nasal colonization in humans was eradicated by the introduction of Corynebacterium spp. into the nares (7). In another study, the serine protease Esp, secreted by Staphylococcus epidermidis, was found to inhibit nasal colonization by S. aureus (8). Furthermore, a study looking at nasal colonization in Dutch children showed a negative association between the presence of Streptococcus pneumoniae and S. aureus (9, 10).
The approach from the host is based on the observation that not all hosts—human and pig—are colonized: some are intermittent carriers, and others never carry S. aureus (11, 12). This difference suggests that there are genetic differences between hosts, e.g., the absence of a receptor or presence of a modified receptor. In Danish pigs, a relationship between a single nucleotide polymorphism (SNP) and carriage of S. aureus has been identified, although the function of the gene in which this SNP is present is unknown (13). If such genetic predisposition for S. aureus colonization is present in purebred pigs used in breeding, this might offer possibilities to select for animals that are genetically less susceptible to colonization with S. aureus and therewith MRSA.
By characterizing the nasal staphylococcal microbiota in defined genetic pig lineages both approaches can be studied. Although the focus for control in livestock is only on MRSA, we investigated carriership of S. aureus in general. The aim of this study was to explore if pigs of different genetic lineages have different susceptibilities for nasal colonization with S. aureus and to investigate possible (negative) associations with colonization of other staphylococci.
Materials and Methods
Sample Collection
Nasal swabs (dry, rayon-tipped, Copan, Italy) were collected for routine screening from 13 groups of pigs, on 11 farms. Pigs were selected randomly. In total, 129 samples were analyzed. These pigs were of seven different high-end lineages: A (n = 20 samples), B (n = 20), C (n = 20), D (n = 10), E (n = 10), F (n = 20), and G (n = 29) (Table 1). Farm owners gave informed consent for using these samples for this study. These are all high-end lineages that are used by Topigs Norsvin (the Netherlands) and are used in pig production on the European continent. Farms were located in the Netherlands (n = 8), France (n = 2), and Germany (n = 1). Samples were obtained on August 2014 and March–April 2015. Ten swabs per lineage were used; in some cases a single farm hosted multiple lineages (see Table 1). Swabs were suspended in 1 mL molecular-grade saline before analysis.
Table 1.
Pig farm details.
| Farm | Country | Lineage | Samples | Lineages on farm |
|---|---|---|---|---|
| 1 | FR | D | 10 | B, D, E |
| 2 | FR | F | 10 | F, G |
| G | 10 | F, G | ||
| 3 | NL | B | 10 | A, B |
| 4 | NL | E | 10 | E |
| 5 | NL | G | 10 | G |
| 6 | NL | C | 10 | C |
| 7 | DE | B | 10 | B |
| 8 | NL | A | 10 | A |
| 9 | NL | F | 10 | F, G |
| G | 9 | F, G | ||
| 10 | NL | C | 10 | C |
| 11 | NL | A | 10 | A |
FR, France; NL, the Netherlands; DE, Germany.
Real-time (RT)-PCR
To perform an accurate quantification of S. aureus in these samples RT-PCR was performed. 200 μL of the swab suspension was used for DNA isolation with the High-Pure PCR Template Preparation kit (Roche, the Netherlands) and eluted in 50 µL. 5 µL of DNA was used for quantitative PCR of S. aureus, targeting the nuc gene (14). Phocine herpes virus was added before DNA isolation and used as an internal amplification control (15). RT-PCR was performed on a LightCycler 480-II system (Roche), and quantification was performed using a standard curve. The theoretical limit of detection for this PCR is 50 CFU/sample.
Bacterial Culture
All staphylococci, including S. aureus, were quantified by bacterial culture. Serial dilutions of suspensions in saline were prepared with 10−4 as the highest dilution. From all dilutions 100 µL was plated onto mannitol salt agar (MSA) for the detection of staphylococci in general. After overnight incubation at 37°C (O/N), the dilution with 20–200 colonies on MSA was used to count and select all morphologically distinct colonies, which were identified with MALDI-TOF MS (Bruker, Germany) after they were subcultured on blood agar. If MALDI-TOF MS was inconclusive, tuf sequencing was performed to identify the staphylococcal species (16).
