Abstract
Genomic information has become a ubiquitous and almost essential aspect of biological research. Over the last 10–15 years, the cost of generating sequence data from DNA or RNA samples has dramatically declined and our ability to interpret those data increased just as remarkably. Although it is still possible for biologists to conduct interesting and valuable research on species for which genomic data are not available, the impact of having access to a high quality whole genome reference assembly for a given species is nothing short of transformational. Research on a species for which we have no DNA or RNA sequence data is restricted in fundamental ways. In contrast, even access to an initial draft quality genome (see below for definitions) opens a wide range of opportunities that are simply not available without that reference genome assembly. Although a complete discussion of the impact of genome sequencing and assembly is beyond the scope of this short paper, the goal of this review is to summarize the most common and highest impact contributions that whole genome sequencing and assembly has had on comparative and evolutionary biology.
Keywords: genome assembly, sequencing, gene family analysis, segmental duplications
Introduction
One basic distinction is critical at the outset. In many circumstances, the phrase “sequencing a new genome” refers to the analysis of an individual, including the subsequent comparison of that one individual’s genome to a reference genome assembly meant to represent that species. When the genome of a human patient with an undiagnosed clinical disorder is sent for “sequencing,” the typical procedure is to generate sufficient raw read data to compare some (the protein coding exome) or (nearly) all of the patient’s genome to a standard human reference and look for differences that may be pathogenic and hence clinically relevant. Although it is likely that in the future it will be possible, and maybe even routine, to produce de novo whole genome assemblies for individual patients at acceptable cost this is not now practical. However, the re-sequencing of individuals (i.e. the analysis of a given individual by comparing that individual’s DNA sequence to a curated and annotated reference genome) is not the focus of this review. Rather, we discuss the methods for and impact of producing de novo whole genome assemblies for species for which such information is not currently available. Thus, the question at issue is not “what do we learn about a specific individual from sequencing the genome of that specific individual” but rather “what do we learn about species X from sequencing an individual or pool of individuals that represent species X.” Producing a new reference genome facilitates analyses of other individuals from the same or closely related species, and this wider meaning and impact is our topic here.
How does one produce a new reference genome assembly?
The technology for sequencing DNA and assembling the raw sequence read data into a continuous representation of chromosomes continues to improve at a rapid pace. For this reason, any review of genomic methods and anticipated results will have a short shelf-life. Nevertheless, some general principles are likely to remain relevant for the foreseeable future. The current methods for producing the raw sequence data from which de novo whole genome assemblies can be constructed fall into two categories. The dominant short read technology comes from Illumina, Inc. and uses well-established chemistry to identify the sequence of nucleotides present in a given short segment of DNA. Current Illumina sequencing platforms produce basepair sequences of lengths up to 250 bp in a given DNA segment, and generally are used to read those sequences from both ends of a DNA fragment. This “next generation” technology was a dramatic improvement over older Sanger sequencing methods that produced longer reads but at much higher cost. The Illumina platforms have become the workhorses of genome sequencing but “third generation” technologies from Pacific Biosciences, Oxford Nanopore and other companies are gaining users and making impact. These “third generation” methods generate longer reads, up to tens of thousands of basepairs per read, but with higher error rates and other disadvantages.
The field of de novo whole genome assembly is currently grappling with the challenges of determining the optimal use of these various and constantly changing data types. As the methods for generating the raw sequence read data have evolved, so have the analytical strategies and software tools designed to assemble the large number of short or long reads into continuous sequences millions of bases (megabases) in length. The ultimate goal of genome assembly is production of error-free continuous sequences that span entire chromosomes. In contrast, currently attainable genome assemblies have tens to hundreds of thousands of gaps, though much of the genome is represented with good sequence quality in the assembled regions. Progress has been rapid and it is not outrageous to envision that essentially complete, highly accurate whole genome assemblies will be practical in the near future.
