Summary
Actinobacillus pleuropneumoniae causes porcine pleuropneumonia and forms biofilms in vitro on abiotic surfaces; however, presence of biofilms during infections has not been documented. The aim of this study was to use a species‐specific fluorescent oligonucleotide probe and confocal microscopy to localize A. pleuropneumoniae in the lungs of two naturally infected pigs. Actinobacillus pleuropneumoniae was detected by fluorescence in situ hybridization and observed to grow as aggregates (~30–45 μm) during a natural infection. As the A. pleuropneumoniae aggregates observed in porcine lungs differed from the biofilms grown on a solid surface obtained in vitro, we designed a new biofilm assay using agarose, a porous substrate, favouring the formation of aggregates. In this study, we described for the first time the mode of growth of A. pleuropneumoniae during a natural infection in pigs. We also propose an in vitro biofilm assay for A. pleuropneumoniae using a porous substrate which allows the formation of aggregates. This assay might be more representative of the in vivo situation, at least in terms of the size of the bacterial aggregates and the presence of a porous matrix, and could potentially be used to test the susceptibility of A. pleuropneumoniae aggregates to antibiotics and disinfectants.
Introduction
Bacterial biofilms are structured clusters of bacterial cells enclosed in a self‐produced polymer matrix that are attached to a biotic or abiotic surface (Costerton et al., 1999; Jacques et al., 2010). This structure protects bacteria from hostile environmental conditions. Bacteria within a biofilm can resist attack from the host immune response, and are less sensitive than planktonic cells to the action of antibiotics and disinfectants.
Actinobacillus pleuropneumoniae is a Gram‐negative bacterium belonging to the Pasteurellaceae family and is the causative agent of porcine pleuropneumonia, a disease causing important economic losses to the swine industry worldwide (Gottschalk, 2012). Several virulence factors of A. pleuropneumoniae have been identified and these include the Apx toxins, iron uptake systems, adhesins and surface polysaccharides (Bossé et al., 2002; Chiers et al., 2010). We have shown that A. pleuropneumoniae is able to produce a dense biofilm on abiotic (plastic and glass; Labrie et al., 2010; Tremblay et al., 2013a) and biotic (cell line; Tremblay et al., 2013b) surfaces. We have also shown that A. pleuropneumoniae biofilm cells were 100–30 000 times more tolerant to ampicillin, florfenicol, tiamulin or tilmicosin than their planktonic counterparts (Archambault et al., 2012). Such decrease in susceptibility was dependent on the isolate. Additionally, we used whole‐genome DNA microarrays to identify differentially expressed genes in A. pleuropneumoniae planktonic cells or cells in biofilms formed under static (microtiter plate) or dynamic (drip‐flow biofilm reactor) conditions (Tremblay et al., 2013a).
Although the use of in vitro assays has greatly increased our understanding of the biology of bacterial biofilms, it is becoming increasingly apparent that many of these methods do not accurately represent in vivo conditions (Roberts et al., 2015). Thus, the purpose of this study was to use a species‐specific fluorescent oligonucleotide probe and confocal microscopy to localize A. pleuropneumoniae in the lungs of infected pigs.
Results and Discussion
For this study, lungs were obtained from two pigs with clinical signs of infection and macroscopic examination consistent with acute porcine pleuropneumonia that were submitted to LEAQ (Laboratoire d’épidémiosurveillance animale du Québec). Microscopic examination of the lungs revealed typical, multiple foci of coagulation necrosis in the pulmonary parenchyma, associated with oedema, fibrin exudation, congestion, haemorrhages, capillary thrombosis, and the presence of microcolonies of small Gram‐negative bacilli. Numerous neutrophils and degenerated macrophages (round and oat cells) were delineating these foci. Fibrin, oedema, and leucocytes also severely distended the interlobular septa and pleura. The latter was covered with a thin layer of fibrin and leucocytes debris (Fig. 1). Actinobacillus pleuropneumoniae were isolated from the lung tissues of both pigs and were identified as APP4294 and APP286 for the isolates from the first and second pig respectively. Serotyping analysis (M. Gottschalk laboratory, Université de Montréal) showed that APP4294 and APP286 belong to serotype 7 and serotype 5, respectively, which are two of the most prevalent serotypes in North America (Gottschalk, 2012). Overall, the diagnostic tests confirmed that the animals were infected by A. pleuropneumoniae and that we were using lungs from natural infections.
Figure 1.

Swine lung, case SHY13‐04294. A large area of coagulation necrosis (N) that spares the interlobular septa, large vessels and airways (S) is present in the parenchyma, associated with oedema, congestion and fibrin exudation. It is surrounded by a rim of neutrophils and degenerated macrophages (L). Fibrin, oedema and leucocytes moderately distend the interlobular septa and pleura. The pleura (P) is covered by a thin layer of fibrin. Haematoxylin‐eosin‐phloxin‐saffron (HEPS), 25× magnification.
