Summary
Partial nitritation‐anammox (PNA) permits energy effective nitrogen removal. Today PNA is used for treatment of concentrated and warm side streams at wastewater treatment plants, but not the more diluted and colder main stream. To implement PNA in the main stream, better knowledge about microbial communities at the typical environmental conditions is necessary. In order to investigate the response of PNA microbial communities to decreasing substrate availability, we have operated a moving bed biofilm reactor (MBBR) at decreasing reactor concentrations (311–27 mg‐N l−1 of ammonium) and low temperature (13°C) for 302 days and investigated the biofilm community using high throughput amplicon sequencing; quantitative PCR; and fluorescence in situ hybridization. The anammox bacteria (Ca. Brocadia) constituted a large fraction of the biomass with fewer aerobic ammonia oxidizing bacteria (AOB) and even less nitrite oxidizing bacteria (NOB; Nitrotoga, Nitrospira and Nitrobacter). Still, NOB had considerable impact on the process performance. The anammox bacteria, AOB and NOB all harboured more than one population, indicating some diversity, and the heterotrophic bacterial community was diverse (seven phyla). Despite the downshifts in substrate availability, changes in the relative abundance and composition of anammox bacteria, AOB and NOB were small and also the heterotrophic community showed little changes in composition. This indicates stability of PNA MBBR communities towards decreasing substrate availability and suggests that even heterotrophic bacteria are integral components of these communities.
Introduction
Autotrophic nitrogen removal from wastewater can be achieved by partial nitritation together with anaerobic ammonium oxidation (anammox). Partial nitritation‐anammox (PNA) saves energy due to a reduced need for aeration by > 50% (Siegrist et al., 2008) and enables a higher utilization of organic carbon for production of valuable products, for example, biogas, compared to conventional nitrogen removal with nitrification‐denitrification. Together, this makes energy positive wastewater treatment plants (WWTPs) possible (Kartal et al., 2010).
Today, PNA is established for treatment of warm and concentrated wastewater side streams (Lackner et al., 2014), where the conditions for growth of aerobic ammonia oxidizing bacteria (AOB) and anammox bacteria are beneficial and inhibition of unwanted aerobic nitrite oxidation by nitrite oxidizing bacteria (NOB) can be effective. However, the side stream nitrogen removal at WWTPs treats only 15–20% of the total nitrogen. To utilize the benefits of PNA in the wastewater main stream is highly desirable and has recently become a prioritized research area. In the main stream, the conditions for PNA are much more challenging (De Clippeleir et al., 2013; Hu et al., 2013; Laureni et al., 2016). The cold and diluted water causes low activity and slow growth rate of particularly the anammox bacteria (Hendrickx et al., 2014; Lotti et al., 2015), even though some adaptations to low temperatures have been observed (Dosta et al., 2008; Hu et al. 2013; Hendrickx et al., 2014). Moreover, the competition between AOB and anammox bacteria with NOB and denitrifying bacteria is challenging at these conditions (see e.g. De Clippeleir et al., 2013; Perez et al., 2014), which necessitates detailed knowledge about the dynamics of these microorganisms in order to understand process performance.
Despite the challenges, maintenance and activity of AOB and anammox bacteria at low temperatures and low substrate concentrations have been demonstrated in biofilm‐ and granular sludge reactors (De Clippeleir et al., 2013; Gustavsson et al., 2014; Lotti et al., 2014a; Gilbert et al., 2015; Ma et al., 2015; Laureni et al., 2016) and in a few studies the major population of AOB, anammox bacteria and NOB have been identified (Gilbert et al., 2014; Lotti et al., 2014a). Little is, however, known about the microbial community structure and dynamics at main stream conditions (Gilbert et al., 2014) and how such communities differ from the communities in the PNA reactors treating concentrated wastewater with higher substrate availability. Furthermore, the heterotrophic bacteria in PNA reactors (Gilbert et al., 2014; Pellicer‐Nàcher et al., 2014; Chu et al., 2015) most likely affect the nitrogen turnover and process performance, but the composition, diversity and roles of these are little investigated, particularly at main stream conditions.