Statistics
All analyses were performed using R statistical software v3.0.2. Proportions of S. aureus-positive samples of different lineages, obtained by RT-PCR, were compared using Fisher’s exact test. As post hoc test the proportions of S. aureus-positive samples from each lineage were compared against the proportions in all other lineages by Fisher’s exact test. Probabilities were corrected for multiple testing by the Holm–Bonferroni method. Quantitative RT-PCR results were compared using the one-way ANOVA test, with Tukey Honestly Significant Differences as post hoc test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to explore associations between the presence of S. aureus and other staphylococci.
Results
Nasal Colonization with S. aureus
Staphylococcus aureus was detected by RT-PCR in 84/129 samples (65%) on all farms and in all different lineages: A: 11/20; B: 16/20; C: 13/20, D: 9/10; E: 8/10; F: 5/20; and G: 22/29 (Figure 1A). None of the RT-PCRs was inhibited.
Figure 1.
Presence of Staphylococcus aureus in different pig lineages. (A) Percentage of S. aureus-positive samples. (B) Quantitative results from S. aureus-specific real-time-PCR (only S. aureus-positive samples). Horizontal bars indicate median of the amount of S. aureus in positive samples. In graphs, “S. aureus per farm per lineage” the x-axis indicates the combination of farm number and lineage name.
For lineage F, the percentage of negative samples was significantly lower than for other lineages (p < 0.01). The amount of S. aureus in the positive samples ranged from 3.5 × 103 to 4.7 × 107 CFU/sample, with an overall mean of 1.1 × 105 CFU/sample (Figure 1B).
The mean amount of S. aureus was also the lowest in pigs from lineage F, although this difference was not statistically significant (one-way ANOVA, Tukey HSD). On farms 2 and 9 were also pigs of lineage G, alongside pigs from lineage F. Although these animals were in the same environment as those of lineage F, 70% of the pigs of lineage G on farm 2 and 78% of the pigs of lineage G on farm 9 were positive for S. aureus, compared to 30 and 20% S. aureus-positive animals of lineage F on each farm, respectively. However, on farm 11 (lineage A) 80% of the samples were S. aureus-negative, while 90% of the samples from lineage A on farm 8 were S. aureus-positive.
Association of S. aureus Colonization with Presence of Other Staphylococci
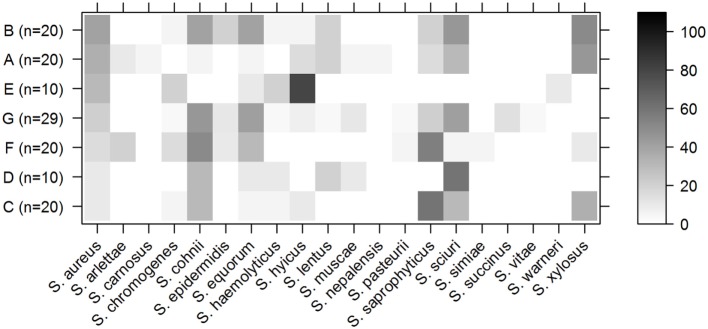
Besides S. aureus, 20 other staphylococcal species were isolated by culture, of which Staphylococcus cohnii was isolated most frequently (41 samples), followed by Staphylococcus sciuri (40 samples), Staphylococcus saprophyticus (36 samples), Staphylococcus equorum (30 samples), and Staphylococcus xylosus (28 samples). S. aureus was isolated by culture from 30 samples. Figure 2 shows the distribution of all the staphylococcal species that were isolated from the different genetic pig lineages.
Figure 2.
Staphylococci in pig lineages. Percentage of samples per lineage from which a specified staphylococcal species was isolated by culture. Rows indicate the different pig lineages with the total number of samples from this lineage between brackets. The columns contain different staphylococcal species. The shading of the cell represents the percentage of samples from that lineage that contained this species (right bar shows scale). Pig lineages are sorted by percentage of S. aureus-positive samples (first column).
Staphylococcus aureus was cultured from 40% of the animals of lineage B, but only 20% of the animals from this lineage were positive for S. saprophyticus (Figure 2). Conversely, S. aureus was cultured from only 10% of the animals of lineage C, but 60% were positive for S. saprophyticus.
