Summary
Almost one‐third of crop yields are lost every year due to microbial alterations and diseases. The main control strategy to limit these losses is the use of an array of chemicals active against spoilage and unwanted pathogenic microorganisms. Their massive use has led to extensive environmental pollution, human poisoning and a variety of diseases. An emerging alternative to this chemical approach is the use of microbial biocontrol agents. Biopesticides have been used with success in several fields, but a better understanding of their mode of action is necessary to better control their activity and increase their use. Very few studies have considered that biofilms are the preferred mode of life of microorganisms in the target agricultural biotopes. Increasing evidence shows that the spatial organization of microbial communities on crop surfaces may drive important bioprotection mechanisms. The aim of this review is to summarize the evidence of biofilm formation by biocontrol agents on crops and discuss how this surface‐associated mode of life may influence their biology and interactions with other microorganisms and the host and, finally, their overall beneficial activity.
Introduction
Approximately 30% of crop yields are lost every year worldwide, mostly due to diseases caused by pests, weeds or pathogenic microorganisms (Teng and Krupa, 1980; Teng, 1987; Oerke, 1999, 2006; Savary et al., 2012). The microbiological control of agricultural products along the food chain is still mainly ensured by the extensive use of chemical pesticides, preservatives and synthetic drugs (Horrigan et al., 2002). Environmental pollution and associated human diseases caused by this excessive use of chemicals during last century has led many agencies and governments worldwide to support an alternative route, where agriculture can be productive and economically viable, while still addressing societal and environmental concerns (Anonymous, 1999; Hazell and Wood, 2008; Aktar et al., 2009). Biological protection strategies are used and encouraged from farm to forks to prevent pathogen contaminations and livestock or crop diseases (Pal and McSpadden Gardener, 2006; Sundh and Melin, 2010; Jordan et al., 2014). Biological control, or ‘biocontrol’, consists in the removal of the harmful activity of one organism via one or more organisms or natural products extracted from microorganisms, plants, animals or minerals (Pal and McSpadden Gardener, 2006).
The relationship between survival, persistence and virulence of pathogenic microorganisms with their biofilm mode of life have been clearly established since the early 1980s (Costerton et al., 1978; Lam et al., 1980). According to the National Institute of Health, 80% of human infections involves microbial biofilms (NIH, 2002). Biofilm‐associated infections have also been reported in agricultural settings, e.g., in crops and animal diseases (Davey and O'toole, 2000; Prigent‐Combaret et al., 2012; Li et al., 2015). Indeed, the sessile mode is the preferential lifestyle of microorganisms, regardless of their biotope (Davey and O'toole, 2000; Morris and Monier, 2003). A biofilm can be described as a spatially structured community of microorganisms, generally embedded in an extracellular matrix, and adhering to a living or inert surface (Costerton et al., 1999; O'Toole et al., 2000). Biofilm formation is generally favoured in harsh environmental conditions, such as low nutritive or toxic media (Rendueles and Ghigo, 2015) and most bacteria can form biofilms in various environments (Morris and Monier, 2003; Aparna and Yadav, 2008). Staphylococcus aureus and Pseudomonas aeruginosa are two opportunistic pathogenic bacteria that cause a diverse set of diseases and are the most highly used model bacteria for biofilm studies. They can colonize the human nasopharynx and form biofilms when specific environmental conditions are met, causing invasive diseases, such as chronic pneumonia. These infections are difficult to treat because of the persistence of biofilms and their high resistance to antimicrobials (Blanchette and Orihuela, 2012; Ding et al., 2016a). Bacteria can colonize and form biofilms on stems, leaves and the rhizosphere of plants, as well as soil particles, mushrooms or organic compost (Figs 1A and 2) (Ramey et al., 2004; Prigent‐Combaret et al., 2012). For example, Dickeya dadantii, the causal agent of soft rot disease in a wide range of plant species, can colonize and form biofilms on chicory leaves, causing disease due to the production of degradative enzymes (Prigent‐Combaret et al., 2012; Pandin et al., 2016).
Figure 1.
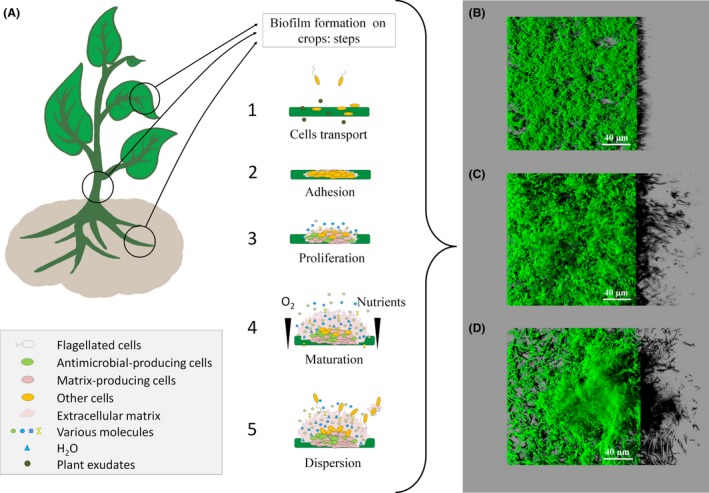
Biofilm formation on crops and in vitro: (A): On crops: The first step involves deposition on the substratum (1) followed by adhesion (2) to the support through cell wall decorations and extracellular appendages. Once attached, a proliferation phase (3) and the diversification of cell types initiate the spatial organization of the biostructure, leading to biofilm maturation (4). Biofilm ageing or environmental conditions unfavourable for the maintenance of the biofilm results in regulated dispersion of the biofilm (5), disseminating free cells and cell clusters that will start a new biofilm cycle on a new surface. B–D. In vitro: Structural diversity of three biocontrol agents as observed in vitro (24 h of axenic culture in microplates at 25°C) by confocal laser scanning microscopy (Leica SP8); (B) Bacillus amyloliquefaciens FZB42 expressing a green fluorescent protein (GFP), forming flat undifferentiated architecture, (C) Bacillus amyloliquefaciens SQR9 expressing a GFP and (D) Bacillus subtilis QST 713 (labelled in green with syto 9, Invitrogen, France) forming differentiated 3D biostructures.
Figure 2.
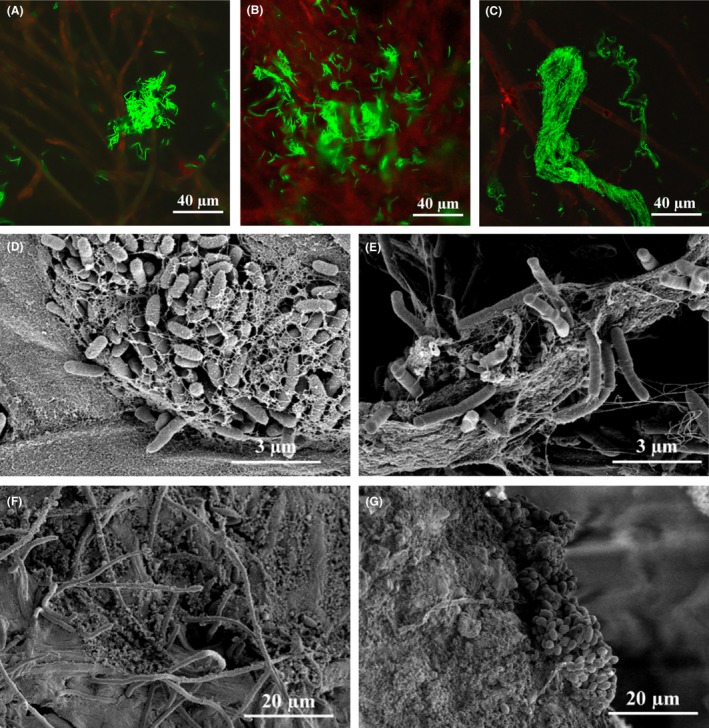
Microbial biofilms on the carpophore and culture compost of Agaricus bisporus. A–C. Confocal laser scanning microscopy of Agaricus bisporus carpophore (red autofluorescent hyphae), harbouring Bacillus amyloliquefaciens FZB42 expressing GFP and forming (A) clusters, (B) biofilm features and (C) bundles. Agaricus bisporus carpophores were immersed under axenic conditions in TSB (Tryptone Soy Broth, Sigma‐Aldrich, France) inoculated with Bacillus amyloliquefaciens FZB42 (GFP tagged) and incubated for 48 h at 17°C. Observations were performed using a Leica SP8 (Leica Microsystems, Danaher, Germany). D–G. Scanning electron microscopy of natural biofilms formed on Agaricus bisporus carpophore and compost protected with Bacillus subtilis QST 713, a biocontrol agent used at the French Mushroom Centre (Distré, France). Samples were fixed in 0.10 M cacodylate buffer containing 2.5% (v/v) glutaraldehyde (pH 7.4) and post‐fixed in 1% osmium tetroxide. Samples were then dehydrated with increasing concentrations of ethanol at room temperature (50–100%). After drying, samples were mounted on grids, sputter‐coated in argon plasma with platinum (Polaron SC7640, Elexience, France) and observed using a FE‐SEM S4500 (Hitachi, Japan). (D) Pseudomonas‐like bacteria with extracellular material, (E) Bacillus‐like bacteria, (F) fungi hyphae with extracellular material, (G) bacterial microcolony.
Although less explored, the formation of biofilms by moulds, yeast and algae, alone or in combination, in a variety of biotopes has also been reported (Morris and Monier, 2003; Aparna and Yadav, 2008; Zarnowski et al., 2014; He et al., 2016; Rajendran and Hu, 2016; Sheppard and Howell, 2016). Aspergillus fumigatus, a human pathogen, is a filamentous fungus that can form structured biofilms. The cohesive cement of the fungal biostructure is a polymeric extracellular matrix that protects the hyphae from the host immune system, similar to bacterial biofilms (Breitenbach et al., 2016; Mitchell et al., 2016; Sheppard and Howell, 2016; Shirazi et al., 2016). Fusarium oxysporum f. sp. cucumerinum, the pathogen responsible for cucumber Fusarium wilt, can also grow inter‐ and intracellularly, allowing the rapid colonization of the plant and biofilm formation (Li et al., 2015). Until recently, efforts in biofilm research have focused mainly on the medical field and essentially towards their eradication. With the emergence of biocontrol in agriculture, many microbiological products have been developed and are used in fields (Borriss, 2015). The main way of action of most of these commercial products is the antagonistic effect of antimicrobial molecules secreted by the biocontrol agent (Chowdhury et al., 2015; Mora et al., 2015). However, recent research in this field has made it possible to consider other major biological processes, including biofilm formation of biocontrol agents in crops (Bais et al., 2004; Bogino et al., 2013; De la Fuente et al., 2013).