Here, an experiment was designed to investigate the role of the substrate concentration in shaping the PNA microbial community in a MBBR over 302 days. The influent nitrogen concentration was decreased from 500 to 45 mg‐N l−1 at low temperature (13°C) to stepwise approach main stream conditions. For investigation of the composition and diversity of the total bacterial community (including heterotrophic bacteria), the abundance of key functional groups and their localization in the biofilms, a multiphase approach of high throughput amplicon sequencing (Illumina MiSeq), quantitative PCR (qPCR) and fluorescence in situ hybridization (FISH) in conjunction with confocal laser scanning microscopy (CLSM) of cryosectioned biofilms was used. Reactor performance and potential activity of key functional groups was also monitored.
Results
Reactor performance
The concentration of the influent was decreased from target concentrations of 500 to 45 mg‐N l−1 from period I to period VI (Figure S1) resulting in average ammonium concentrations of 311 to 27 mg‐N l−1 in the reactor. During periods I to V, the nitrogen removal rate (NRR) was rather similar, with a decrease in period VI, at the lowest influent ammonium concentration (Table 1). The biomass weight on the carriers was stable, with insignificant changes during the study period (Figure S3, ANOVA, P > 0.05). The set‐up and the performance of the reactor is summarized in Table 1. Time‐course displays of nitrogen species are found in Figure S1.
Table 1.
Study design and operational data of the MBBR
| Period I | Period II | Period III | Period IV | Period V | Period VI | |
|---|---|---|---|---|---|---|
| Operational set‐up | ||||||
| Time (d) | 1–55 | 62–99 | 106–133 | 136–189 | 195–258 | 262–302 |
| NH4‐N infl (mg l−1) | 496 ± 32 | 249 ± 14 | 170 ± 19 | 129 ± 11 | 86 ± 8 | 43 ± 2 |
| COD infl (mg l−1) | 313 ± 36 | 174 ± 7 | 155 ± 26 | 84 ± 9 | 56 ± 4 | 48 ± 8 |
| HRT (d) | 3.3 ± 0.2 | 3.2 ± 0.0 | 3.2 ± 0.0 | 2.3 ± 0.1 | 1.6 ± 0.0 | 0.8 ± 0.0 |
| NLR (g‐N m‐2 d−1) | 0.75 ± 0.03 | 0.39 ± 0.02 | 0.26 ± 0.03 | 0.29 ± 0.02 | 0.28 ± 0.03 | 0.26 ± 0.02 |
| pH | 7.9 ± 0.12 | 7.7 ± 0.09 | 7.8 ± 0.08 | 7.5 ± 0.19 | 7.1 ± 0.07 | 7.2 ± 0.12 |
| DO (mg l−1) | 0.93 ± 0.08 | 0.67 ± 0.04 | 0.64 ± 0.07 | 0.82 ± 0.25 | 0.49 ± 0.06 | 0.48 ± 0.10 |
| Reactor performance | ||||||
| NH4‐N effl (mg l−1) | 311 ± 70 | 136 ± 21 | 119 ± 12 | 63 ± 22 | 31 ± 10 | 27 ± 5 |
| NO2‐N effl (mg l−1) | 12.5 ± 5.6 | 6.0 ± 5.7 | 2.2 ± 1.0 | 2.8 ± 1.4 | 1.6 ± 0.7 | 1.8 ± 0.3 |
| NO3‐N effl (mg l−1) | 98 ± 41 | 33 ± 23 | 8 ± 4 | 17 ± 13 | 21 ± 7 | 9 ± 6 |
| COD effl (mg l−1) | 281 ± 15 | 153 ± 19 | 118 ± 8 | 67 ± 14 | 53 ± 8 | 40 ± 6 |
| ARR (g‐N m−2 d−1) | 0.28 ± 0.10 | 0.18 ± 0.04 | 0.11 ± 0.07 | 0.14 ± 0.05 | 0.18 ± 0.04 | 0.10 ± 0.04 |
| NRR (g‐N m−2 d−1) | 0.11 ± 0.06 | 0.12 ± 0.03 | 0.09 ± 0.07 | 0.10 ± 0.03 | 0.10 ± 0.03 | 0.03 ± 0.03 |
| NO3 production (%) | 51 ± 19 | 28 ± 16 | 15 ± 8 | 23 ± 11 | 35 ± 12 | 58 ± 26 |
HRT = hydraulic retention time, NLR = nitrogen loading rate, DO = dissolved oxygen, ARR = ammonia removal rate, NRR = nitrogen removal rate. COD and nitrogen species are dissolved (filtered, 0.45 μm).