The presence of S. aureus was negatively associated with the presence of three other staphylococci species, namely, S. sciuri (OR: 0.11, CI: 0.03–0.51), S. cohnii (OR: 0.18, CI: 0.05–0.63), and S. saprophyticus (OR: 0.22, CI: 0.06–0.79). The prevalence of these bacteria is displayed in Figure S1 in Supplementary Material.
Discussion
Staphylococcus aureus was detected in all pig lineages on all farms, but not in all individual pigs. As S. aureus is transmitted via direct contact and through dust, it may be assumed that this organism is present throughout the entire farm and all pigs are exposed to this organism (17). Despite this general exposure, S. aureus was only detected in 65% of the animals. This favors the hypothesis that certain factors prevent S. aureus from colonizing in all animals.
Lineage F was the only lineage with less than 50% S. aureus-positive pigs, which was also observed in pigs of lineage F from both individual farms (farms 2 and 9). Lineage A on farm 11 also had more S. aureus-negative than positive animals, but the pigs on farm 8 had more S. aureus-positive than negative samples suggesting that other factors on a farm also influence colonization, as described elsewhere (18). Pigs of lineage G were also present on both farms (mixed housing), and on both of these farms pigs from lineage G were more often S. aureus-positive than negative. Also, the mean amount of S. aureus in the positive samples from lineage F was the lowest among all lineages. This suggests that pigs of lineage F have a reduced susceptibility to S. aureus colonization. A recent study in pigs in Denmark indicated that genetic variation in pigs influences the ability of S. aureus to colonize, and a locus was identified that was associated with nasal colonization of S. aureus (13). Currently, this is the only association study of nasal colonization of S. aureus in different pig lineages. Because the pig production chain in Denmark uses different pig lineages than the lineages described in our study these SNPs are not necessarily present in the pigs in our study. The concept of host genetics influencing the ability of bacteria to colonize is not new. A study in China identified SNPs in HEG1, an uncharacterized protein, and ITGB5, which plays a role in the innate immune system and influenced the susceptibility of pigs to enterotoxigenic Escherichia coli (19, 20). However, none of these studies report a 100% association between gene presence and the colonization of S. aureus or E. coli and indicates that this is most likely determined by an interplay of multiple factors.
The other factor we studied is if S. aureus colonization is associated with the presence of other staphylococcal species. A previous study showed that a serine protease called Esp from S. epidermidis inhibits growth of S. aureus in vitro. When this strain was introduced into the nares of S. aureus carriers, it was able to eliminate the presence of S. aureus (8). Staphylococcus lugdunensis produces lugdunin, which is encoded by the lugD gene, was shown to outcompete S. aureus in an in vitro experiment. Also, when rats were cocolonized with S. lugdunensis and S. aureus in the nose, the S. aureus strain was outcompeted (21). In this study, we did not find any S. lugdunensis, so these findings could not be confirmed in pigs. Alternatively, colonization with S. aureus can be associated with other bacterial species. In a study in Japan, both MRSA and MSSA were successfully replaced by Corynebacterium spp. in almost all volunteers by nasal administration of corynebacteria (7). Other studies showed a negative association between the presence of S. aureus and nasal carriage of pneumococci (9, 10). In the present study, we found S. aureus together with S. sciuri, S. cohnii, or S. saprophyticus in only eight samples (from six different farms), while 22 samples contained S. aureus without any of these three species. Seventy-five samples did not contain S. aureus, but at least one of the other three species that were mentioned. It is possible that the presence of one of these staphylococcal species can prevent—or at least inhibit—colonization by S. aureus; but intervention studies are required to prove this.
In our study, we also observed an influence of between-farm differences in lineage A: many pigs on farm 8 were colonized with S. aureus, while only a few of the pigs of the same lineage on farm 11 were colonized. The existence of other factors that may have a role in determining S. aureus colonization in pigs cannot be excluded, but our study was not designed to investigate those. We did not collect information on other potential factors of relevance at the farm, and the number of farms in this study was not sufficient to allow proper analysis of those factors.