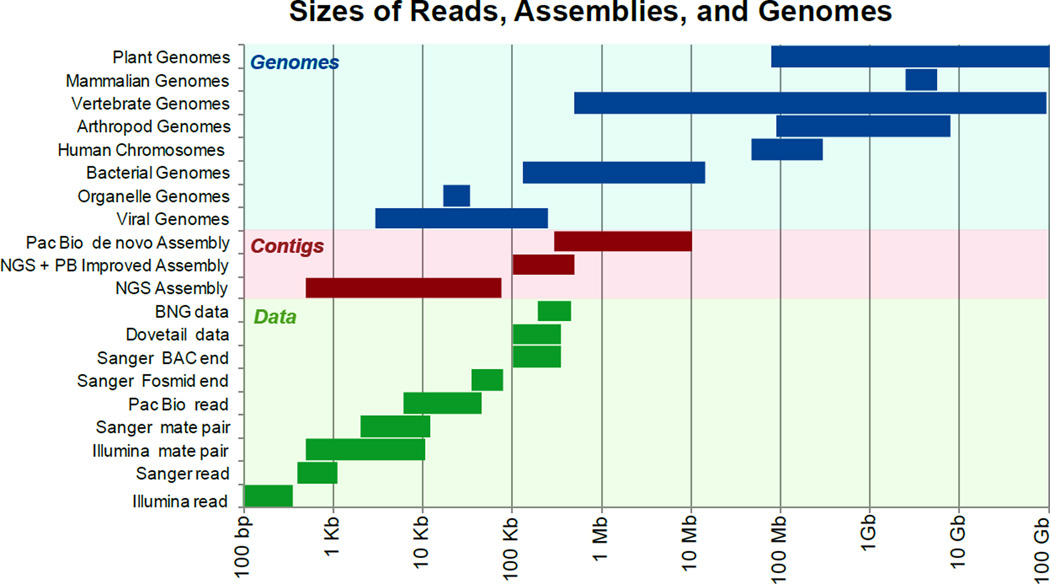
The data, problem and current results are illustrated in Figure 1 where the size of elements are represented on the logarithmic scale range from 100 basepairs to 100 billion basepairs (100 Gb). At the top in Figure 1 are the sizes of the targets of genome assembly (genomes and chromosomes). The sequence read lengths and mate-pair distances for the different technologies are on the bottom.
Figure 1.
This figure shows the challenges of creating complete genome representations.
The relative sizes of different genomes, genome assembly contigs, and sequencing technologies are shown with the logarithmic scale across the bottom. At the top in blue are the sizes of genomes for different clades of organisms. Vertebrate genome sizes vary over 5 orders of magnitude, while mammalian genome sizes are more similarly sized (~3Gb). In the middle, in red, are the size ranges for the contigs (measure by contig N50) for current sequencing technologies (Illumina next generation or NGS assemblies, NGS assemblies with PacBio improvement, and PacBio de novo assemblies). The lengths of sequence reads, mate-pair distances and mapping fragments of different technologies are shown in the green bars at the bottom.
In the middle of Figure 1, the lengths of the pieces in a de novo assembly are indicated using the contig N50 values. Contig N50 is a statistic commonly used to compare different genome assembly results in terms of their contiguity (the length of continuous DNA sequences without gaps, called contigs). Contig N50 is calculated by sorting all the contigs within an assembly from largest to smallest, then determining the size of the contig at which half of the total genome is in pieces bigger than that N50 value. Larger contig N50 values correlate with improved recovery of genomic features (see below). Note that the genome sizes and chromosome sizes are 10 to 1,000 times longer than the contig N50 values of current de novo assemblies. Improving the contiguity by adding some long read data to an NGS assembly or using a de novo long read method is beneficial. Often it is genomic features such as repeats that limit the contiguity attained in a genome assembly and these methods resolve many of these repeats and recover missing data. As an example, the de novo PacBio gorilla genome recovered 87% of the exons missing in earlier assemblies[1]. There remains room for improvement, because even easier to assemble haploid samples need directed finishing efforts to address the last difficult regions[2, 3], and the extensively studied “finished” human reference genome is still being improved[4]. Despite this potential for further progress, changing reference genome versions is a painful process of transferring genome analysis coordinates (the basepair locations) from the old version to the new, and there will always be sequences that are altered during this transfer that someone prefers as the old version. So even as genomic sequencing costs drop and de novo genome assemblies becomes more achievable, researchers should plan to use the current genome iteration for several years.
Genome Analysis Wish Lists
As a foundational biological tool there are many analyses possible with a new genome (Table 1). We present here a short list of the most common analyses with example results leading to biological insight. First and foremost on this wish list is identification of the protein-coding genes in that genome and the comparative analysis of gene families. Gene family analyses begin with identifying the collection of protein coding genes in the organism, which is compared to the gene complement from other species already sequenced. Differences in the number and types of genes present as well as differences in the sequences of orthologous genes are evaluated.
Table 1.