Abundance of autotrophic nitrogen converting bacteria
The anammox bacteria dominated the total bacterial community in all periods, as measured by qPCR (Fig. 1). The gene copy numbers of AOB (amoA) were about two orders of magnitude lower than the anammox bacteria (16S rRNA). The abundances of the NOB, Nitrobacter and Nitrospira, were even lower. There were no major changes in the relative abundances of the anammox bacteria, AOB, Nitrobacter and Nitrospira over the course of the study, as measured by qPCR (ANOVA, P > 0.05), and correlations between the relative abundances for each sampling occasion (n = 17) and the reactor concentrations of nitrogen species, COD and alkalinity were non‐significant (P > 0.05). The methods of qPCR, FISH and amplicon sequencing, all showed that anammox bacteria was the largest group followed by AOB and NOB (Fig. 1 and Table S4). FISH detected a higher percentage of AOB than the other methods, but it should be noted that only 20 to 40% of the cells detected by a general DNA stain (SYTO62) were detected by the general FISH EUB probe mix (Figure S4).
Figure 1.
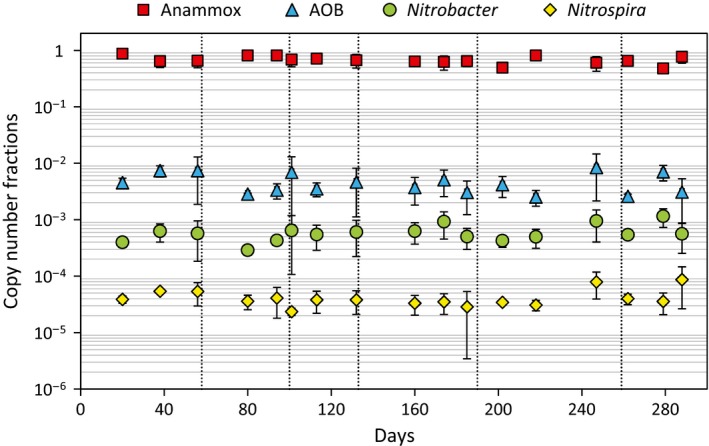
Relative abundances (copy number fractions) of nitrogen converting bacteria measured by qPCR. The periods in the study are separated by vertical dashed lines. Average values. Error bars show standard deviation.
Batch activity tests of nitrogen converting groups of microorganisms
Batch tests (Fig. 2) were performed to assess the potential aerobic ammonium oxidation (AOB), aerobic nitrite oxidation (NOB), anammox and nitrate uptake rate (potential denitrification). Although the variations between samples and sampling occasions were considerable, the data showed a gradual decrease in potential anammox (ANOVA, P < 0.01, F = 5.7) and an increase in potential nitrite oxidation (ANOVA, P < 0.01, F = 4.6). For the aerobic ammonium oxidation and the nitrate uptake rate, the changes between the periods were not significant (P > 0.05).
Figure 2.
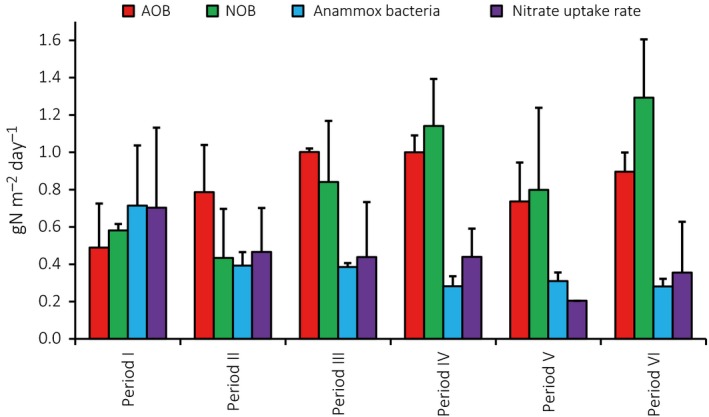
Potential activity of nitrogen converting bacterial guilds measured in batch tests at period I–VI. Bars show average values, error bars show standard deviation.
Composition and diversity of the biofilm communities
High throughput amplicon sequencing of the 16S rRNA gene (V4 region) showed that the bacterial communities at four periods in the MBBR consisted of similar numbers of OTUs (477 to 523), when resampled at 10 000 sequences (Table S2) and of all the 886 OTUs detected, 236 were shared by all samples. The diversity of the sample from the final period (VI) was somewhat higher, indicative of a slightly more even community (Table S2). Pairwise comparisons of the biofilm communities (Bray‐Curtis) in periods I, III and V resulted in coefficients of 0.10–0.11, while period VI differed slightly more (0.15–0.18). However, the dissimilarity coefficients were generally low, indicating similar community structure of all samples.