It was shown that for pig farmers the most important determinants for becoming an MRSA carrier are the exposure to high amounts of MRSA in the air of the barn and the hours that are spent in the barn (17). That study did not find an association between MRSA persistence in farmers and the proportion of MRSA-positive animals on the farm. However, on veal calf farms (where an all-in-all-out regime with or without cleaning before the next herd is housed) the MRSA prevalence in calves increased over time during a production cycle. This led to higher probabilities for human MRSA carriage and indicates that the level of MRSA contamination in the farm needs time to accumulate (18). In pig farming in the Netherlands an all-in-all-out regime is largely applied, but not with carrying sows on reproduction farms, so contamination of the environment by MRSA-shedding pigs is continuous on the latter type of farms. Further research should focus on the question if a reduction in the number of MRSA-positive pigs by introducing pigs with a genetic background that makes them less susceptible to S. aureus colonization will lead to a reduced or slower contamination of the environment. This would reduce the MRSA exposure of individuals working on these farms.
Real-time-PCR to detect S. aureus in this study was found to be more sensitive than culture on MSA because low amounts of S. aureus may be overgrown when other staphylococci are present in larger quantities. Therefore, the RT-PCR results were used when only S. aureus results were considered (i.e., comparison of number of positive samples), while the culture results for S. aureus were used when comparing the results from all staphylococci.
Most of the lineages were sampled on more than one farm, except for lineages D and E. Although samples from multiple farms in different countries were included in the analysis, farm characteristics may still contribute to the amount of S. aureus in the pig’s nose. However, the only lineage that had less S. aureus-positive samples was lineage F. Samples from this lineage were obtained from two farms: one in France and one in the Netherlands. But given the small number of tested pigs these data should be confirmed in a larger study.
In conclusion: (i) pigs from lineage F are less often S. aureus-positive than pigs from other lineages. (ii) S. aureus was rarely found in the same samples as S. sciuri, S. cohnii, or S. saprophyticus, which indicates a possible interaction between these species. (iii) The farm environment seems to influence the presence of S. aureus in pigs.
Ethics Statement
Samples were collected as part of a routine screening procedure and not for the purpose of this project. Therefore, approval of an ethics committee was not mandatory.
Author Contributions
EW, AF, BD, MM, JW, and KV designed the experiments. EW collected samples, and KV performed analysis. All the authors were involved in the interpretation of the results. KV drafted the manuscript; EW, AF, BD, MM, and JW revised the manuscript. All the authors agreed with the publication of this manuscript.
Conflict of Interest Statement
Coauthors MM and EW are employees of Topigs Norsvin. The other authors declare no conflict of interest.
Acknowledgments
We are grateful to Dr. Haitske Graveland for her help with the statistical analyses.
Footnotes
Funding. This article is the result of self-funded work.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fvets.2017.00097/full#supplementary-material.
Venn-diagram indicating samples containing Staphylococcus aureus, Staphylococcus sciuri, Staphylococcus cohnii, or Staphylococcus saprophyticus. Most samples only contained one of these four organisms, except for 13 samples containing both S. sciuri and S. cohnii.