The formation of biofilms by microbial biocontrol agents
Evidence of biofilm formation on crops by biocontrol agents
There is ongoing research to identify new biocontrol agents from environmental isolates and numerous biocontrol products have been developed and put on the agricultural market, mostly in Europe and North America (Borriss, 2015). Various products are in use and are effective on a wide range of plants. These include biofungicides, bactericides and biofertilizers based on Bacillus subtilis QST 713 or Bacillus amyloliquefaciens FZB42 (Borriss, 2015). These biocontrol products have an antagonistic effect towards unwanted microbes due to their secretion of antimicrobials, such as surfactin, fengycin or iturin (Ongena et al., 2005; Ongena and Jacques, 2008; Cawoy et al., 2014, 2015; Saravanakumar et al., 2016). However, their precise mechanisms of action in fields are still unknown. Few studies have focused on the determinants of effective bioprotection. The surface colonization step and biofilm formation by biocontrol agents are highlighted in the publications cited in Table 1. These reports demonstrate that many biocontrol agents can form biofilms on crops and in the rhizosphere. It has also been shown that biofilm formation by biopesticides can be stimulated by plant root exudates (Espinosa‐Urgel et al., 2002; Timmusk et al., 2005; Haggag and Timmusk, 2008; Khezri et al., 2011; Chen et al., 2013; Sang and Kim, 2014; Zhang et al., 2015), or by exposure of the microorganisms to antimicrobial products or stress (Bais et al., 2004; Selin et al., 2010; Fan et al., 2011; Xu et al., 2014; Chi et al., 2015; Wu et al., 2015; Zhou et al., 2016), but only a few studies have focused on biocontrol mechanisms that may be related to the properties of the mature biofilm itself, rather than the secretion of antimicrobials. Bacillus are ubiquitous spore forming bacteria predominantly found in soil. They are frequently used as biocontrol agents because they can sporulate and be stored for long periods (Branda et al., 2004; Borriss, 2015). Bacillus amyloliquefaciens FZB42 forms biofilms with little spatial organization in vitro (Fig. 1B), but exhibits a strong swarming capacity allowing a rapid surface colonization. For example, this strain can form biofilms on the fruiting body of Agaricus bisporus by forming bacterial clusters surrounded by extracellular matrix in contact with the mycelium of the carpophore (Fig. 2A and B), as well as cell bundles (Fig. 2C). Fan et al. (2011) reported the induction of biofilm formation of B. amyloliquefaciens FZB42 by root exudates of maize and surfactin. Similarly, surfactin triggers biofilm formation by B. subtilis UMAF6614 on the melon phylloplane (Zeriouh et al., 2014). Root exudate of cucumber also drives the chemotaxis of Bacillus amyloliquefaciens SQR9 and induces the production of bacillomycin D that triggers biofilm formation in the rhizosphere (Xu et al., 2014). Similarly, stem lesions of rice induce the production of GltB, leading to the production of bacillomycin L and surfactin, both involved in the biofilm formation of B. subtilis Bs916 (Zhou et al., 2016). Other biocontrol agents, such as endophytes, can also form biofilms. For example, some bacteria of the genus Paenibacillus form biofilms in wheat seeds and protect them from the invasion of Fusarium graminearum (Díaz Herrera et al., 2016).
Table 1.
Biocontrol agent reported to form biofilms and the described associated biocontrol mechanisms
| Biocontrol strain | Host/Location | Biofilm induction | Biocontrol mechanism | References |
|---|---|---|---|---|
| Bacillus atrophaeus 176s | Lettuce, sugar beet, tomato | Surfactin triggers biofilm formation |
Induced systemic resistance (ISR) antimicrobial‐producing biofilm (fengycin, surfactin) |
(Aleti et al., 2016) |
| Bacillus subtilis | Wheat seeds | Root exudates, death or lysis of cortex cells | Biofilm formation, antimicrobial, volatile compounds decrease mycelial growth | (Khezri et al., 2011) |
| Bacillus subtilis 3610 | Tomato roots | Root exudates induce matrix | Antimicrobial‐producing biofilm (surfactin) | (Chen et al., 2013) |
| Bacillus subtilis 6051 | Arabidopsis thaliana | Surfactin triggers biofilm formation | Antimicrobial‐producing biofilm (surfactin) | (Bais et al., 2004) |
| Bacillus subtilis Bs916 | Rice stem | Stem lesions induce GltB production triggering bacillomycin L and, surfactin production involved in biofilm formation | Antimicrobial‐producing biofilm (fengycin) | (Zhou et al., 2016) |
| Bacillus subtilis UMAF6614 | Melon phylloplane | Surfactin triggers biofilm formation | Antimicrobial‐producing biofilm (bacillomycin, fengycin) | (Zeriouh et al., 2014) |
| Bacillus amyloliquefaciens SQR9 | Cucumber roots | Root exudates induce chemotaxis and enhance bacillomycin D production | Antimicrobial‐producing biofilm (bacillomycin) | (Xu et al., 2014) |
| Bacillus amyloliquefaciens SQR9 | Maize roots | Root exudates induce the expression of genes related to extracellular matrix production | Promote plant growth | (Zhang et al., 2015) |
| Bacillus amyloliquefaciens SQY 162 | Tobacco roots | Pectin enhances surfactin production, increasing biofilm biomass | May trigger induced systemic resistance (ISR) antimicrobial‐producing biofilm (surfactin) | (Wu et al., 2015) |
| Bacillus amyloliquefaciens FZB42 | Maize roots | Root exudates and surfactin trigger biofilm formation | Likely not linked with the production of antibiotic or biofilm formation | (Fan et al., 2011) |
| Paenibacillus polymyxa | Arabidopsis thaliana | Root exudates induce matrix synthesis | Niche exclusion and mechanical protection | (Timmusk et al., 2005) |
| Paenibacillus polymyxa A26 | Wheat seeds | Not mentioned | Niche exclusion of pathogens | (Abd El Daim et al., 2015) |
| Paenibacillus polymyxa B5 | Arabidopsis thaliana | Root exudates | Niche exclusion of pathogens | (Haggag and Timmusk, 2008) |
| Pseudomonas corrugata CCR04 and CCR80 | Pepper roots | Root exudates | Competitive colonization, such as swimming and swarming activities, biofilm formation, antimicrobial activity | (Sang and Kim, 2014) |
| Pseudomonas chlororaphis PA23 | Canola roots | Phenazine enhances biofilm formation | Antimicrobial‐producing biofilm (pyrrolnitrin) | (Selin et al., 2010) |
| Pseudomonas putida 06909 | Citrus roots | Phytophthora exudates as attractants and growth substrates for bacteria | Biofilm formation and mycelial colonization of the pathogen Phytophtora | (Steddom et al., 2002; Ahn et al., 2007) |
| Pseudomonas putida KT2440 | Corn roots Arabidopsis thaliana | Root exudates | Promote plant growth and induced systemic resistance (ISR) | (Espinosa‐Urgel et al., 2002; Matilla et al., 2010) |
| Pichia kudriavzevii | Pear fruit | Oxidative stress | Greater activation of the antioxidant system in the biofilm form | (Chi et al., 2015) |
| Kloeckera apiculate | Citrus fruit | Phenylethanol promotes filamentous adhesion and biofilm formation | Niche exclusion and mechanical protection | (Pu et al., 2014) |
Another family of biocontrol agents consists of the Gram‐negative Pseudomonas, ubiquitous bacteria found in many plant rhizospheres (Table 1) (Espinosa‐Urgel et al., 2002; Steddom et al., 2002; Matilla et al., 2010; Selin et al., 2010). Biofilm formation by Pseudomonas putida 06909 on citrus roots is induced by exudates of the phytopathogen Phytophthora parasitica. The bacteria colonize the mycelium of the fungi by feeding on its exudates and then form a protective biofilm on the citrus roots, which prevents new growth of the pathogen (Steddom et al., 2002; Ahn et al., 2007).
Living in a biofilm profoundly alters microbial properties relative to the planktonic mode of life (Whiteley et al., 2001; Shemesh et al., 2007; Vlamakis et al., 2008, 2013; Bridier et al., 2011b). Ongoing research is currently deciphering the molecular mechanisms involved in biofilm formation and their repercussions on biocontrol efficacy.
Molecular mechanisms involved in biofilm formation of biocontrol agents
Until recently, few studies in the biocontrol field have considered that the preferred lifestyle of microorganisms in the environment is the biofilm mode of life. The main features associated with biofilm formation are a diversification of cell types and increased tolerance to the fluctuation of environmental factors, boosting microbial persistence in the environment (Vlamakis et al., 2008, 2013; Flemming et al., 2016). Bacteria and fungi can form biofilms on crops (as illustrated by the cultivated mushroom microbiota in Fig. 2), and in both cases, biofilm formation is composed of five major steps described in Fig. 1A (Costerton et al., 1999; O'Toole et al., 2000; Bianciotto et al., 2001; Davies, 2003; Triveni et al., 2012; Vlamakis et al., 2013; Pu et al., 2014; Haagensen et al., 2015; Li et al., 2015; Gulati and Nobile, 2016; Sheppard and Howell, 2016). Bacillus subtilis is the most highly documented bacterial model currently used to study the regulatory molecular mechanisms that govern biofilm formation. One specificity of the biofilm mode of life is the diversification of cell types. The presence of several bacterial subpopulations within the biofilm of B. subtilis has been clearly demonstrated, suggesting the spatiotemporal regulation of gene expression within such 3D structures (Vlamakis et al., 2008, 2013). Matrix‐producing cells, surfactin‐producing cells, flagellated motile cells and sporulated cells coexist in the same community (Fig. 1A) and are spatially and temporally organized, differentially expressing specific sets of genes (Vlamakis et al., 2008; van Gestel et al., 2015; Mielich‐Süss and Lopez, 2015; Wang et al., 2016). Indeed, the combination of surfactin‐ and matrix‐producing cells enables the organization of cells into bundles (Fig. 2C). These interfacial microbial cables allow bacteria to visit surrounding spaces to increase the biofilm surface area for nutrient and oxygen intake (van Gestel et al., 2015). Several genes involved in the phenotypic heterogeneity have been identified and extensively analysed in this species. For example, hag, encoding a flagellar protein and expressed by a subpopulation of motile cells; tasA, eps, blsA expressed by matrix‐producing cells; sfrA, involved in the production of surfactin lipopeptide; sigF, involved in cell sporulation; swr, involved in swarming motility; and the com genes, involved in genetic competence (Kearns et al., 2004; Verhamme et al., 2007; López and Kolter, 2010; Vlamakis et al., 2013; van Gestel et al., 2015; Mielich‐Süss and Lopez, 2015). All these genes are directly or indirectly regulated by various regulators (e.g. Spo0A, DegU, ComA, SinI, SinR, AbrB), which can thus play a role in the regulation of plant bioprotection by B. subtilis (López and Kolter, 2010; Vlamakis et al., 2013; Cairns et al., 2014; Mielich‐Süss and Lopez, 2015; Romero et al., 2016). Indeed, a mutation in a gene coding for a positive regulator (e.g. SinI) will decrease plant colonization and protection by diminishing attachment of cells to the roots, while mutations in a gene coding for a repressor (e.g. SinR, AbrB) will increase plant protection by an increased numbers of root‐attached cells and the formation of hyper‐robust biofilms (Chen et al., 2013).