The composition at the phylum level showed that the majority of the sequences belonged to Planctomycetes, with Chloroflexi and Proteobacteria also having large contributions to the community (Fig. 3). In addition, Bacteroidetes, Acidobacteria, Chlorobi, the candidate phylum BRC1, Actinobacteria and Deinococcus‐Thermus were present in relative sequence abundances > 0.5%.
Figure 3.
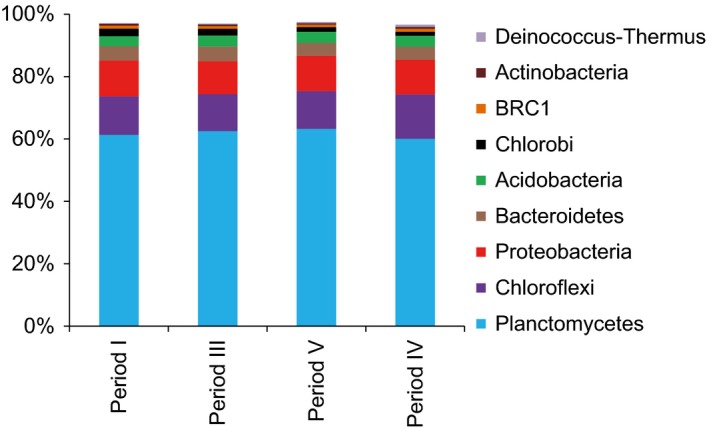
Major bacterial phyla (> 0.5% relative sequence contribution) in the biofilm communities, as revealed by high throughput amplicon sequencing.
The anammox bacteria were all affiliated to the genus Ca. Brocadia in one single OTU that dominated the bacterial biofilm community in all reactor periods (Table 2). Further subdivision of the sequences within Ca. Brocadia, by applying a more stringent criterion of 99% sequence similarity of OTUs, revealed that most anammox bacteria were similar to Ca. Brocadia sp. 40, with a smaller population of Ca. Brocadia fulgida‐like bacteria (Table S3). This population was also detected by FISH (0.5–2.0% of total bacteria).
Table 2.
Autotrophic nitrogen converting bacteria in the biofilm communities, as revealed by high throughput amplicon sequencing. OTUs clustered at 97% sequence similarity. BLAST analysis was used for classification
| OTU | Classification | Similarity | Period I | Period III | Period V | Period IV |
|---|---|---|---|---|---|---|
| 2 | Ca. Brocadia sp. 40/Ca. B. caroliniensis 20b | 98 | 56% | 56% | 57% | 50% |
| 85 | Nitrosomonas europaea/N. eutropha | 99 | 0.32% | 0.27% | 0.14% | 0.25% |
| 3 | Nitrosomonas sp. JL21 | 100 | 0.038% | 0.026% | 0.032% | 0.014% |
| 2049 | Nitrosospira multiformis | 97 | 0.017% | 0.010% | 0.012% | 0.028% |
| 930 | Ca. Nitrotoga sp. clone JS16NT08 | 100 | 0.13% | 0.051% | 0.071% | 0.047% |
| 354 | Nitrospirales 4‐29a | N.A. | 0.11% | 0.18% | 0.059% | 0.079% |
No described species with > 90% similarity from BLAST analysis. Classification by the Greengenes taxonomy.
All identified AOB were affiliated to Betaproteobacteria and the major OTU (OTU 85) was affiliated to the Nitrosomonas europaea/eutropha cluster (Table 2). Separate populations of Nitrosomonas europaea and Nitrosomonas eutropha within this cluster could be detected at 99% sequence similarity (Table S3). OTUs similar to Nitrosomonas sp. JL21 and Nitrosospira multiformis were also observed (Table 2), but at very low relative abundances. The presence of Nitrosomonas europaea/eutropha, Nitrosomonas oligotropha (JL21) and Nitrosospira was confirmed by FISH (Fig. 5) at relative abundances of 2.4–6.7%, 0–0.2% and 0.06–0.6% respectively.
Figure 5.