Genome Analysis Wish List
| Protein coding genes | Structure and complete sequence |
| Gene families | Expansions and contractions |
| Rapidly evolving genes | Positively selected |
| Non-coding RNA genes | Structure and complete sequence |
| Ancestral gene content | |
| Repetitive elements | Transposable elements |
| Segmental duplications | |
| Population history | Phylogenetics, population size |
| Genomic history | Lateral gene transfer, admixture |
The genome assemblies first produced by large-scale sequencing efforts allowed investigation of lineage-specific gene duplication and gene loss[5]. Expanding gene families are thought to be a birthplace for molecular innovation[6] and have repeatedly provided biological insight into for example photoreception[7] and rumination[8]. The sheep genome highlighted genes in the epidermal development complex involved in keratinized structures, expressed in the rumen in sheep and cattle, and that appear to function in this ruminate-specific adaptation. Gene losses can also be important. The T1R1 gene is inactivated in the panda, creating an inactive umami taste receptor, which may be related to this carnivore’s herbivorous diet[9]. These innovations may be targets for therapeutics or pesticides, such as the secreted proteins in blood feeding pests [10, 11]. The list of genes may be interesting for what is missing, suggesting that the organism can do without some genes that are required in similar organisms. For example, in the pea aphid, a number of metabolic pathways were incomplete because the functions were carried out by its intracellular mutualistic bacterium[12]. More commonly, there will be gene families that are expanded or present in a lineage where they were not anticipated. In the honey bee and other eusocial bees genetic pathways that control DNA methylation are found that are not present in drosophila, and DNA methylation is used to regulate the reproductive suppression of worker bees[13–15]. Combinations in these innovations may indicate a shift in species ecology such as the kiwi, where opsin genes related to color vision are inactivated and the odorant receptor repertoire is expanded, consistent with increased reliance on olfaction for nocturnal foraging[16]. Perhaps the most dramatic example of massive expansion and diversification is that of animal parasitic effector genes to avoid recognition by the plant immune system. There are more than1,000 effector genes in the gall forming pest Hessian fly [17] and similar massive expansions are in the infamous Irish potato famine pathogen [18]. Independent gene family expansions of three gene families in the mammalian platypus and the reptile snake lineages have led to the convergent development of venoms[19]. The convergence of similar genomic strategies across such divergent taxa is remarkable.
Each new genome identifies genes that are very similar to genes in other sequenced species. Genes involved in fundamental highly conserved cellular functions such as DNA and RNA processing are commonly in this category. Analysis also identifies genes that are more divergent, often genes involved in interactions with the environment such as detoxification genes, immunobiology and sexual selection. Gene family analyses often focus on these genes that are rapidly evolving, and show signatures of positive selection. In the giraffe, over half of the genes identified as showing multiple signs of adaption are involved in developmental patterning functional pathways. These include unique changes predicted to be functional in homeobox genes consistent with the growth required to produce and support the long giraffe neck[20]. Killer whale ecotypes show positively selected genes associated with cold adaptation and feeding behavior (mammal or fish predation)[21].
Genes identified as having been subjected to positive selection on the lineage being studied are found to have associations with diseases, or are enriched in pathways related to phenotypes of the species under study. For example, a quarter of the positively selected genes in the genome of the nocturnal, large-eyed tarsier have been implicated in eye development or visual disorders[22]. And the genome of the sheep identified a gene found in wool follicles positively selected in the sheep vs. cattle lineages that is potentially associated with wool development[8].
As more genomes become available, it is becoming possible to predict the ancestral gene content of a clade of species and to analyze the biology of that ancestor by genomic proxy. The report of two acorn worm genomes identified shared traits inherited from the last common deuterostome ancestor such as transcription factor genes associated with gill slits, as well as potential lateral gene transfer from marine microbes[23].
A related important focus of attention is the identification of the complement of non-coding genes including miRNA, lncRNA, and other regulatory RNA genes. These are also often different between closely related species. The analysis of the marmoset genome sequence discovered differences in miRNA genes and targets relative to human orthologs that raised more questions then could be immediately answered, primarily because these genes had not been extensively studied[24]. In bumblebee genomes, miRNAs appear to have a role in their social behavior[14].