Major components of biofilm structure that ensure its cohesion are the extracellular polymeric substances (EPS) that are mostly composed of water and extracellular biopolymers (polysaccharides, proteins, DNA, lipids) (Flemming and Wingender, 2010). Many microbial EPS have a backbone composed of various biomolecules forming gels with various cohesive and viscoelastic properties. Trapping a high amount of water is important for microbial survival against desiccation on plant surfaces (Abdian and Zorreguieta, 2016). This organic slime also protects their inhabitants from the action of environmental pollutants and toxic compounds (Sutherland, 2001; Sheppard and Howell, 2016). Another important component of the biofilm structure are amyloid fibres formed by the protein TasA. These filaments bind cells together, leading to formation of complex structures in biofilms that can hold and concentrate molecules (e.g. quorum sensing signalling molecules), and may also form aggregates to defend cells within the biofilm (de Jong et al., 2009; Romero et al., 2010; Flemming et al., 2016).
Several studies have recently highlighted various physiological behaviours of Bacillus within biofilm communities, demonstrating the high level of complexity of their interactions (Mitri et al., 2011; Houry et al., 2012; Liu et al., 2015; Prindle et al., 2015; Flemming et al., 2016). Prindle et al. (2015) described a new function for ion channels in biofilms in which they conduct electrical signals via spatial propagation of potassium waves which depolarize adjoining cells and coordinate the state of the exterior and interior cells of the biofilm. In addition, Liu et al. (2015) discovered a ‘collective oscillation’ phenomenon involved in toxic chemical tolerance, based on metabolic codependency between exterior and interior cells of the biofilm, and consisting of cyclic pauses during biofilm growth which increase the availability of nutrients in the deepest layers. Houry et al. (2012) also demonstrated that motile bacilli, expressing a bactericide, can kill a heterologous biofilm population and then occupy the newly created space (Houry et al., 2012). Altogether, these cellular traits show the complexity of living associated with a surface in a spatially organized microbial community. They also give an overview of the protection that biofilms can provide to their inhabitants on plant surfaces. Those basic insights into biofilm development and interaction might pave our way towards various applications in the field of crop protection.
Biofilm‐specific properties that should be considered in biocontrol mechanisms
Only a few published studies have considered the possibility of interspecies and microbial–host interactions in spatially organized plurimicrobial biofilms involved in agricultural biocontrol (De la Fuente et al., 2013; Triveni et al., 2015) (Table 1). The biofilm‐associated properties to be considered can be divided into five classes (Fig. 3): (i) antagonism by niche exclusion orchestrated by spatial and nutritive competition (Timmusk et al., 2005; Haggag and Timmusk, 2008; Pu et al., 2014; Abd El Daim et al., 2015), (ii) microbial communication, e.g. cooperation/interference (Hogan et al., 2004; Audrain et al., 2015; Chen et al., 2015), (iii) production of antimicrobials by biofilm cells (Bais et al., 2004; Selin et al., 2010; Chen et al., 2013; Sang and Kim, 2014; Xu et al., 2014; Zeriouh et al., 2014; Wu et al., 2015; Zhou et al., 2016), (iv) stress tolerance (Timmusk et al., 2005; Harriott and Noverr, 2009; Pu et al., 2014) and (v) direct effects on plant physiology, e.g. the induction of plant defences (Wu et al., 2015) and/or stimulation of plant growth (Espinosa‐Urgel et al., 2002; Zhang et al., 2015). This new vision could significantly change our understanding of the interactions involved in biocontrol by considering them in terms of spatial/nutritive competition (Habimana et al., 2011), tolerance/resistance (Bridier et al., 2011a) or their physiology, as microorganisms in a biofilm differ greatly from their planktonic homologues (Stewart and Franklin, 2008). These local processes are described in the following sections, using illustrative examples from other fields, if they have not been explored yet in the microbial biocontrol area.
Figure 3.
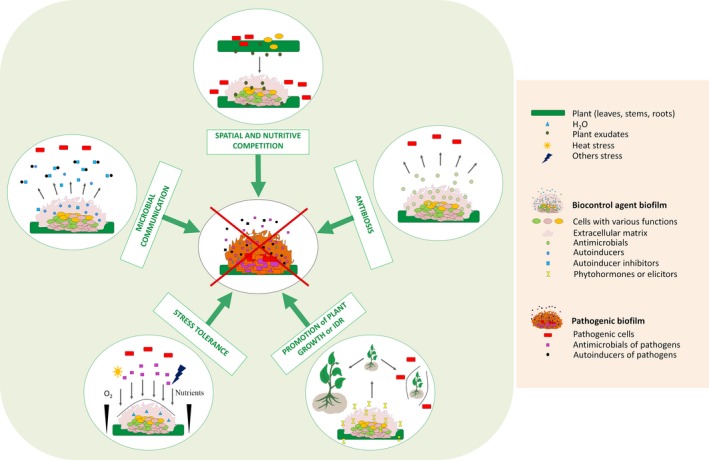
Proposed mechanisms of plant interactions with biocontrol agents and pathogenic strains. (IDR: induced disease resistance).
Spatial and nutritive competition
The spatial organization of biocontrol agent biofilms on crop surfaces varies depending on their genetic potential and the environmental conditions. For example, the biofilms of Bacillus biocontrol agents display wide architectural diversity between strains. In vitro, biofilms of B. amyloliquefaciens SQR9 and B. subtilis QST 713 exhibit the classical thick and highly organized 3D structure of bacilli (Fig. 1C and D). In contrast, B. amyloliquefaciens FZB42 forms only thin structures of a few cell layers (Fig. 1B). However, this strain outcompetes the other two due to its swarming activity, leading to rapid coverage of the entire surface. This ability to rapidly colonize a niche (Fig. 3) has been described previously as a potential biocontrol mechanism and could be called upon for the strain B. amyloliquefaciens FZB42 (Timmusk et al., 2005; Haggag and Timmusk, 2008; Fan et al., 2011; Abd El Daim et al., 2015). For Paenibacillus polymyxa, root exudates of plants induce invasive root colonization and biofilm formation that invades sites that could be potentially occupied by pathogens, thus preventing them from settling onto the surface by forming a protective biofilm (Timmusk et al., 2005; Haggag and Timmusk, 2008; Abd El Daim et al., 2015). In an organized, 3D community, nutrients may be consumed faster than they can diffuse throughout the matrix (Breugelmans et al., 2008; Stewart and Franklin, 2008). Growth and survival in such a dense community is frequently associated with spatial competition. Habimana et al. (2011) explained the inhibition of Listeria monocytogenes by Lactococcus lactis on surfaces by considering the 3D race between the two species. Using a simplified individual‐based model approach, they demonstrated that the differences in the growth parameters (lag phase and growth rate) of the two species could explain the observed inhibition of the pathogenic cells. Lactococcus lactis cells rapidly formed layers on the mixed community and completely saturated the interface in contact with the nutrient, limiting nutrient access to the pathogen. This example illustrates that part of a biofilm population can be starved within the bulk of the biostructure, even in a very rich environment. In addition, Liu et al. (2016) underlined that the specific interactions between species, such as strong or weak cooperation, exploitation or competition, contribute mostly to the spatial organization of biofilms, as these interactions create fitness effects in multispecies biofilms. Taking spatial organization and interspecies interactions within multispecies biofilms into account could increase our understanding of the interactions that take place in agrosystems that use biocontrol agents.
Antibiosis
The production of secondary metabolites by selected organisms is one of the best described mechanisms of agricultural microbial biocontrol (Ongena et al., 2005; Ongena and Jacques, 2008; Khezri et al., 2011; Cawoy et al., 2014, 2015; Chen et al., 2015; Aleti et al., 2016; Raza et al., 2016; Saravanakumar et al., 2016). Bacillus genomes contain many genes involved in the production of secondary metabolites, recently compiled in an exhaustive classification of known and putative antimicrobial compounds (Zhao and Kuipers, 2016). Indeed, 4–5% of the genome of B. subtilis is allocated to the production of antibiotics and 8.5% of the genome of B. amyloliquefaciens FZB42 is allocated to the production of secondary metabolites with antimicrobial properties (Stein, 2005; Chen et al., 2009; Zhao and Kuipers, 2016). Many exhibit interesting antibacterial properties (e.g. difficidin), antifungal properties (e.g. bacillomycin D, fengycin and surfactin), or both (e.g. bacilysin) (Ongena and Jacques, 2008; Chen et al., 2009; Guo et al., 2014; Guo et al., 2015; Chowdhury et al., 2015; Luo et al., 2015; Kröber et al., 2016). Most of the studies that have analysed the profile of antimicrobial production have relied on experiments using planktonic laboratory cultures. However, in B. amyloliquefaciens FZB42, the genes involved in bacilysin synthesis are overexpressed in biofilms, suggesting that the bacteria have a stronger antagonistic effect in their sessile mode of life (Fig. 3) (Kröber et al., 2016). Similarly, in B. subtilis, the regulator NtdR controls the expression of the ntdABC operon, encoding enzymes involved in the biosynthesis of the antibiotic kanosamine (Inaoka et al., 2004; Vetter et al., 2013). A global transcriptomic study that compared gene expression of B. subtilis in various modes of life showed that this operon is strongly overexpressed in biofilms (Nicolas et al., 2012), suggesting the possible involvement of kanosamine in interspecies interactions in plurimicrobial biofilms. Volatile organic compounds (VOCs) can also trigger antimicrobial activity (Khezri et al., 2011; Audrain et al., 2015; Raza et al., 2016). Raza et al. (2016) demonstrated that VOCs of B. amyloliquefaciens SQR9 inhibited the growth of Ralstonia solanacearum on agar medium or in soil. Altogether, these studies show that secondary metabolites with antimicrobial activity can be overproduced (or simply produced) in the biofilm lifestyle, improving antagonistic biocontrol activity. The presence of EPS or amyloid fibres in biofilms can also locally concentrate these molecules and prevent their dilution into the ambient aqueous environment, and thus presumably increase the virulence of biocontrol agents against pathogens in agrosystems (Bianciotto et al., 2001; Romero et al., 2010; Xu et al., 2014; Flemming et al., 2016). Previous studies highlighted effects of antimicrobials secreted by one producer on crop protection. Santhanam et al. (2015) have also shown that in certain cases, a consortium of different antimicrobial producers is required for optimal plant bioprotection.