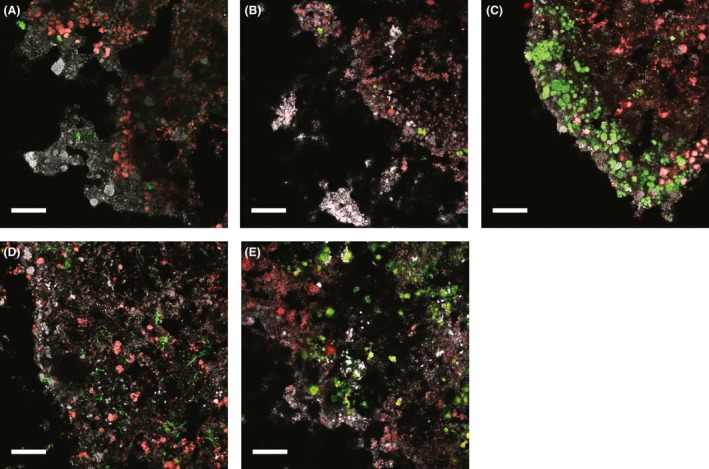
FISH‐CLSM of biofilm cryosections. The water‐biofilm interface is oriented to the lower left. In all images, anammox bacteria (Amx820) are in red and nonspecific bacteria (EUB‐mix) are in white‐grey.
A. In green: AOB within the Nitrosomonas europaea/eutropha cluster (Nse1472).
B. In green: AOB within Nitrosospira (Nsv443).
C. In green: NOB within Nitrospira (Ntspa662).
D. In green: Bacteria within Chloroflexi (CFX123 + GNSB941).
E. In green: Ca. Brocadia fulgida (Bfu613). Scale bar: 25 μm.
Among the NOB, one OTU belonged to the order Nitrospirales (Table 2) but could not be affiliated to any described bacterium. However, BLAST analysis showed high similarities to unclassified sequences from other nitrogen converting wastewater reactors (OTU1534‐1535 in Table S3), suggesting that these non‐described Nitrospirales converted nitrogen. In addition, the presence of Nitrospira was demonstrated by qPCR and FISH (Table S4), but no FISH signal was seen with comammox probes for Nitrospira nitrosa and N. nitrificans (Table S1). One OTU highly similar to Nitrotoga was detected in all biofilm communities (Table 2) and Nitrotoga cells were also detected by FISH (Table S4). No OTU was assigned to Nitrobacter, although Nitrobacter was detected by qPCR and FISH (Table S4). However, OTUs with high similarity to Nitrobacter sp., but also to other species within Bradyrhizobiaceae, were revealed by BLAST (OTU 0176, 0226 in Table S3). Hence, the sequence information in the V4 region of the 16S rRNA gene was not sufficient for Nitrobacter identification.
In the MBBR, 43–50% of the sequences were affiliated to putative heterotrophic bacteria. These bacteria were subdivided into 25 orders with a sequence contribution > 0.5% in any of the samples (Fig. 4).
Figure 4.
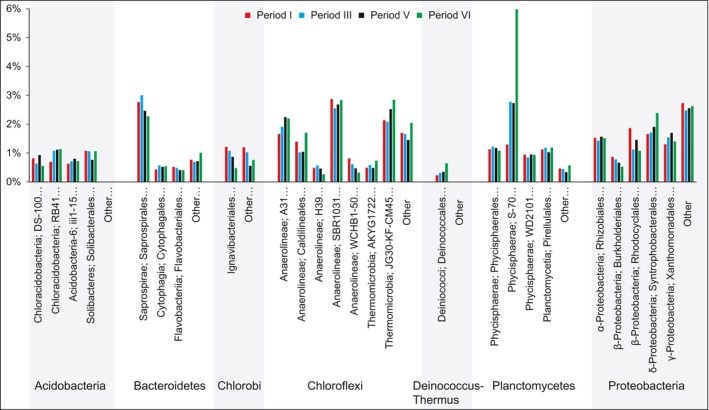
Major orders (> 0.5% relative sequence contribution) of bacteria not involved in autotrophic nitrogen conversion in the biofilm communities, as revealed by high throughput amplicon sequencing.
Localization of key bacterial groups in the biofilm
FISH‐CLSM of biofilm cryosections was used to show the localization of key bacterial groups (for FISH probes, see Table S1). Clusters of AOB (Nitrosomonas europaea/eutropha, Fig. 5A) and Nitrosospira, Fig. 5B), and clusters of NOB (Nitrospira, Fig. 5C) were detected near the biofilm–water interface. Anammox bacteria were observed in high numbers deeper in the biofilm (Fig. 5), with two populations present closer to the biofilm–water interface (Fig. 5E). Bacteria within the phylum Chloroflexi were also detected both near the biofilm–water interface and deeper in the biofilm (Fig. 5D).