The analysis of a new whole genome assembly inevitably provides new insights into repetitive elements and sequences within genomes. Retrotransposons, pseudogenes, segmental duplications and other repeated sequences make up a large fraction of eukaryotic genomes and hence are an important aspect of comparative genomic analysis. The movement and duplication of transposable elements can generate copies of genes or parts of genes and creating substrates for gene innovations. An extreme example is found in the gibbon, where the gibbon-specific LAVA element inserted in a number of genes involved in chromosome segregation. This appears to disturb the gene regulation enough to drive the excessive chromosome rearrangements in the genome[25]. Gene families can be expanded via retrotransposition duplication as in in the dinoflagellate coral endosymbiote [26]. And changes in low copy repeats can increase risk of disease-causing deletions[27].
The evolutionary insertion of new mobile elements allows researchers to distinguish ancestral from derived haplotypes, and this information about ancestral states has also been used to investigate population history[28]. An improved tarsier genome assembly was recently published [22] and allowed comparisons between this primate and other species phylogenetically closer to humans including the New World monkeys, Old World monkeys, and the great apes (which include humans). The new genome sequence allowed a more complete description of the history of the repeat elements showing when different types of elements were active in the different lineages.
Sex chromosomes[29], centromeres[30], telomeres, and segmental duplications are all technically difficult to sequence and assemble and contain functional elements that have interesting biology[3, 5, 6, 28–38]. Such recently or repeatedly duplicated sequences are hotbeds for gene family expansions. Hominoid lineage specific gene expansions are frequently found in pericentromeric and subtelomeric genomic regions[5].
Comparisons to other sequenced genomes are used to identify syntenic relationships as well as population history, introgression and admixture. Introgression has been identified in a number of species complexes including mosquitos[39] and butterfly mimics[40]. Moreover, recent introgression can complicate the analysis of older introgression events as in the diversity study of Darwin’s finches with evidence of both ancient and modern hybridization[41]. And of course, there has been substantial interest in the finding that the modern human genome contains significant amounts of DNA sequence introgressed from both Neanderthals and Denisovans[42, 43].
Although we think of genomic history as being from inherited sequence, either chromosomal or mitochondrial, genomic studies have highlighted the role of lateral gene transfer. This includes insertions from mitochondrial genomes already present in the species (numts), and lateral gene transfer (LGT) from symbiotic bacteria. The latter can include almost the entire bacterial genome as in the case of Wolbachia in Drosophila ananassae [44]. In some cases such as the bedbug, the genome seems to contain the degraded remains of older bacterial LGTs from multiple sources [45]. LGTs can lead to ecologically important functional innovation as in the Asian Longhorned beetle genome which harbors bacterially derived glycoside hydrolases enabling the lignin and cellulose digestion that makes it a serious pest of forests[46]. Finally, genome references can also show where LGTs have not occurred. The pea aphid symbiont Buchnera has a dramatically reduced genome size due to loss of genes that are functionally replaced by the host aphid. This is in contrast to the functions that have been transferred to the aphid genome from other bacterial species[12]. Overall LGT is a significant evolutionary force.
Complementary Genomic Data that synergistically enhance the analyses
Other types of genomic data have been used to study new genomes, either as a cost-saving approach or to provide added dimension to the genome that allows better interpretation. These data include expressed sequences, genomic DNA sequences from related individuals [47], and sequencing data that samples epigenetic regulation[48] and three-dimensional chromatin structure[49]. Expressed RNA sequences were initially used as a cost-saving method to assess the gene complement without sequencing the rest of the genome. As sequencing costs have dropped these analyses have become integral analyses to improve the identification of genes in the genome sequence[8]. Gonadal and brain tissue expression are often the first sequenced as these tissues are richer in expressed sequences that are not found in other tissues. Using RNAseq data from a number of different tissues is necessary to more completely sample the genes that are not universally expressed[50]. The GC content of the honey bee genome is bimodal and the initial annotation was more complete in regions with higher GC content. Once additional RNAseq data became available from different tissues, it became clear that this was an annotation bias and not due to gene deserts or regions where genes were not found[51]. More recently RNAseq data has been used to sample a variety of tissues and developmental states to understand genome regulation and function[52].
Beyond understanding the gene changes described above, comparisons to the genomes of other species that are closely or distantly related will produce novel insights[53]. Comparisons among individuals within the same species are used to understand the extent of variation within the species[54, 55]. The data for these analyses can be WGS or sample sequences like genotyping assays using chips designed for other species. The deeper sequencing data improves the resolution, but the sampling methods can provide an initial interpretation that can be expanded and refined in subsequent studies. The amount of variation between individuals within a species varies widely[56]. An extreme example is the cheetah with very low single nucleotide variation [57]. The type of variation can include larger events such as chromosome inversions, which in stickleback play a role in the repeated evolution of marine and freshwater adaptations[58].