Microbial communication
Biofilms are dense, spatially organized communities of microorganisms with extensive forms of social life. They can use specific signalling molecules (autoinducers) that allow them to sense and communicate with the local surrounding populations (Fuqua et al., 1994). This quorum sensing (QS) is involved in various biological processes, such as swarming, stress tolerance (pH, antimicrobials, etc.), the production of secondary metabolites, horizontal gene transfer, colonization, biofilm maturation and the synthesis of virulence factors (Fuqua et al., 1994; Von Bodman et al., 2003). These signalling pathways are widely used in bacteria–bacteria and bacteria–eukaryote associations to regulate and coordinate their interactions. For example, N‐acylhomoserine lactones (AHL) in Gram‐negative bacteria, oligopeptides in Gram‐positive bacteria and gamma‐butyrolactones in species of the genus Streptomyces are autoinducers (Danhorn and Fuqua, 2007). In Pseudomonas aeruginosa, QS controls the expression of many bacterial functions. The LasI‐LasR QS system, with the autoinducer synthase LasI and the signal receptor LasR, is involved in biofilm maturation and the organization of its 3D structure. A lasI mutant can initiate biofilm formation but is unable to form a mature biofilm, suggesting that LasI is involved in the late stages of biofilm development (Davies et al., 1998; De Kievit et al., 2001). Kavanaugh and Horswill (2016) demonstrated that the Staphylococci QS system, agr, is involved in biofilm disruption and dispersal.
In the field of biocontrol, it was shown that the protective activity of Pseudomonas fluorescens 2P24 on wheat was mainly controlled by the PcoI‐PcoR QS system that governs biofilm formation, and not directly by the production of antimicrobial metabolites (Wei and Zhang, 2006). Such social behaviour has been shown to also govern intermicrobial and interkingdom interactions, such as communication interference represented in Fig. 3 or cooperative communication (Zhang and Dong, 2004; Kalia, 2013). For example, P. aeruginosa secretes 3‐oxo‐C12‐HSL that affects the growth of C. albicans hyphae and inhibits its biofilm formation (Hogan et al., 2004). Amyloid fibres of the matrix form aggregates that can act as QS inhibitors by binding QS signalling molecules, and thus locally concentrate these molecules that can reach a 1000‐fold higher concentration in the matrix than in planktonic cell environments (Charlton et al., 2000; Hense et al., 2007; Romero et al., 2010; Flemming et al., 2016). Other types of molecules can quench or degrade QS signalling molecules of another species (Zhang and Dong, 2004). Indeed, AHL‐lactonase of Bacillus thuringiensis hinders the accumulation of AHL of Erwinia carotovora, thus decreasing the virulence of this bacterium on potatoes (Dong et al., 2004). In Bacillus, the lipopeptide surfactin, in addition to its antibiotic properties, can act like a signalling molecule to promote biofilm formation of the other relative Bacillus (López et al., 2009; Aleti et al., 2016). Volatile organic compounds emitted by prokaryotic and eukaryotic microbes are also part of their communication repertoire and can trigger global reprogramming of gene expression of their perceivers (Farag et al., 2013; Audrain et al., 2015; Raza et al., 2016). For example, acetic acid emitted by B. subtilis biofilms can promote biofilm formation of other physically separated B. subtilis cells and thus act as an important pathway of cell–cell communication (Audrain et al., 2015; Chen et al., 2015). These communication phenomena specific to biofilms, or amplified in biofilms, could be used to improve biocontrol in agrosystems.
Stress tolerance: adaptability properties and matrix as a protective shield
Bacteria in biofilms exhibit specific physiologies associated with increased tolerance/resistance of the overall community to harsh conditions (Costerton et al., 1999; Whiteley et al., 2001; Shemesh et al., 2007; Bridier et al., 2011b). Physiological differences between sessile and planktonic cells are mostly related to differential patterns of gene expression associated with the two modes of life (Whiteley et al., 2001; Shemesh et al., 2007). Transcriptomic analysis of Streptococcus mutans, a bacterium associated with tooth decay, showed that 12% of the genome was differentially expressed in biofilm communities relative to their single‐cell homologues. The differentially expressed genes coding for known functions are involved in transport, signalling, metabolism, protein and antimicrobial synthesis (Shemesh et al., 2007). Mark et al. (2005) evaluated the influence of exudates of two varieties of sugar beets on the transcriptomic profile of Pseudomonas aeruginosa PAO1. They showed that the expression of 516 genes was altered in response to one exudates and 451 to the other, and 134 genes responded to both. They found that genes coding for the synthesis of alginate, a major component of the biofilm matrix, were upregulated. These results suggest that P. aeruginosa PAO1 forms a biofilm in response to sugar beet exudates. They also showed that the transcriptomic profile of Pseudomonas aeruginosa PAO1 in response to exudates is variety dependent. Similarly, Matilla et al. (2007) compared the transcriptome profiles of Pseudomonas putida KT2440 in the planktonic exponential growth phase, the planktonic stationary growth phase, the sessile form, in sand microcosms and in the rhizosphere. They showed that transcriptomic profile of the planktonic mode of life in the stationary growth phase was the most different from that of the rhizosphere, whereas that of the biofilm lifestyle was more comparable. Indeed, they found that the gene involved in the synthesis of alginate was upregulated in the rhizosphere (Matilla et al., 2007; : additional data file). These ‘omics’ studies confirm the presence of biofilm formation in the rhizosphere or in response to plant exudates. These techniques should be increasingly considered in the study of microbial interactions in agrosystems and extended to metagenomics, metaproteomics and metatranscriptomic approaches as successfully performed in other fields (Blottière et al., 2013; Kaul et al., 2016).
Other cellular variations can occur during biofilm formation. In the early stages, Pseudomonas aeruginosa shows enhanced genetic diversification. Resulting phenotypes vary from cells involved in accelerated biofilm formation to those with enhanced dispersion properties. In the first case, biofilms exhibited pronounced spatial differentiation leading to rough and wrinkled colonies on agar. In the second case, the biofilms showed little spatial organization resulting in small and flat colonies (Boles et al., 2004). These differences illustrate the genetic plasticity of cells within a biofilm that enables them to cope with harsh environmental conditions. Stewart and Franklin (2008) also reported the existence of nutrient and oxygen gradients within biofilms creating a stratification of local microenvironments associated with a diversification of cell physiologies (Fig. 3). Population heterogeneity can generate multiple regulatory pathways leading, for example, to the phenomenon of competence in a subpopulation of cells, which coupled with the spatial proximity, facilitates horizontal gene transfer between biofilm cells. This can include the acquisition of plasmids carrying antimicrobial resistance genes (Witte, 2000; Abraham, 2011; Kung and Almeida, 2014; Liu et al., 2016). This diversification of cell types in biofilms strongly suggests that the biofilm lifestyle of biocontrol agents enables them to better adapt to, and resist, the hostile conditions encountered in agrosystems (the so‐called insurance effects in Boles et al., 2004) than their planktonic counterparts.
Biofilms are ubiquitous and subject to harsh conditions, such as the presence of antimicrobials or desiccation. Stewart (2015) recently performed a meta‐analysis of the literature from which he proposed that biofilm tolerance to antimicrobials depends neither on the size or chemistry of the antimicrobials nor the composition of the microbial biofilm or the material to which it adheres (Stewart, 2015). Based on his analysis, biofilm‐associated tolerance is primarily related to the nature and composition of the biofilm matrix. Indeed, the composition of the matrix creates various meshes within the biofilm leading to a diffusion–reaction limitation that can reduce antimicrobial penetration and local biodisponibility (Fig. 3) (Stewart et al., 2001; Stewart and Franklin, 2008; Flemming and Wingender, 2010; Bridier et al., 2011b; Stewart, 2015). Stewart (2015) also stressed that the presence of the matrix and the associated 3D organization renders slow‐growth populations less sensitive to certain stresses than their planktonic counterparts.
The matrix also plays a central role in interspecies and interkingdom interactions. Staphylococcus aureus and Candida albicans are often associated in human diseases, where they form a polymicrobial biofilm (Harriott and Noverr, 2009; Lindsay and Hogan, 2014). This association allows S. aureus to resist vancomycin, an antibiotic that is usually efficient against the planktonic form of S. aureus. The biofilm of C. albicans serves as the backbone of S. aureus microcolonies that form on their surface with the matrix of C. albicans covering and protecting the cells of S. aureus from the action of the antibiotic (Harriott and Noverr, 2009). Other reports have described the protection of Staphylococcus epidermidis by C. albicans (Adam et al., 2002) or of S. aureus by B. subtilis (Sanchez‐Vizuete et al., 2015). In the latter case, the authors identified a single gene of the B. subtilis NDmed whose disruption suppressed the protective effect. This gene (ypqP) is likely involved in the production of matrix exopolysaccharides (Sanchez‐Vizuete et al., 2015). Non‐polysaccharidic components of the matrix can also contribute to the matrix shield; the amphiphilic protein BlsA produced by B. subtilis prevents the penetration of biocides by forming a hydrophobic ‘raincoat’ layer at the biofilm–air interface (Epstein et al., 2011; Kobayashi and Iwano, 2012). Biocontrol agents likely benefit from this protective shield on crop surfaces to avoid invasion by aggressive detrimental flora and protect crops.