Discussion
Substrate availability is a main factor that determines microbial competitive interactions and thereby shapes the structure of microbial communities (Hibbing et al., 2010; Litchman et al., 2015). PNA is used for treatment of highly concentrated as well as diluted streams of wastewater and the substrate concentrations vary a lot (e.g. Hu et al., 2013; Lackner et al., 2014; Lotti et al., 2014a), but very few systematic studies of the community response to changes in substrate availability have been made. Here, we test the hypothesis that a reduction in substrate availability influences the PNA microbial community structure and function in a MBBR at low temperature, gradually approaching main stream conditions. The stepwise diluted influent resulted in decreased reactor ammonium concentrations, from 311 to 27 mg‐N l−1, as well as decreased concentrations of COD (Table 1). Despite these decreases in substrate concentrations, no major effects on the reactor biomass were observed. The biofilm weight did not change significantly (Table S3). The anammox bacteria dominated the bacterial community with AOB and NOB being considerably fewer (Fig. 1, Table 2, S4), located near the biofilm–water interface (Fig. 5). Changes in the relative abundances of anammox bacteria, AOB and NOB (qPCR) were non‐significant between test periods and could not be related to the reactor concentrations of nitrogen species, COD and alkalinity. Changes in potential activity of the anammox bacteria and NOB were observed (Fig. 2) and reflected the nitrogen conversions in the reactor (Table 1), but in general reactor operation was stable (Table 1), suggesting similar functionality of the microbial community.
The estimation of the relative abundances of anammox bacteria, AOB and NOB generally agreed between the methods (Table S4). The largest deviation was the higher AOB percentage assessed by FISH, but smaller differences among the methods were observed also for anammox bacteria and NOB. Discrepancies between rRNA‐based FISH and DNA‐based qPCR and sequencing can be expected. A large fraction of the microbial community had a ribosomal content below the FISH detection limit (Figure S4). However, AOB are known to retain their ribosomes even at challenging conditions such as starvation (Morgenroth et al., 2000), which may help to explain their particularly high relative abundance observed by FISH. Also, the DNA extraction methodology, the choice of PCR primers and the cell copy numbers of target genes influence relative abundances (see e.g. Acinas et al., 2004; Albertsen et al., 2015). Quantification by high throughput amplicon sequencing is furthermore technically challenging (Zhou et al., 2015). Although sequence processing has improved significantly with pipelines, such as Mothur used here (Kozich et al., 2013), relative abundances, especially of rare OTUs (e.g. the NOB), have to be interpreted with caution. Hence, all methods suffer from limitations and multiple methods provide important complementary information. The methods also vary in response time. Specific populations can be estimated by qPCR and FISH on suspended biofilms within a day or two, which is useful for routine monitoring. High throughput amplicon sequencing and FISH biofilms cryosections provide more detailed information, but takes considerably longer time.
The major anammox population was highly similar to Ca. Brocadia sp. 40 (Table 2, Table S3), which has previously been observed in several anammox reactors (van der Star et al., 2008; Park et al., 2010; Costa et al., 2014; Gilbert et al., 2014). Only one population of anammox bacteria is usually observed in PNA reactors (Hu et al., 2013; Gilbert et al., 2014; Laureni et al., 2015). However, using 99% similarity for sequence clustering (Table S3) and a competitor probe to improve the FISH specificity (Table S1, Persson et al. (2014)), a second, closely related, Ca. Brocadia fulgida‐like population was detected (Fig. 5, Table S3). It is likely that the two populations have different niches, just as described for closely related Nitrospira strains (Gruber‐Dorninger et al., 2014). This was supported by the localization of the smaller Ca. Brocadia fulgida‐like population near the biofilm–water interface while the Ca. Brocadia sp. 40 population was detected throughout the biofilm. Furthermore, despite studies showing that Ca. Kuenenia have higher substrate affinity than Ca. Brocadia, and hence would be selected for at low substrate concentrations (van der Star et al., 2008; Oshiki et al., 2011), Ca. Brocadia fulgida and/or Ca. Brocadia sp. 40, rather than Ca. Kuenenia have dominated the anammox guild in this and other main stream PNA studies (Gilbert et al., 2014; Lotti et al., 2014a,b).