We first evaluated the capacity of these isolates to form biofilms in vitro in a standard microtiter plate assay routinely used in our laboratory. Both isolates formed a robust biofilm on the plastic surface after an incubation of 24 h (Fig. 2A). These biofilms were then visualized by confocal laser scanning microscopy. The biofilms completely covered the plastic surface and their thickness was evaluated to be around 50 μm (Fig. 2B).
Figure 2.

Biofilm formation by Actinobacillus pleuropneumoniae isolates APP286 and APP4294 in microtiter plates after 24 h of incubation. (A) OD 590 nm after crystal violet staining. (B) Confocal laser scanning microscopic image of biofilm of isolate APP286 stained with FilmTracer FM 1‐43. Stack of sections through the X‐Z plane is shown.
We then performed fluorescence in situ hybridization (FISH) with an apxIV probe on the lung samples. The use of species‐specific oligonucleotide probe allowed the localization of A. pleuropneumoniae cells (in red) within the lung tissue (stained with DAPI, in blue) (Fig. 3). It is clear from the images obtained from the lung of the pig infected with isolate of serotype 5 (Fig. 3A) that A. pleuropneumoniae seems to grow as aggregates (~30–45 μm) during a natural infection. An identical image was obtained from the lung of the other pig infected with a serotype 7 isolate (Fig. 3B). Controls (e.g. hybridization without probe, hybridization with probe of an A. pleuropneumoniae negative lung) (Fig. 3C) were performed to confirm the specificity of the FISH assay.
Figure 3.
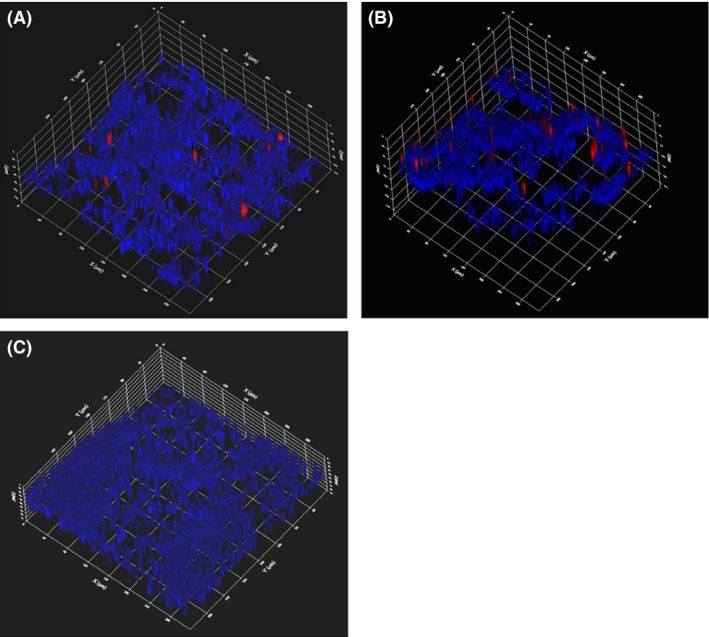
Fluorescence in situ hybridization (FISH) with an apxIV probe on the Actinobacillus pleuropneumoniae‐infected lung samples. CLSM images of A. pleuropneumoniae aggregates (red) within the lung tissue (blue) obtained from pigs of clinical cases SHY14‐286 (A) and SHY13‐04294 (B). (C) FISH on a lung sample from a control pig that was negative for A. pleuropneumoniae.
These aggregates are in agreement with recent reports in the literature indicating that the in vivo biofilms differ from their in vitro counterpart, in both size and shape (Roberts et al., 2015). Indeed, a meta‐analysis on the size of in vivo biofilms from human chronic infections by Bjarnsholt et al. (2013) showed the presence of bacterial aggregates ranging from ~5 to 200 μm in diameter; the median for the smallest and largest biofilm diameters were 5 and 50 μm respectively. Furthermore, these aggregates are not necessarily attached to a surface. Bjarnsholt et al. (2013) concluded that in vivo biofilms are smaller in physical dimensions, lack mushroom‐like structures, are embedded in host material and are continuously exposed to host defence reactions. More recently, Kragh et al. (2016) suggested that current models of biofilm formation should be reconsidered to incorporate the role of aggregates in biofilm initiation. In the case of A. pleuropneumoniae infections, lesion is known to contain blood, fibrin and leucocytes (dead or alive). Furthermore, neutrophils could release DNA during NET (neutrophil extracellular traps) formation and the extracellular DNA could help bacterial aggregation. We have previously observed that extracellular DNA increases biofilm formation and autoagregation in A. pleuropneumaniae (Hathroubi et al., 2015). Overall, these host components, combined to the lung architecture, could provide the porous substrate that supports the formation aggregates.