The direct response of crops to biocontrol agents
Plants can develop natural defence systems against pathogenic microorganisms during their interactions with their environment (biotic and abiotic) (Pieterse and Wees, 2015). Several induced diseases resistance (IDR) mechanisms have been described, including induced systemic resistance (ISR) that is an innate defence mechanism of the plant (Choudhary and Johri, 2009; Pieterse and Wees, 2015) elicited by various environmental stimuli, such as VOCs and QS signalling molecules (Farag et al., 2006; Choudhary and Johri, 2009; Matilla et al., 2010; Wu et al., 2015; Aleti et al., 2016). Various stimuli, such as VOCs, QS signals and phytohormones produced and concentrated in the biofilm matrix, can stimulate plant growth, analogous to ISR (Fig. 3) (Espinosa‐Urgel et al., 2002; Farag et al., 2006; Han et al., 2006; Spaepen, 2015; Zhang et al., 2015; Díaz Herrera et al., 2016; Ding et al., 2016b). Thus, VOCs, originating from various sources, can induce ISR and plant growth (Yi et al., 2009; Farag et al., 2013; Audrain et al., 2015; Raza et al., 2016; Sharifi and Ryu, 2016). These host responses can also be induced by products coming from plant growth‐promoting rhizobacteria that have already colonized the root surface, or endophyte colonization of its host (Whipps, 2001; Farag et al., 2006; Borriss, 2015; Díaz Herrera et al., 2016). The plant growth‐promoting rhizobacteria B. subtilis GB03 and B. amyloliquefaciens IN937a can produce 2,3‐butanediol and acetoin on plant roots and promote both plant growth and ISR by eliciting these phenomena (Ryu et al., 2003, 2004; Farag et al., 2006). Han et al. (2006) also showed that the surface area of tobacco leaves increased when they were exposed to 2,3‐butanediol secreted by Pseudomonas chlororaphis O6 or exposed to the strain itself, even without physical contact. Phytohormones (auxins, cytokinins, gibberellins, abscisic acid and ethylene) are elicitors, which can be produced by bacteria and play a role in promoting plant growth (Spaepen, 2015; Zhang et al., 2015). The auxin, indole‐3‐acetic acid, is produced by B. amyloliquefaciens SQR9 and B. amyloliquefaciens FZB42 biofilms and promotes the growth of maize and Lemna minor (Chen et al., 2007; Idris et al., 2007; Zhang et al., 2015). Endophytes can promote growth of wheat and protect it from Fusarium graminearum (Díaz Herrera et al., 2016).
Perspectives for sustainable agroecological approaches
Biocontrol mechanisms triggered by biological control agents in agriculture are not yet well understood, and even unknown in certain cases. A single biocontrol agent can use a combination of various biocontrol processes, best described for the strain B. amyloliquefaciens FZB42. The use of this bacilli can lead to antagonism, spatial and nutritional competition, antimicrobial production, the stimulation of plant growth and the induction of plant resistance (Timmusk et al., 2005; Haggag and Timmusk, 2008; Babalola, 2010; Fan et al., 2011; Xu et al., 2011; Kröber et al., 2014; Chowdhury et al., 2015; Kröber et al., 2016; Abd El Daim et al., 2015). The biofilm mode of life is still poorly taken into account in biocontrol, although it clearly plays a role in agrosystems and governs some of the observed beneficial effects. It would be informative, in the near future, to include phenotypic screening of the ability of strains to form biofilms as a rapid selection criterion of biocontrol agents. Several high‐throughput screening tests that could be used for this application are described in the literature (Azeredo et al., 2016). Better genetic knowledge of the various cell functions in biofilms will also open doors to selection criteria based on the presence of specific genes involved in important and specific biocontrol functions (Kaul et al., 2016).
Invoking biofilm formation as a determinant of biocontrol efficacy could be a new attractive strategy to better control its beneficial effects. This could be achieved, for example, using natural biofilm promoting molecules that trigger the biocontrol agent QS response. In the case of B. subtilis, this could be surfactin, VOCs, specific microbial exopolysaccharides or crop extracts (Bais et al., 2004; Chen et al., 2013; Chen et al., 2015; Audrain et al., 2015; Zhou et al., 2016). This effect could also be obtained by adding a second strain (or more) with the ability to stimulate biofilm formation by the initial biocontrol agent, for example through VOC synthesis (Filoche et al., 2004; Audrain et al., 2015; Chen et al., 2015; Figueiredo et al., 2016). Attention should be paid, in this case, to select strains without antagonistic activity against each other or the beneficial action of the biocontrol agent, as previously reported for certain cocktails in the literature (Xu et al., 2011). The benefit of the biofilm mode of life for biocontrol agents could also be obtained using dedicated formulations, as suggested in other areas; for example, the development of new formulas grown as biofilms to orally administer probiotics (e.g. beads of agar or alginates) is under consideration (Rieu et al., 2014). Similarly, a system using bacteria‐containing polymersomes, which permits rapid biofilm growth, has been developed for bioremediation to reduce the toxicity of environmental selenium contamination (Barlow et al., 2016).
Increasing evidence, based on available data from the agrosystem and biofilm fields, strongly suggests that the combination of features associated with the 3D biofilm mode of life should be considered when evaluating the performance of biocontrol organisms.
Conflict of interest
None declared.
Acknowledgements
This project was supported by the Région Ile‐de‐France, DIM ASTREA. We thank the MIMA2 imaging platform and J. Deschamps (INRA) for assistance in microscopy. We thank R. Borriss and R. Zhang for providing the Bacillus amyloliquefaciens FZB42 and SQR9 strains respectively. Finally, we warmly thank R. Védie and T. Rousseau from the French Mushroom Centre (Centre Technique du Champignon, Distré) for helpful discussions, and Alex Edelman and Associates for English revision of the manuscript.
Microbial Biotechnology (2017) 10(4), 719–734
References
- Abd El Daim, I. , Häggblom, P. , Karlsson, M. , Stenström, E. and Timmusk, S. (2015) Paenibacillus polymyxa A26 Sfp‐type PPTase inactivation limits bacterial antagonism against Fusarium graminearum but not of F. culmorum in kernel assay. Front Plant Sci 6, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdian, P. and Zorreguieta, A. (2016) Extracellular factors involved in biofilm matrix formation by Rhizobia In The Perfect Slime – Microbial Extracellular Polymeric Substances (EPS). Flemming H.‐C., Neu T.R. and Wingender J. (eds). London: IWA Publishing, pp. 227–247. [Google Scholar]
- Abraham, W.‐R. (2011) Megacities as sources for pathogenic bacteria in rivers and their fate downstream. Int J Microbiol 2011: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, B. , Baillie, G.S. , and Douglas, L.J. (2002) Mixed species biofilms of Candida albicans and Staphylococcus epidermidis . J Med Microbiol 51: 344–349. [DOI] [PubMed] [Google Scholar]
- Ahn, S.‐J. , Yang, C.‐H. , and Cooksey, D.A. (2007) Pseudomonas putida 06909 genes expressed during colonization on mycelial surfaces and phenotypic characterization of mutants. J Appl Microbiol 103: 120–132. [DOI] [PubMed] [Google Scholar]
- Aktar, M.W. , Sengupta, D. , and Chowdhury, A. (2009) Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol 2: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleti, G. , Lehner, S. , Bacher, M. , Compant, S. , Nikolic, B. , Plesko, M. , et al (2016) Surfactin variants mediate species‐specific biofilm formation and root colonization in Bacillus . Environ Microbiol 18: 2634–2645. [DOI] [PubMed] [Google Scholar]
- Anonymous . (1999). Killer environment. Environ Health Perspect 107, A62–A63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparna, M.S. , and Yadav, S. (2008) Biofilms: microbes and disease. Braz J Infect Dis 12: 526–530. [DOI] [PubMed] [Google Scholar]
- Audrain, B. , Farag, M.A. , Ryu, C.‐M. , and Ghigo, J.‐M. (2015) Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol Rev 39: 222–233. [DOI] [PubMed] [Google Scholar]
- Azeredo, J. , Azevedo, N.F. , Briandet, R. , Cerca, N. , Coenye, T. , Costa, A.R. , et al (2016) Critical review on biofilm methods. Crit Rev Microbiol [Epub ahead of print]. doi:10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- Babalola, O.O. (2010) Beneficial bacteria of agricultural importance. Biotechnol Lett 32: 1559–1570. [DOI] [PubMed] [Google Scholar]
- Bais, H.P. , Fall, R. , and Vivanco, J.M. (2004) Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, J. , Gozzi, K. , Kelley, C.P. , Geilich, B.M. , Webster, T.J. , Chai, Y. , et al (2016) High throughput microencapsulation of Bacillus subtilis in semi‐permeable biodegradable polymersomes for selenium remediation. Appl Microbiol Biotechnol 101: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto, V. , Andreotti, S. , Balestrini, R. , Bonfante, P. , and Perotto, S. (2001) Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol Plant‐Microbe Interact MPMI 14: 255–260. [DOI] [PubMed] [Google Scholar]
- Blanchette, K.A. , and Orihuela, C.J. (2012) Future perspective on host‐pathogen interactions during bacterial biofilm formation within the nasopharynx. Future Microbiol 7: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blottière, H.M. , de Vos, W.M. , Ehrlich, S.D. , and Doré, J. (2013) Human intestinal metagenomics: state of the art and future. Curr Opin Microbiol 16: 232–239. [DOI] [PubMed] [Google Scholar]
- Bogino, P.C. , Oliva, M.de.las.M. , Sorroche, F.G. and Giordano, W. (2013) The role of bacterial biofilms and surface components in plant‐bacterial associations. Int J Mol Sci 14, 15838–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles, B.R. , Thoendel, M. , and Singh, P.K. (2004) Self‐generated diversity produces ‘insurance effects’ in biofilm communities. Proc Natl Acad Sci USA 101: 16630–16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriss, R. (2015) Bacillus, a plant‐beneficial bacterium In Princ Plant‐Microbe Interact. Lugtenberg B. (ed.). Cham, Switzerland: Springer International Publishing, pp. 379–391. [Google Scholar]