Finally, in addition to the expressed data and other genomes, epigenetic data from methylation sequencing or chromatin conformation studies can enhance the understanding of genomic regulation. A comprehensive review of these types of data is available [59].
Resources are always limited so one must set priorities and decide what is most important when approaching a genome sequencing project. Associated data such as RNA-seq and data to support annotation can compensate for a lower quality genome assembly. In some situations, additional data to study within species variability may be more important than a more complete and accurate reference genome assembly. Initial directed efforts to capture and sequence protein sequences similar to those in the genome of a sequenced related species or to limit de novo genome analyses to protein coding genes can conserve resources. Wider scope with analyses including non-coding RNAs, regulatory sequences, methylation and other epigenetic marks are more resource intensive than just the sequencing required to sample the genome.
The Larger Audiences for Genomes
There are a number of constituencies beyond genomics researchers with different requirements for genome analysis. Researchers working on the physiology or metabolism of a given species want a catalog of protein-coding genes and noncoding RNAs with complete and accurate sequences for each. These resources are also required to define peptide mass expectations for proteomics mass spec assays. In addition, researchers working on cellular and molecular mechanisms of gene regulation want high quality sequences of upstream and downstream regulatory elements and a deep transcriptome to allow analysis of tissue-specific expression. For evolutionary genomics, well-justified orthology lists are also needed to know which genes in a given species are orthologous to what genes in another species. Population geneticists need whole genome sequences and variant lists from additional individuals in the species. Phylogenetics studies require whole or partial genome sequences to compare sequence divergence among groups of species. Microbiome metagenomic studies leverage existing genome sequences to interpret the functional capacity and composition of the populations they study.
Biomedical researchers do not necessarily require information for all of the genes in a model organism, sequence and expression data for genes known or suspected to be involved in a particular human disease may suffice. Getting the genomic structure information needed for these studies may still be difficult, particularly for genes in regions such as segmental duplications[33]. Association studies designed to locate the relevant genes also benefit from a well-annotated genome[60]. Often a research inquiry starts with a less expensive technique and then when the answer proves elusive, the analysis expands to more extensive (and usually more expensive) methods.
Despite the expectation that having a genome sequence will improve many future analyses, the translational impact of a genome sequence may be hard to predict. One unexpected finding in the bovine genome, is how much it changed production agricultural practice. Prior to the genome sequence, there were prize bulls (chosen based upon the milk production of their daughters) that contributed large fractions of the chromosome complement of the Australian Holstein population. More recent methods have been developed to make genomic predictions using DNA marker information early in the life of an animal, thus avoiding the need to wait until a sire can be evaluated based on his daughter’s phenotype[61].
Genome engineering is no longer limited to a few model organisms [62]. RNA interference has been used for agricultural pest control[63]. As CRISPR genome editing techniques move genetic modification to the individual instead of requiring breeding experiments, these methods are becoming ubiquitous. Combining these modifications with gene drive mechanisms proposed to control mosquito vectors for infectious diseases[64]. Genomic sequence is the necessary substrate for these experiments.
Concluding Thoughts
In conclusion, a whole genome reference assembly is foundational to biological studies. With the newer genomic techniques for studying gene expression and function, genome regulation and for engineering genomes, this is more true today than when the earlier Sanger-based genome assemblies were published. Any genome sequence assembled today will be used to open up new avenues of research, even in well-studied species.
Acknowledgments
Funding acknowldegement: This work was supported by the National Institutes of Health (U54 HG003273 awarded to Richard Gibbs).