As the A. pleuropneumoniae aggregates observed in porcine lungs differed from the biofilms grown on a solid surface obtained in vitro, we designed an in vitro assay using agarose, a porous substrate. The control, uninoculated agarose, showed a homogenous appearance when observed by differential interference contrast microscopy (Fig. 4A). Actinobacillus pleuropneumoniae grew as microcolonies or aggregates of approximately 20–30 μm when embedded in agarose (Fig. 4B), which is roughly similar in size to those observed in the porcine lungs. Furthermore, these aggregates were stained by the fluorescent lectin WGA‐Oregon Green 488 that binds to the poly‐N‐acetylglucosamine, a known component of the A. pleuropneumoniae biofilm matrix (Fig. 4C).
Figure 4.

CLSM images of Actinobacillus pleuropneumoniae embedded in agarose after 24 h of incubation. (A) Agarose control, not inoculated with bacteria observed in the differential interference contrast mode. (B) A. pleuropneumoniae isolate APP286 aggregates as observed in the differential interference contrast mode, stack of ~10 images. (C) A. pleuropneumoniae isolate APP286 aggregates stained with WGA‐Oregon Green and observed in the fluorescence mode, stack of ~10 images. Bars = 30 μm.
In this study, we described for the first time the mode of growth of A. pleuropneumoniae during a natural infection in pigs. We also propose an in vitro biofilm assay for A. pleuropneumoniae using a porous substrate which allows the formation of aggregates. This assay might be more representative of the in vivo situation, at least in terms of the size of the bacterial aggregates and the presence of a porous matrix, and could potentially be used to test the susceptibility of A. pleuropneumoniae aggregates to antibiotics and disinfectants.
Experimental procedures
Porcine lung samples and Actinobacillus pleuropneumoniae isolates
Two pigs with clinical signs and macroscopic and microscopic examinations consistent with acute porcine pleuropneumonia were submitted to LEAQ and used in the present study (clinical cases SHY13‐04294 and SHY14‐286). Actinobacillus pleuropneumoniae was isolated from the lung tissues of both pigs. The isolates were identified as APP4294 and APP286.
In vitro biofilm assays
The two A. pleuropneumoniae isolates were grown on brain heart infusion agar or broth (BHI; Oxoid Ltd, Basingstoke, Hampshire, UK) supplemented with 5 μg ml−1 NAD (broth) or 15 μg ml−1 NAD (agar) (BHI‐NAD) at 37°C with 5% CO2. We evaluated their capacity to form biofilms in vitro in a standard microtiter plate assay routinely used in our laboratory (Labrie et al., 2010; Tremblay et al., 2013a). The biofilms were also visualized by confocal laser scanning microscopy (CLSM; FV1000 IX81; Olympus, Markham, ON, Canada) using FilmTracer FM 1‐43 (Molecular Probes, Eugene, OR, USA).
We also designed an in vitro assay using a porous substrate. Actinobacillus pleuropneumoniae isolates were grown overnight on BHI‐NAD plates as described above. A colony was transferred into 5 ml of BHI with 5 μg ml−1 NAD and incubated at 37°C overnight with agitation and then mixed (1:100) with agarose (autoclaved and placed in a water bath at 48°C) to obtain a final concentration of 0.5%. Aliquots of 1 ml were deposited in wells of a 24‐well microtiter plate; 1 ml of BHI‐NAD was added to each well once the agarose had solidified. The plates were then incubated at 37°C with 5% CO2 for 24 h. The agarose pellets were stained with Wheat Germ Agglutinin‐Oregon Green 488 (Molecular Probes) for 30 min at room temperature in the dark, then washed once and covered with PBS prior to observation by CLSM.
Fluorescence in situ hybridization
We performed FISH with an apxIV probe on the lung samples as described by Loera‐Muro et al. (2013) with some modifications. Before the hybridization step, lung tissues were prepared and fixed as described by Niscito et al. (2009). Briefly, lungs were cut in small pieces (3 mm × 10 mm) and fixed with a solution of 4% paraformaldehyde‐PBS at 4°C for 6 h. Lungs were then washed two times for 10 min with PBS and kept at −20°C in 50% ethanol‐PBS until the hybridization. Lungs were put in Petri dishes and covered with 5–10 ml of hybridization solution. The hybridization solution contained 900 mM NaCl, 20 mM Tris‐Cl, 0.01% (v/v) SDS, 12.15% (v/v) formamide and 1.5 μM of Alexa Fluor 633‐labelled probe APXIVAN‐forward (GGGGACGTAACTCGGTGATT) (Invitrogen, Burlington, ON, Canada). Lung pieces were incubated for 18 h at 50°C and washed for 15 min at the same temperature with a solution containing 15 mM NaCl and 5 mM Trisma Base. The lung pieces were then rinsed in distilled water (50°C) and incubated for 5 min in a solution containing 0.1% (v/v) Triton X‐100. Lung pieces were washed three times with PBS and stained for 30 min at room temperature with DAPI. Two additional washes were done with PBS and the lungs pieces were transferred to a new Petri dish. PBS was added to fully submerged lung pieces and a glass slide was added on top to immobilize the lung pieces. The stained lung pieces were visualized by CLSM and images were acquired using the Fluoview software (Olympus).