- Branda, S.S. , González‐Pastor, J.E. , Dervyn, E. , Ehrlich, S.D. , Losick, R. , and Kolter, R. (2004) Genes involved in formation of structured multicellular communities by Bacillus subtilis . J Bacteriol 186: 3970–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach, R. , Toepel, J. , Dementyeva, P. , Knabe, N. and Gorbushina, A. (2016) Snapshots of fungal extracellular matrices In The Perfect Slime – Microbial Extracellular Polymeric Substances (EPS). Flemming H.‐C., Neu T.R. and Wingender J. (eds). London: IWA Publishing, pp. 269–299. [Google Scholar]
- Breugelmans, P. , Barken, K.B. , Tolker‐Nielsen, T. , Hofkens, J. , Dejonghe, W. , and Springael, D. (2008) Architecture and spatial organization in a triple‐species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol Ecol 64: 271–282. [DOI] [PubMed] [Google Scholar]
- Bridier, A. , Briandet, R. , Thomas, V. , and Dubois‐Brissonnet, F. (2011a) Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27: 1017–1032. [DOI] [PubMed] [Google Scholar]
- Bridier, A. , Dubois‐Brissonnet, F. , Greub, G. , Thomas, V. , and Briandet, R. (2011b) Dynamics of the action of biocides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55: 2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, L.S. , Hobley, L. , and Stanley‐Wall, N.R. (2014) Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol 93: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy, H. , Mariutto, M. , Henry, G. , Fisher, C. , Vasilyeva, N. , Thonart, P. , et al (2014) Plant defence stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol Plant‐Microbe Interact MPMI 27: 87–100. [DOI] [PubMed] [Google Scholar]
- Cawoy, H. , Debois, D. , Franzil, L. , De Pauw, E. , Thonart, P. , and Ongena, M. (2015) Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens . Microb Biotechnol 8: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton, T.S. , de Nys, R. , Netting, A. , Kumar, N. , Hentzer, M. , Givskov, M. , and Kjelleberg, S. (2000) A novel and sensitive method for the quantification of N‐3‐oxoacyl homoserine lactones using gas chromatography‐mass spectrometry: application to a model bacterial biofilm. Environ Microbiol 2: 530–541. [DOI] [PubMed] [Google Scholar]
- Chen, X.H. , Koumoutsi, A. , Scholz, R. , Eisenreich, A. , Schneider, K. , Heinemeyer, I. , et al (2007) Comparative analysis of the complete genome sequence of the plant growth‐promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25: 1007–1014. [DOI] [PubMed] [Google Scholar]
- Chen, X.H. , Koumoutsi, A. , Scholz, R. , Schneider, K. , Vater, J. , Süssmuth, R. , et al (2009) Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140: 27–37. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Yan, F. , Chai, Y. , Liu, H. , Kolter, R. , Losick, R. , and Guo, J. (2013) Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol 15: 848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Gozzi, K. , Yan, F. and Chai, Y. (2015) Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio 6, e00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, M. , Li, G. , Liu, Y. , Liu, G. , Li, M. , Zhang, X. , et al (2015) Increase in antioxidant enzyme activity, stress tolerance and biocontrol efficacy of Pichia kudriavzevii with the transition from a yeast‐like to biofilm morphology. Biol Control 90: 113–119. [Google Scholar]
- Choudhary, D.K. , and Johri, B.N. (2009) Interactions of Bacillus spp. and plants – with special reference to induced systemic resistance (ISR). Microbiol Res 164: 493–513. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S.P. , Hartmann, A. , Gao, X. , and Borriss, R. (2015) Biocontrol mechanism by root‐associated Bacillus amyloliquefaciens FZB42 ‐ a review. Front Microbiol 6: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton, J.W. , Geesey, G.G. , and Cheng, K.J. (1978) How bacteria stick. Sci Am 238: 86–95. [DOI] [PubMed] [Google Scholar]
- Costerton, J.W. , Stewart, P.S. , and Greenberg, E.P. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- Danhorn, T. , and Fuqua, C. (2007) Biofilm formation by plant‐associated bacteria. Annu Rev Microbiol 61: 401–422. [DOI] [PubMed] [Google Scholar]
- Davey, M.E. and O'toole, G.A. (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D. (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2: 114–122. [DOI] [PubMed] [Google Scholar]
- Davies, D.G. , Parsek, M.R. , Pearson, J.P. , Iglewski, B.H. , Costerton, J.W. , and Greenberg, E.P. (1998) The involvement of cell‐to‐cell signals in the development of a bacterial biofilm. Science 280: 295–298. [DOI] [PubMed] [Google Scholar]
- De Kievit, T.R. , Gillis, R. , Marx, S. , Brown, C. , and Iglewski, B.H. (2001) Quorum‐sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 67: 1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente, M. , Vidal, J.M. , Miranda, C.D. , González, G. , and Urrutia, H. (2013) Inhibition of Flavobacterium psychrophilum biofilm formation using a biofilm of the antagonist Pseudomonas fluorescens FF48. SpringerPlus 2: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Herrera, S. , Grossi, C. , Zawoznik, M. , and Groppa, M.D. (2016) Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum . Microbiol Res 186–187: 37–43. [DOI] [PubMed] [Google Scholar]
- Ding, C. , Yang, Z. , Wang, J. , Liu, X. , Cao, Y. , Pan, Y. , et al (2016a) Prevalence of Pseudomonas aeruginosa and antimicrobial‐resistant Pseudomonas aeruginosa in patients with pneumonia in mainland China: a systematic review and meta‐analysis. Int J Infect Dis 49: 119–126. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Cao, J. , Duan, Y. , Li, J. , Yang, Y. , Yang, G. , and Zhou, Y. (2016b) Proteomic and physiological responses of Arabidopsis thaliana exposed to salinity stress and N‐acyl‐homoserine lactone. Physiol Plant 158: 414–434. [DOI] [PubMed] [Google Scholar]
- Dong, Y.‐H. , Zhang, X.‐F. , Xu, J.‐L. , and Zhang, L.‐H. (2004) Insecticidal Bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl Environ Microbiol 70: 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, A.K. , Pokroy, B. , Seminara, A. , and Aizenberg, J. (2011) Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci 108: 995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa‐Urgel, M. , Kolter, R. , and Ramos, J.‐L. (2002) Root colonization by Pseudomonas putida: love at first sight. Microbiology 148: 341–343. [DOI] [PubMed] [Google Scholar]
- Fan, B. , Chen, X.H. , Budiharjo, A. , Bleiss, W. , Vater, J. , and Borriss, R. (2011) Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J Biotechnol 151: 303–311. [DOI] [PubMed] [Google Scholar]
- Farag, M.A. , Ryu, C.‐M. , Sumner, L.W. , and Paré, P.W. (2006) GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67: 2262–2268. [DOI] [PubMed] [Google Scholar]
- Farag, M.A. , Zhang, H. , and Ryu, C.‐M. (2013) Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J Chem Ecol 39: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, M.do.V.B. , Bonifacio, A. , Rodrigues, A.C. , de Araujo, F.F. and Stamford, N.P. (2016) Beneficial microorganisms: current challenge to increase crop performance In Bioformulations Sustain Agric. Arora N.K., Mehnaz S. and Balestrini R. (eds). New Delhi: Springer India, pp. 53–70. [Google Scholar]
- Filoche, S.K. , Anderson, S.A. , and Sissons, C.H. (2004) Biofilm growth of Lactobacillus species is promoted by Actinomyces species and Streptococcus mutans . Oral Microbiol Immunol 19: 322–326. [DOI] [PubMed] [Google Scholar]
- Flemming, H.‐C. , and Wingender, J. (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Flemming, H.‐C. , Wingender, J. , Szewzyk, U. , Steinberg, P. , Rice, S.A. , and Kjelleberg, S. (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14: 563–575. [DOI] [PubMed] [Google Scholar]
- Fuqua, W.C. , Winans, S.C. , and Greenberg, E.P. (1994) Quorum sensing in bacteria: the LuxR‐LuxI family of cell density‐responsive transcriptional regulators. J Bacteriol 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel, J. , Vlamakis, H. , and Kolter, R. (2015) From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13: doi:10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati, M. , and Nobile, C.J. (2016) Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect 18: 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Q. , Dong, W. , Li, S. , Lu, X. , Wang, P. , Zhang, X. , et al (2014) Fengycin produced by Bacillus subtilis NCD‐2 plays a major role in biocontrol of cotton seedling damping‐off disease. Microbiol Res 169: 533–540. [DOI] [PubMed] [Google Scholar]
- Guo, S. , Li, X. , He, P. , Ho, H. , Wu, Y. , and He, Y. (2015) Whole‐genome sequencing of Bacillus subtilis XF‐1 reveals mechanisms for biological control and multiple beneficial properties in plants. J Ind Microbiol Biotechnol 42: 925–937. [DOI] [PubMed] [Google Scholar]
- Haagensen, J.A.J. , Hansen, S.K. , Christensen, B.B. , Pamp, S.J. , and Molin, S. (2015) Development of spatial distribution patterns by biofilm‐cells. Appl Environ Microbiol 81: 6120–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habimana, O. , Guillier, L. , Kulakauskas, S. , and Briandet, R. (2011) Spatial competition with Lactococcus lactis in mixed‐species continuous‐flow biofilms inhibits Listeria monocytogenes growth. Biofouling 27: 1065–1072. [DOI] [PubMed] [Google Scholar]
- Haggag, W.M. , and Timmusk, S. (2008) Colonization of peanut roots by biofilm‐forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J Appl Microbiol 104: 961–969. [DOI] [PubMed] [Google Scholar]
- Han, S.H. , Lee, S.J. , Moon, J.H. , Park, K.H. , Yang, K.Y. , Cho, B.H. , et al (2006) GacS‐dependent production of 2R, 3R‐butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol Plant Microbe Interact 19: 924–930. [DOI] [PubMed] [Google Scholar]
- Harriott, M.M. , and Noverr, M.C. (2009) Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53: 3914–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazell, P. , and Wood, S. (2008) Drivers of change in global agriculture. Philos Trans R Soc Lond B Biol Sci 363: 495–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X. , Wang, J. , Abdoli, L. , and Li, H. (2016) Mg(2+)/Ca(2+) promotes the adhesion of marine bacteria and algae and enhances following biofilm formation in artificial seawater. Colloids Surf B Biointerfaces 146: 289–295. [DOI] [PubMed] [Google Scholar]