The AOB community was dominated by two populations within the Nitrosomonas europaea/eutropha cluster (Table 2, S3), which are commonly found in PNA reactors (Park et al., 2010; Vlaeminck et al., 2010; Pellicer‐Nàcher et al., 2014). Minor OTUs similar to N. oligotropha (sp. JL21) and Nitrosospira multiformis were also detected. This diverse AOB community was confirmed by FISH.
NOB were constantly present (Fig. 1) and were active during all periods, as seen by the production of nitrate (Table 1, Figure S1) and batch activity tests (Fig. 2). In particular, they had large impact on the process performance in period VI, resulting in low nitrogen removal efficiency (11%). Strategies to abate NOB include careful control of DO‐ and substrate concentrations (Perez et al., 2014) as well as intermittent periods of anoxic and aerated phases, either at high‐or low‐DO concentrations (Wett et al., 2013; Ma et al., 2015). Despite maintained ammonium concentrations and careful DO control, NOB could not be repressed in the MBBR biofilms. Unwanted NOB activity in main stream PNA reactors is frequently reported (De Clippeleir et al., 2013; Gilbert et al., 2014; Lotti et al., 2014a) and the strategy for NOB repression depending on the aggregation state of the biomass (suspended, granular, biofilm) and the ecophysiology of AOB and NOB.
The NOB consisted of Nitrobacter, Nitrospira and Nitrotoga. In PNA reactors, the low bulk concentrations of nitrite would select for Nitrospira, rather than Nitrobacter, due to their higher substrate affinity (Isanta et al., 2015). Nitrospira is furthermore hard to outcompete using low DO concentrations due to their high oxygen affinity (Isanta et al., 2015). Nitrospira has, in fact, been the only NOB observed in some main stream PNA reactors (De Clippeleir et al., 2013; Gilbert et al., 2014), but as shown here and elsewhere (Liu et al., 2012), even Nitrobacter can sustain at such conditions. Interestingly, the long‐term operation of the MBBR at 13°C allowed the establishment of a small Nitrotoga population (Table 2, S4). Nitrotoga has been shown to be important in activated sludge communities at 7–16°C (Lücker et al., 2015), but so far little is known about their ecophysiology, except for their low temperature requirements. Nitrotoga has, to the best of our knowledge, not been previously detected in PNA reactors and this finding may have implications for NOB suppression strategies.
In the MBBR, 43–50% of the sequences were affiliated to putative heterotrophic bacteria. Also in other studies of PNA systems, significant fractions of the microbial communities have been heterotrophs, although little is known about their composition, dynamics and roles (Chu et al., 2015; Gilbert et al., 2014; Pellicer‐Nàcher et al., 2014). The relative abundances of the major contributors of the heterotrophic community in the MBBR (from seven phyla) were stable in relative abundance throughout the study (Fig. 4). This implies that the decreasing COD concentrations in the reactor (Table 1) had little impact on the heterotrophic community and suggests that even heterotrophic bacteria were an integral part of the community, possibly with defined roles in the PNA biofilm. Heterotrophic bacteria may contribute to nitrogen removal via denitrification, but their competition with AOB and anammox bacteria for space and electron acceptors can also be detrimental (Kumar and Lin, 2010). Furthermore, they can utilize soluble microbial products (SMP) from the biofilm (Ni et al., 2012) and aid in biofilm formation (Cho et al., 2010). A minor fraction of the influent COD was consistently removed in the MBBR (Table 1) and batch activity tests showed anoxic nitrate uptake (Fig. 2), which suggests some denitrification. We found members of Rhodocyclales, Burkholderiales, Rhizobiales and Xanthomonadales (Fig. 4), which all are important contributors to wastewater denitrification (Baytshtok et al., 2009; McIlroy et al., 2016) and have been detected in PNA reactors treating organic‐free wastewater (Pellicer‐Nàcher et al., 2014; Chu et al., 2015), suggesting SMP utilization. SMP may also have sustained the biofilm population of the non‐denitrifying, protein degrading Saprospirales (Xia et al., 2008). Chloroflexi were abundant here (Figs. 3 and 4), as well as in other PNA communities (Gilbert et al., 2014; Chu et al., 2015). Chloroflexi can provide biofilm structural integrity (Cho et al., 2010) and metabolize SMP from autotrophs at both aerobic and anoxic conditions (Okabe et al., 2005; Kindaichi et al., 2012), which would explain their distribution throughout the biofilm (Fig. 5). Phycisphaerae, Ignavibacteriales, Deinococcales (Fig. 4) harbour bacteria with mostly undefined ecophysiologies, but their presence here and in other PNA‐ and anammox communities (Costa et al., 2014; Chu et al., 2015) suggests defined functions.