Abbreviations
- NGS
next generation sequencing
- LGT
lateral gene transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon D, Huddleston J, Chaisson MJ, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352(6281):aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider VA, Lindsay TG, Howe K, Bouk N, Chen H-C, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. 2016 doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg KM, Schneider VA, Graves-Lindsay TA, Fulton RS, Agarwala R, Huddleston J, Shiryev SA, Morgulis A, Surti U, Warren WC, et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014;24(12):2066–2076. doi: 10.1101/gr.180893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church DM, Schneider VA, Steinberg KM, Schatz MC, Quinlan AR, Chin CS, Kitts PA, Aken B, Marth GT, Hoffman MM, et al. Extending reference assembly models. Genome Biol. 2015;16:13. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2(7):E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorente-Galdos B, Bleyhl J, Santpere G, Vives L, Ramirez O, Hernandez J, Anglada R, Cooper GM, Navarro A, Eichler EE, et al. Accelerated exon evolution within primate segmental duplications. Genome Biol. 2013;14(1):R9. doi: 10.1186/gb-2013-14-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies WI, Tamai TK, Zheng L, Fu JK, Rihel J, Foster RG, Whitmore D, Hankins MW. An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 2015;25(11):1666–1679. doi: 10.1101/gr.189886.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344(6188):1168–1173. doi: 10.1126/science.1252806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anstead CA, Korhonen PK, Young ND, Hall RS, Jex AR, Murali SC, Hughes DS, Lee SF, Perry T, Stroehlein AJ, et al. Lucilia cuprina genome unlocks parasitic fly biology to underpin future interventions. Nat Commun. 2015;6:7344. doi: 10.1038/ncomms8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Aphid Genomics C. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honeybee Genome Sequencing C. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P, Elsik CG, Gadau J, Grimmelikhuijzen CJ, Hasselmann M, Lozier JD, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16(1):76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapheim KM, Pan H, Li C, Salzberg SL, Puiu D, Magoc T, Robertson HM, Hudson ME, Venkat A, Fischman BJ, et al. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science. 2015;348(6239):1139–1143. doi: 10.1126/science.aaa4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Duc D, Renaud G, Krishnan A, Almen MS, Huynen L, Prohaska SJ, Ongyerth M, Bitarello BD, Schioth HB, Hofreiter M, et al. Kiwi genome provides insights into evolution of a nocturnal lifestyle. Genome Biol. 2015;16:147. doi: 10.1186/s13059-015-0711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Escalante LN, Chen H, Benatti TR, Qu J, Chellapilla S, Waterhouse RM, Wheeler D, Andersson MN, Bao R, et al. A Massive Expansion of Effector Genes Underlies Gall-Formation in the Wheat Pest Mayetiola destructor. Current biology : CB. 2015 doi: 10.1016/j.cub.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 18.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 19.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453(7192):175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agaba M, Ishengoma E, Miller WC, McGrath BC, Hudson CN, Bedoya Reina OC, Ratan A, Burhans R, Chikhi R, Medvedev P, et al. Giraffe genome sequence reveals clues to its unique morphology and physiology. Nat Commun. 2016;7:11519. doi: 10.1038/ncomms11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foote AD, Vijay N, Avila-Arcos MC, Baird RW, Durban JW, Fumagalli M, Gibbs RA, Hanson MB, Korneliussen TS, Martin MD, et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun. 2016;7:11693. doi: 10.1038/ncomms11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz J, Noll A, Raabe CA, Churakov G, Voss R, Kiefmann M, Rozhdestvensky T, Brosius J, Baertsch R, Clawson H, et al. Genome sequence of the basal haplorrhine primate Tarsius syrichta reveals unusual insertions. Nat Commun. 2016;7:12997. doi: 10.1038/ncomms12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simakov O, Kawashima T, Marletaz F, Jenkins J, Koyanagi R, Mitros T, Hisata K, Bredeson J, Shoguchi E, Gyoja F, et al. Hemichordate genomes and deuterostome origins. Nature. 2015 doi: 10.1038/nature16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmoset Genome Sequencing and Analysis Consortium. The common marmoset genome provides insight into primate biology and evolution. Nat Genet. 2014;46(8):850–857. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513(7517):195–201. doi: 10.1038/nature13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Cheng S, Song B, Zhong X, Lin X, Li W, Li L, Zhang Y, Zhang H, Ji Z, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350(6261):691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 27.Yuan B, Liu P, Gupta A, Beck CR, Tejomurtula A, Campbell IM, Gambin T, Simmons AD, Withers MA, Harris RA, et al. Comparative Genomic Analyses of the Human NPHP1 Locus Reveal Complex Genomic Architecture and Its Regional Evolution in Primates. PLoS genetics. 2015;11(12):e1005686. doi: 10.1371/journal.pgen.1005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing J, Wang H, Zhang Y, Ray DA, Tosi AJ, Disotell TR, Batzer MA. A mobile element-based evolutionary history of guenons (tribe Cercopithecini) BMC biology. 2007;5:5. doi: 10.1186/1741-7007-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner BM, Sargent CA, Churcher C, Hunt T, Herrero J, Loveland JE, Dunn M, Louzada S, Fu B, Chow W, et al. The pig X and Y Chromosomes: structure, sequence, and evolution. Genome Res. 2016;26(1):130–139. doi: 10.1101/gr.188839.