- Hense, B.A. , Kuttler, C. , Müller, J. , Rothballer, M. , Hartmann, A. , and Kreft, J.‐U. (2007) Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol 5: 230–239. [DOI] [PubMed] [Google Scholar]
- Hogan, D.A. , Vik, A. , and Kolter, R. (2004) A Pseudomonas aeruginosa quorum‐sensing molecule influences Candida albicans morphology. Mol Microbiol 54: 1212–1223. [DOI] [PubMed] [Google Scholar]
- Horrigan, L. , Lawrence, R.S. , and Walker, P. (2002) How sustainable agriculture can address the environmental and human health harms of industrial agriculture. Environ Health Perspect 110: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry, A. , Gohar, M. , Deschamps, J. , Tischenko, E. , Aymerich, S. , Gruss, A. , and Briandet, R. (2012) Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc Natl Acad Sci USA 109: 13088–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris, E.E. , Iglesias, D.J. , Talon, M. , and Borriss, R. (2007) Tryptophan‐dependent production of indole‐3‐acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact 20: 619–626. [DOI] [PubMed] [Google Scholar]
- Inaoka, T. , Takahashi, K. , Yada, H. , Yoshida, M. , and Ochi, K. (2004) RNA polymerase mutation activates the production of a dormant antibiotic 3,3′‐neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis . J Biol Chem 279: 3885–3892. [DOI] [PubMed] [Google Scholar]
- de Jong, W. , Wösten, H.A.B. , Dijkhuizen, L. , and Claessen, D. (2009) Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol 73: 1128–1140. [DOI] [PubMed] [Google Scholar]
- Jordan, K. , Dalmasso, M. , Zentek, J. , Mader, A. , Bruggeman, G. , Wallace, J. , et al (2014) Microbes versus microbes: control of pathogens in the food chain. J Sci Food Agric 94: 3079–3089. [DOI] [PubMed] [Google Scholar]
- Kalia, V.C. (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31: 224–245. [DOI] [PubMed] [Google Scholar]
- Kaul, S. , Sharma, T. and K Dhar, M. (2016) “Omics” tools for better understanding the plant‐endophyte interactions. Front Plant Sci 7, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh, J.S. , and Horswill, A.R. (2016) Impact of environmental cues on Staphylococcal quorum‐sensing and biofilm development. J Biol Chem 24: 12556–12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, D.B. , Chu, F. , Rudner, R. , and Losick, R. (2004) Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52: 357–369. [DOI] [PubMed] [Google Scholar]
- Khezri, M. , Ahmadzadeh, M. , Jouzani, G.S. , Behboudi, K. , Ahangaran, A. , Mousivand, M. , and Rahimian, H. (2011) Characterization of some biofilm‐forming Bacillus subtilis strains and evaluation of their biocontrol potential against Fusarium culmorum . J Plant Pathol 93: 373–382. [Google Scholar]
- Kobayashi, K. , and Iwano, M. (2012) BslA (YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85: 51–66. [DOI] [PubMed] [Google Scholar]
- Kröber, M. , Wibberg, D. , Grosch, R. , Eikmeyer, F. , Verwaaijen, B. , Chowdhury, S.P. , et al (2014) Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol 5: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröber, M. , Verwaaijen, B. , Wibberg, D. , Winkler, A. , Pühler, A. , and Schlüter, A. (2016) Comparative transcriptome analysis of the biocontrol strain Bacillus amyloliquefaciens FZB42 as response to biofilm formation analyzed by RNA sequencing. J Biotechnol 231: 212–223. [DOI] [PubMed] [Google Scholar]
- Kung, S.H. , and Almeida, R.P.P. (2014) Biological and genetic factors regulating natural competence in a bacterial plant pathogen. Microbiol Read Engl 160: 37–46. [DOI] [PubMed] [Google Scholar]
- Lam, J. , Chan, R. , Lam, K. , and Costerton, J.W. (1980) Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun 28: 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Pu, X. , Feng, B. , Yang, Q. , Shen, H. , Zhang, J. , and Lin, B. (2015) FocVel1 influences asexual production, filamentous growth, biofilm formation, and virulence in Fusarium oxysporum f. sp. cucumerinum . Front Plant Sci 6: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A.K. , and Hogan, D.A. (2014) Candida albicans: molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus . Fungal Biol Rev 28: 85–96. [Google Scholar]
- Liu, J. , Prindle, A. , Humphries, J. , Gabalda‐Sagarra, M. , Asally, M. , Lee, D.D. , et al (2015) Metabolic codependence gives rise to collective oscillations within biofilms. Nature 523: 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Røder, H.L. , Madsen, J.S. , Bjarnsholt, T. , Sørensen, S.J. , and Burmølle, M. (2016) Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol 7: 1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López, D. , and Kolter, R. (2010) Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis . FEMS Microbiol Rev 34: 134–149. [DOI] [PubMed] [Google Scholar]
- López, D. , Vlamakis, H. , Losick, R. , and Kolter, R. (2009) Paracrine signaling in a bacterium. Genes Dev 23: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C. , Zhou, H. , Zou, J. , Wang, X. , Zhang, R. , Xiang, Y. , and Chen, Z. (2015) Bacillomycin L and surfactin contribute synergistically to the phenotypic features of Bacillus subtilis 916 and the biocontrol of rice sheath blight induced by Rhizoctonia solani . Appl Microbiol Biotechnol 99: 1897–1910. [DOI] [PubMed] [Google Scholar]
- Mark, G.L. , Dow, J.M. , Kiely, P.D. , Higgins, H. , Haynes, J. , Baysse, C. , et al (2005) Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe‐plant interactions. Proc Natl Acad Sci USA 102: 17454–17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Espinosa‐Urgel, M. , Rodríguez‐Herva, J.J. , Ramos, J.L. , and Ramos‐González, M.I. (2007) Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8: R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Ramos, J.L. , Bakker, P.A.H.M. , Doornbos, R. , Badri, D.V. , Vivanco, J.M. , and Ramos‐González, M.I. (2010) Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ Microbiol Rep 2: 381–388. [DOI] [PubMed] [Google Scholar]
- Mielich‐Süss, B. , and Lopez, D. (2015) Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, K.F. , Zarnowski, R. , and Andes, D.R. (2016) Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog 12: e1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri, S. , Xavier, J.B. , and Foster, K.R. (2011) Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108: 10839–10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora, I. , Cabrefiga, J. , and Montesinos, E. (2015) Cyclic lipopeptide biosynthetic genes and products, and inhibitory activity of plant‐associated Bacillus against phytopathogenic bacteria. PLoS ONE 10: e0127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, C.E. , and Monier, J.‐M. (2003) The ecological significance of biofilm formation by plant‐associated bacteria. Annu Rev Phytopathol 41: 429–453. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health (NIH) . (2002) NIH guide: research on microbial biofilms. URL https://grants.nih.gov/grants/guide/pa-files/PA-03-047.html [Google Scholar]
- Nicolas, P. , Mäder, U. , Dervyn, E. , Rochat, T. , Leduc, A. , Pigeonneau, N. , et al (2012) Condition‐dependent transcriptome reveals high‐level regulatory architecture in Bacillus subtilis . Science 335: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Oerke, E.‐C. (1999) Estimated crop losses due to pathogens, animal pests and weeds Crop production and Crop Protection. Amsterdam: Elsevier, pp. 72–741. [Google Scholar]
- Oerke, E.‐C. (2006) Crop losses to pests. J Agric Sci 144: 31–43. [Google Scholar]
- Ongena, M. , and Jacques, P. (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16: 115–125. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , Jacques, P. , Touré, Y. , Destain, J. , Jabrane, A. , and Thonart, P. (2005) Involvement of fengycin‐type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis . Appl Microbiol Biotechnol 69: 29–38. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. , Kaplan, H.B. , and Kolter, R. (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54: 49–79. [DOI] [PubMed] [Google Scholar]
- Pal, K.K. and McSpadden Gardener, B. (2006) Biological control of plant pathogens. Plant Health Instr doi:10.1094/PHI‐A‐2006‐1117‐02. [Google Scholar]
- Pandin, C. , Caroff, M. , and Condemine, G. (2016) Antimicrobial peptide resistance genes in the plant pathogen Dickeya dadantii . Appl Environ Microbiol 82: 6423–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. and Wees, S.C.M.V. (2015) Induced disease resistance In Princ Plant‐Microbe Interact. Lugtenberg B. (ed.). Cham, Switzerland: Springer International Publishing, pp. 123–133. [Google Scholar]
- Prigent‐Combaret, C. , Zghidi‐Abouzid, O. , Effantin, G. , Lejeune, P. , Reverchon, S. , and Nasser, W. (2012) The nucleoid‐associated protein Fis directly modulates the synthesis of cellulose, an essential component of pellicle–biofilms in the phytopathogenic bacterium Dickeya dadantii . Mol Microbiol 86: 172–186. [DOI] [PubMed] [Google Scholar]
- Prindle, A. , Liu, J. , Asally, M. , Ly, S. , Garcia‐Ojalvo, J. , and Suel, G.M. (2015) Ion channels enable electrical communication in bacterial communities. Nature 527: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu, L. , Jingfan, F. , Kai, C. , Chao‐an, L. , and Yunjiang, C. (2014) Phenylethanol promotes adhesion and biofilm formation of the antagonistic yeast Kloeckera apiculata for the control of blue mold on citrus. FEMS Yeast Res 14: 536–546. [DOI] [PubMed] [Google Scholar]
- Rajendran, A. , and Hu, B. (2016) Mycoalgae biofilm: development of a novel platform technology using algae and fungal cultures. Biotechnol Biofuels 9: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey, B.E. , Koutsoudis, M. , von Bodman, S.B. , and Fuqua, C. (2004) Biofilm formation in plant‐microbe associations. Curr Opin Microbiol 7: 602–609. [DOI] [PubMed] [Google Scholar]