References
- 1.Verkade E, Kluytmans-van den Bergh M, van Benthem B, van Cleef B, van Rijen M, Bosch T, et al. Transmission of methicillin-resistant Staphylococcus aureus CC398 from livestock veterinarians to their household members. PLoS One (2014) 9:e100823. 10.1371/journal.pone.0100823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Cleef BAGL, van Benthem BHB, Verkade EJM, van Rijen MML, Kluytmans-van den Bergh MFQ, Graveland H, et al. Livestock-associated MRSA in household members of pig farmers: transmission and dynamics of carriage, a prospective cohort study. PLoS One (2015) 10:e0127190. 10.1371/journal.pone.0127190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bootsma MCJ, Wassenberg MWM, Trapman P, Bonten MJM. The nosocomial transmission rate of animal-associated ST398 meticillin-resistant Staphylococcus aureus. J R Soc Interface (2011) 8:578–84. 10.1098/rsif.2010.0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassenberg MWM, Bootsma MCJ, Troelstra A, Kluytmans JAJW, Bonten MJM. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin Microbiol Infect (2011) 17:316–9. 10.1111/j.1469-0691.2010.03260.x [DOI] [PubMed] [Google Scholar]
- 5.Larsen J, Petersen A, Sørum M, Stegger M, van Alphen L, Valentiner-Branth P, et al. Meticillin-resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro Surveill (2015) 20:1–9. 10.2807/1560-7917.ES.2015.20.37.30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen J, Stegger M, Andersen PS, Petersen A, Larsen AR, Westh H, et al. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis (2016) 63:1349–52. 10.1093/cid/ciw532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium spp. J Hosp Infect (2000) 44:127–33. 10.1053/jhin.1999.0680 [DOI] [PubMed] [Google Scholar]
- 8.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature (2010) 465:346–9. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- 9.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet (2004) 363:1871–2. 10.1016/S0140-6736(04)16357-5 [DOI] [PubMed] [Google Scholar]
- 10.Lijek RS, Luque SL, Liu Q, Parker D, Bae T, Weiser JN. Protection from the acquisition of Staphylococcus aureus nasal carriage by cross-reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci U S A (2012) 109:13823–8. 10.1073/pnas.1208075109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev (1997) 10:505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Cleef BAGL, van Benthem BHB, Verkade EJM, van Rijen M, Kluytmans-van den Bergh MFQ, Schouls LM, et al. Dynamics of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus carriage in pig farmers: a prospective cohort study. Clin Microbiol Infect (2014) 20:O764–71. 10.1111/1469-0691.12582 [DOI] [PubMed] [Google Scholar]
- 13.Skallerup P, Espinosa-Gongora C, Jørgensen CB, Guardabassi L, Fredholm M. Genome-wide association study reveals a locus for nasal carriage of Staphylococcus aureus in Danish crossbred pigs. BMC Vet Res (2015) 11:290. 10.1186/s12917-015-0599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilic A, Muldrew KL, Tang Y-W, Basustaoglu AC. Triplex real-time polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci and determination of methicillin resistance directly from positive blood culture bottles. Diagn Microbiol Infect Dis (2010) 66:349–55. 10.1016/j.diagmicrobio.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 15.Niesters HG. Quantitation of viral load using real-time amplification techniques. Methods (2001) 25:419–29. 10.1006/meth.2001.1264 [DOI] [PubMed] [Google Scholar]
- 16.Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol (2005) 43:2286–90. 10.1128/JCM.43.5.2286-2290.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bos MEH, Verstappen KM, van Cleef BAGL, Dohmen W, Dorado-García A, Graveland H, et al. Transmission through air as a possible route of exposure for MRSA. J Expo Sci Environ Epidemiol (2014) 26:263–9. 10.1038/jes.2014.85 [DOI] [PubMed] [Google Scholar]
- 18.Dorado-García A, Bos MEH, Graveland H, van Cleef BAGL, Verstappen KM, Kluytmans JAJW, et al. Risk factors for persistence of livestock-associated MRSA and environmental exposure in veal calf farmers and their family members: an observational longitudinal study. BMJ Open (2013) 3:e003272. 10.1136/bmjopen-2013-003272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W-X, Liu Y, Lu X, Niu X-Y, Ding X-D, Liu J-F, et al. A genome-wide association study identifies two novel promising candidate genes affecting Escherichia coli F4ab/F4ac susceptibility in swine. PLoS One (2012) 7:e32127. 10.1371/journal.pone.0032127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Python P, Jörg H, Neuenschwander S, Asai-Coakwell M, Hagger C, Bürgi E, et al. Inheritance of the F4ab, F4ac and F4ad E. coli receptors in swine and examination of four candidate genes for F4acR. J Anim Breed Genet (2005) 122(Suppl 1):5–14. 10.1111/j.1439-0388.2005.00490.x [DOI] [PubMed] [Google Scholar]
- 21.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature (2016) 535:511–6. 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn-diagram indicating samples containing Staphylococcus aureus, Staphylococcus sciuri, Staphylococcus cohnii, or Staphylococcus saprophyticus. Most samples only contained one of these four organisms, except for 13 samples containing both S. sciuri and S. cohnii.