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 2016;26(10):1301–1311. doi: 10.1101/gr.206706.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17(4):224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller ZL, Haynes GD, Richards S, Schaeffer SW. Genomics of Natural Populations: How Differentially Expressed Genes Shape the Evolution of Chromosomal Inversions in Drosophila pseudoobscura. Genetics. 2016;204(1):287–301. doi: 10.1534/genetics.116.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gazave E, Darre F, Morcillo-Suarez C, Petit-Marty N, Carreno A, Marigorta UM, Ryder OA, Blancher A, Rocchi M, Bosch E, et al. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res. 2011;21(10):1626–1639. doi: 10.1101/gr.117242.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marques-Bonet T, Girirajan S, Eichler EE. The origins and impact of primate segmental duplications. Trends Genet. 2009;25(10):443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miga KH. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res. 2015;23(3):421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- 36.She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431(7011):927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 37.Young RS, Hayashizaki Y, Andersson R, Sandelin A, Kawaji H, Itoh M, Lassmann T, Carninci P, Consortium F. Bickmore WA, et al. The frequent evolutionary birth and death of functional promoters in mouse and human. Genome Res. 2015;25(10):1546–1557. doi: 10.1101/gr.190546.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Landback P, Gschwend AR, Shen B, Long M. New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 2015;16:202. doi: 10.1186/s13059-015-0772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heliconius Genome C. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamichhaney S, Berglund J, Almen MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerova M, Rubin CJ, Wang C, Zamani N, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 42.Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, Patterson N, Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507(7492):354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352(6282):235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunning Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Munoz Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317(5845):1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 45.Benoit JB, Adelman ZN, Reinhardt K, Dolan A, Poelchau M, Jennings EC, Szuter EM, Hagan RW, Gujar H, Shukla JN, et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. 2016;7:10165. doi: 10.1038/ncomms10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna DD, Scully ED, Pauchet Y, Hoover K, Kirsch R, Geib SM, Mitchell RF, Waterhouse RM, Ahn S-J, Arsala D, et al. The Asian longhorned beetle (Anoplophora glabripennis) genome reveals key functional and evolutionary innovations at the beetle-plant interface. In Preparation. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bovine HapMap C, Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, Gill CA, Green RD, Hamernik DL, Kappes SM, et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324(5926):528–532. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, Boyle AP, Zhang QC, Zakharia F, Spacek DV, et al. Extensive variation in chromatin states across humans. Science. 2013;342(6159):750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng X, Thierry-Mieg J, Thierry-Mieg D, Nishida A, Pipes L, Bozinoski M, Thomas MJ, Kelly S, Weiss JM, Raveendran M, et al. Tissue-specific transcriptome sequencing analysis expands the non-human primate reference transcriptome resource (NHPRTR) Nucleic Acids Res. 2015;43(Database issue):D737–D742. doi: 10.1093/nar/gku1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535(7612):367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The 1000 Genomes Project C. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, Dahdouli M, Rio Deiros D, Below JE, Salerno W, et al. The population genomics of rhesus macaques (Macaca mulatta) based on whole genome sequences. Genome Res. doi: 10.1101/gr.204255.116. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13(4):e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobrynin P, Liu S, Tamazian G, Xiong Z, Yurchenko AA, Krasheninnikova K, Kliver S, Schmidt-Kuntzel A, Koepfli KP, Johnson W, et al. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 2015;16:277. doi: 10.1186/s13059-015-0837-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shendure J, Lieberman Aiden E. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30(11):1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayward JJ, Castelhano MG, Oliveira KC, Corey E, Balkman C, Baxter TL, Casal ML, Center SA, Fang M, Garrison SJ, et al. Complex disease and phenotype mapping in the domestic dog. Nat Commun. 2016;7:10460. doi: 10.1038/ncomms10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayes BJ, Lewin HA, Goddard ME. The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 2013;29(4):206–214. doi: 10.1016/j.tig.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Reed RD. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 2016;7:11769. doi: 10.1038/ncomms11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hajeri S, Killiny N, El-Mohtar C, Dawson WO, Gowda S. Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphorina citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing) J Biotechnol. 2014;176:42–49. doi: 10.1016/j.jbiotec.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 64.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112(49):E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]