- Raza, W. , Ling, N. , Yang, L. , Huang, Q. , and Shen, Q. (2016) Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR‐9. Sci Rep 6: 24856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendueles, O. , and Ghigo, J.‐M. (2015) Mechanisms of competition in biofilm communities. Microbiol Spectr 3: doi:10.1128/microbiolspec.MB‐0009‐2014. [DOI] [PubMed] [Google Scholar]
- Rieu, A. , Aoudia, N. , Jego, G. , Chluba, J. , Yousfi, N. , Briandet, R. , et al (2014) The biofilm mode of life boosts the anti‐inflammatory properties of Lactobacillus . Cell Microbiol 16: 1836–1853. [DOI] [PubMed] [Google Scholar]
- Romero, D. , Aguilar, C. , Losick, R. , and Kolter, R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci 107: 2230–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, F.M. , Marina, M. , and Pieckenstain, F.L. (2016) Novel components of leaf bacterial communities of field‐grown tomato plants and their potential for plant growth promotion and biocontrol of tomato diseases. Res Microbiol 167: 222–233. [DOI] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.‐H. , Reddy, M.S. , Wei, H.‐X. , Paré, P.W. , and Kloepper, J.W. (2003) Bacterial volatiles promote growth in Arabidopsis . Proc Natl Acad Sci USA 100: 4927–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.‐H. , Reddy, M.S. , Kloepper, J.W. , and Paré, P.W. (2004) Bacterial volatiles induce systemic resistance in Arabidopsis . Plant Physiol 134: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Vizuete, P. , Le Coq, D. , Bridier, A. , Herry, J.‐M. , Aymerich, S. , and Briandet, R. (2015) Identification of ypqP as a new Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed‐species communities. Appl Environ Microbiol 81: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, M.K. , and Kim, K.D. (2014) Biocontrol activity and root colonization by Pseudomonas corrugata strains CCR04 and CCR80 against Phytophthora blight of pepper. Biocontrol 59: 437–448. [Google Scholar]
- Santhanam, R. , Luu, V.T. , Weinhold, A. , Goldberg, J. , Oh, Y. , and Baldwin, I.T. (2015) Native root‐associated bacteria rescue a plant from a sudden‐wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA 112: E5013–E5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanakumar, K. , Yu, C. , Dou, K. , Wang, M. , Li, Y. , and Chen, J. (2016) Synergistic effect of Trichoderma‐derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of Fusarium oxysporum f. sp. cucumerinum . Biol Control 94: 37–46. [Google Scholar]
- Savary, S. , Ficke, A. , Aubertot, J.‐N. , and Hollier, C. (2012) Crop losses due to diseases and their implications for global food production losses and food security. Food Secur 4: 519–537. [Google Scholar]
- Selin, C. , Habibian, R. , Poritsanos, N. , Athukorala, S.N.P. , Fernando, D. , and de Kievit, T.R. (2010) Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol Ecol 71: 73–83. [DOI] [PubMed] [Google Scholar]
- Sharifi, R. , and Ryu, C.‐M. (2016) Are bacterial volatile compounds poisonous odors to a fungal pathogen Botrytis cinerea, alarm signals to Arabidopsis seedlings for eliciting induced resistance, or both? Front Microbiol 7: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh, M. , Tam, A. , and Steinberg, D. (2007) Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 153: 1307–1317. [DOI] [PubMed] [Google Scholar]
- Sheppard, D.C. , and Howell, P.L. (2016) Biofilm exopolysaccharides of pathogenic fungi: lessons from bacteria. J Biol Chem 291: 12529–12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi, F. , Ferreira, J.A.G. , Stevens, D.A. , Clemons, K.V. , and Kontoyiannis, D.P. (2016) Biofilm filtrates of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients inhibit preformed Aspergillus fumigatus biofilms via apoptosis. PLoS ONE 11: e0150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. (2015) Plant hormones produced by microbes In Princ Plant‐Microbe Interact. Lugtenberg B. (ed.). Cham, Switzerland: Springer International Publishing, pp. 247–256. [Google Scholar]
- Steddom, K. , Menge, J.A. , Crowley, D. , and Borneman, J. (2002) Effect of repetitive applications of the biocontrol bacterium Pseudomonas putida 06909‐rif/nal on citrus soil microbial communities. Phytopathology 92: 857–862. [DOI] [PubMed] [Google Scholar]
- Stein, T. (2005) Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56: 845–857. [DOI] [PubMed] [Google Scholar]
- Stewart, P.S. (2015) Antimicrobial tolerance in biofilms. Microbiol Spectr 3. doi:10.1128/microbiolspec.MB‐0010‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, P.S. , and Franklin, M.J. (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6: 199–210. [DOI] [PubMed] [Google Scholar]
- Stewart, P.S. , Rayner, J. , Roe, F. , and Rees, W.M. (2001) Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J Appl Microbiol 91: 525–532. [DOI] [PubMed] [Google Scholar]
- Sundh, I. , and Melin, P. (2010) Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. Antonie Van Leeuwenhoek 99: 113–119. [DOI] [PubMed] [Google Scholar]
- Sutherland, I.W. (2001) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147: 3–9. [DOI] [PubMed] [Google Scholar]
- Teng, P.S. (1987) Crop Loss Assessment and Pest Management. St Paul: APS press. American Phytopathological Society, p. 370. [Google Scholar]
- Teng, P.S. and Krupa, S.V. (1980) Assessment of losses which constrain production and crop improvement in agriculture and forestry: proceedings of the E.C. Stakman commemorative symposium, University of Minnesota. Department of Plant Pathology, 327. [Google Scholar]
- Timmusk, S. , Grantcharova, N. , and Wagner, E.G.H. (2005) Paenibacillus polymyxa invades plant roots and forms biofilms. Appl Environ Microbiol 71: 7292–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triveni, S. , Prasanna, R. , and Saxena, A.K. (2012) Optimization of conditions for in vitro development of Trichoderma viride‐based biofilms as potential inoculants. Folia Microbiol (Praha) 57: 431–437. [DOI] [PubMed] [Google Scholar]
- Triveni, S. , Prasanna, R. , Kumar, A. , Bidyarani, N. , Singh, R. , and Saxena, A.K. (2015) Evaluating the promise of Trichoderma and Anabaena based biofilms as multifunctional agents in Macrophomina phaseolina‐infected cotton crop. Biocontrol Sci Technol 25: 656–670. [Google Scholar]
- Verhamme, D.T. , Kiley, T.B. , and Stanley‐Wall, N.R. (2007) DegU co‐ordinates multicellular behaviour exhibited by Bacillus subtilis . Mol Microbiol 65: 554–568. [DOI] [PubMed] [Google Scholar]
- Vetter, N.D. , Langill, D.M. , Anjum, S. , Boisvert‐Martel, J. , Jagdhane, R.C. , Omene, E. , et al (2013) A previously unrecognized kanosamine biosynthesis pathway in Bacillus subtilis . J Am Chem Soc 135: 5970–5973. [DOI] [PubMed] [Google Scholar]
- Vlamakis, H. , Aguilar, C. , Losick, R. , and Kolter, R. (2008) Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis, H. , Chai, Y. , Beauregard, P. , Losick, R. , and Kolter, R. (2013) Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bodman, S.B. , Bauer, W.D. , and Coplin, D.L. (2003) Quorum sensing in plant‐pathogenic bacteria. Annu Rev Phytopathol 41: 455–482. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Koehler, S.A. , Wilking, J.N. , Sinha, N.N. , Cabeen, M.T. , Srinivasan, S. , et al (2016) Probing phenotypic growth in expanding Bacillus subtilis biofilms. Appl Microbiol Biotechnol 100: 4607–4615. [DOI] [PubMed] [Google Scholar]
- Wei, H.‐L. , and Zhang, L.‐Q. (2006) Quorum‐sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89: 267–280. [DOI] [PubMed] [Google Scholar]
- Whipps, J.M. (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52: 487–511. [DOI] [PubMed] [Google Scholar]
- Whiteley, M. , Bangera, M.G. , Bumgarner, R.E. , Parsek, M.R. , Teitzel, G.M. , Lory, S. , and Greenberg, E.P. (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864. [DOI] [PubMed] [Google Scholar]
- Witte, W. (2000) Ecological impact of antibiotic use in animals on different complex microflora: environment. Int J Antimicrob Agents 14: 321–325. [DOI] [PubMed] [Google Scholar]
- Wu, K. , Fang, Z. , Guo, R. , Pan, B. , Shi, W. , Yuan, S. , et al (2015) Pectin enhances bio‐control efficacy by inducing colonization and secretion of secondary metabolites by Bacillus amyloliquefaciens SQY 162 in the rhizosphere of tobacco. PLoS ONE 10: e0127418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.‐M. , Jeffries, P. , Pautasso, M. , and Jeger, M.J. (2011) Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 101: 1024–1031. [DOI] [PubMed] [Google Scholar]
- Xu, Z. , Zhang, R. , Wang, D. , Qiu, M. , Feng, H. , Zhang, N. , and Shen, Q. (2014) Enhanced control of cucumber wilt disease by Bacillus amyloliquefaciens SQR9 by altering the regulation of Its DegU phosphorylation. Appl Environ Microbiol 80: 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H.‐S. , Heil, M. , Adame‐Álvarez, R.M. , Ballhorn, D.J. , and Ryu, C.‐M. (2009) Airborne induction and priming of plant defenses against a bacterial pathogen. Plant Physiol 151: 2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski, R. , Westler, W.M. , Lacmbouh, G.A. , Marita, J.M. , Bothe, J.R. , Bernhardt, J. , et al (2014) Novel entries in a fungal biofilm matrix encyclopedia. mBio 5, e01333–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeriouh, H. , de Vicente, A. , Pérez‐García, A. , and Romero, D. (2014) Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ Microbiol 16: 2196–2211. [DOI] [PubMed] [Google Scholar]
- Zhang, L.‐H. , and Dong, Y.‐H. (2004) Quorum sensing and signal interference: diverse implications. Mol Microbiol 53: 1563–1571. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Yang, D. , Wang, D. , Miao, Y. , Shao, J. , Zhou, X. , et al (2015) Whole transcriptomic analysis of the plant‐beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genom 16: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , and Kuipers, O.P. (2016) Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom 17: 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Luo, C. , Fang, X. , Xiang, Y. , Wang, X. , Zhang, R. , and Chen, Z. (2016) Loss of gltb inhibits biofilm formation and biocontrol efficiency of Bacillus subtilis Bs916 by altering the production of γ‐polyglutamate and three lipopeptides. PLoS ONE 11: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