Conflict of interest
The authors declare no conflict of interests.
Acknowledgements
This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.J.
Microbial Biotechnology (2017) 10(4), 756–760
References
- Archambault, M. , Harel, J. , Gouré, J. , Tremblay, Y.D. , and Jacques, M. (2012) Antimicrobial susceptibilities and resistance genes of Canadian isolates of Actinobacillus pleuropneumoniae . Microb Drug Resist 18: 198–206. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt, T. , Alhede, M. , Eickhardt‐Sorensen, S.R. , Moser, C. , Kühl, M. , Jensen, P.O. , and Hoiby, N. (2013) The in vivo biofilm. Trends Microbiol 21: 466–474. [DOI] [PubMed] [Google Scholar]
- Bossé, J.T. , Janson, H. , Sheehan, B.J. , Beddek, A.J. , Rycroft, A.N. , Kroll, J.S. , and Langford, P.R. (2002) Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect 4: 225–235. [DOI] [PubMed] [Google Scholar]
- Chiers, K. , De Waele, T. , Pasmans, F. , Ducatelle, R. , and Haesebrouck, F. (2010) Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet Res 41: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton, J.W. , Stewart, P.S. , and Greenberg, E.P. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- Gottschalk, M. (2012) Actinobacillosis In Diseases of Swine, 10th edn Karriker L., Ramirez A., Schwartz K., Stevenson G., and Zimmerman J. (eds). Hoboken, NJ: Wiley, pp. 653–669. [Google Scholar]
- Hathroubi, S. , Fontaine‐Gosselin, S.E. , Tremblay, Y.D.N. , Labrie, J. , and Jacques, M. (2015) Sub‐inhibitory concentrations of penicillin G induce biofilm formation by field isolates of Actinobacillus pleuropneumoniae . Vet Microbiol 179: 277–286. [DOI] [PubMed] [Google Scholar]
- Jacques, M. , Aragon, V. , and Tremblay, Y.D. (2010) Biofilm formation in bacterial pathogens of veterinary importance. Anim Health Res Rev 11: 97–121. [DOI] [PubMed] [Google Scholar]
- Kragh, K.N. , Hutchison, J.A. , Melaugh, G. , Rodesney, C. , Roberts, A.E.L. , Irie, Y. , et al (2016) Role of multicellular aggregates in biofilm formation. mBio 7: e00237–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie, J. , Pelletier‐Jacques, G. , Deslandes, V. , Ramjeet, M. , Auger, E. , Nash, J.H. , and Jacques, M. (2010) Effects of growth conditions on biofilm formation by Actinobacillus pleuropneumoniae . Vet Res 41: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loera‐Muro, V.M. , Jacques, M. , Tremblay, Y.D. , Avelar‐Gonzalez, F.J. , Loera‐Muro, A. , Ramirez‐Lopez, E.M. , et al (2013) Detection of Actinobacillus pleuropneumoniae in drinking water from pig farms. Microbiology 159: 536–544. [DOI] [PubMed] [Google Scholar]
- Niscito, L. , Gieseke, A. , Stoodley, P. , Hall‐Stoodley, L. , Kerschner, J.E. , and Ehrlich, G.D. (2009) Fluorescence “in situ” hybridization for the detection of biofilm in the middle ear and upper respiratory tract mucosa. Methods Mol Biol 493: 191–213. [DOI] [PubMed] [Google Scholar]
- Roberts, A.E. , Kragh, K.N. , Bjarnsholt, T. , and Diggle, S.P. (2015) The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol 427: 3646–3661. [DOI] [PubMed] [Google Scholar]
- Tremblay, Y.D. , Deslandes, V. , and Jacques, M. (2013a) Actinobacillus pleuropneumoniae genes expression in biofilms cultured under static conditions and in a drip‐flow apparatus. BMC Genom 14: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, Y.D. , Lévesque, C. , Segers, R.P. , and Jacques, M. (2013b) Method to grow Actinobacillus pleuropneumoniae biofilm on a biotic surface. BMC Vet Res 9: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]