There are several possible explanations for the observed stability and maintained diversity of the microbial community at the decreasing substrate concentrations. The main anammox bacteria (Ca. Brocadia sp. 40 and Ca. Brocadia fulgida) and AOB (Nitrosomonas europaea/eutropha) have been detected at high relative abundances at different conditions, including a wide range of substrate concentrations (Park et al., 2010; Vlaeminck et al., 2010; Almstrand et al., 2014; Gilbert et al., 2014; Lotti et al., 2014a; Pellicer‐Nàcher et al., 2014), which indicates broad ecophysiologies and a competitiveness at all tested concentrations. Furthermore, the presence of numerous micro‐environments in the thick biofilms with gradients of substrate and electron acceptors likely promoted diversity and permitted the coexistence of competing as well as of commensal community members. As mentioned, the community had both active and non‐active bacteria; the low ribosomal content of a large fraction of the bacteria indicated inactivity (Figure S4). The protected environment in the biofilm carriers may offer a refuge site for active and inactive cells, which may slow down community changes, as would the low temperature. Although the time between subsequent test periods may have been too short for major community changes to occur, for the entire study, spanning 302 days, time was likely sufficient. In municipal wastewater, the continuous variations over time in influent composition of substrates and suspended bacteria, are factors that may affect the stability of the microbial community, but these were not addressed here. Very few studies have been performed on PNA using real main stream wastewater, and the impact of these factors is yet not valuated. Maintenance and activity of anammox and AOB populations for at least 240 days in PNA MBBRs receiving pre‐treated municipal wastewater was recently shown (Laureni et al., 2016), indicating that, at least for biofilm systems, the influence of these factors for the stability of the key functional populations is manageable.
In conclusion, the bacterial community in a PNA MBBR system was stable during decreasing concentrations of substrate, approaching main stream conditions. Within the guilds of AOB, anammox bacteria and NOB, composition and diversity was maintained at all tested concentrations. The composition was largely stable even for the diverse heterotrophic community, suggesting that they were an integral part of the community.
Experimental procedures
The pilot moving bed biofilm reactor
A 200 l MBBR was filled with biofilm carriers (Kaldnes K1) at 40% filling degree. It received reject water after anaerobic digestion from the Himmerfjärden WWTP in Stockholm, diluted with tap water. The study period (302 days) was divided into six periods with stepwise decreased influent concentrations of ammonium from 500 mg‐N l−1, representative of reject water, to 45 mg‐N l−1, representative of main stream wastewater, with concomitant decreases in nitrogen loading rate and hydraulic retention time (Table 1). The MBBR was operated at 13°C corresponding to low main stream temperatures in moderate climates.
Redox, pH and DO was measured using online sensors (Cerlic AB, Segeltorp, Sweden). Air supply was provided from the bottom of the reactor and was controlled, via the DO, by a PID controller. The temperature was monitored and controlled by a compact controller (JUMO GmbH & Co. KG, Fulda, Germany) and a cooler (JULABO GmbH, Seelbach, Germany). Mixing of the bulk water and biofilm carriers was achieved by a two‐blade stirrer (50 rpm).
For analysis of inorganic nitrogen species and COD in the influent and effluent (filtered 0.45 μm), Dr. LANGE cuvettes were used on a XION 500 Spectrophotometer (HACH LANGE GmbH, Düsseldorf, Germany).
Batch tests for potential activity measurements
Batch tests were performed to measure potential microbial activities at 25°C. The activity measurements of AOB and NOB was based on the oxidation rate of ammonium (present in the test medium) and nitrite (formed by the AOB during the test) by measuring the oxygen uptake rate (OUR) using a DO probe (YSI 5905; YSI Inc. Yellow Springs, OH, USA). The method was adopted from Surmacz‐Górska et al. (1996). At the start of each measurement, ammonium was available at 100 mg N l−1. First, the total OUR was measured. After 5 minutes, NaClO3 (17 mM) was added for the inhibition of NOB. After 10 minutes, allylthiourea (43 μM) was added for the inhibition of AOB. The activity of NOB and AOB was obtained from the OUR before minus the OUR after the addition of NaClO3 and allylthiourea respectively. The remaining OUR, after addition of both inhibitors, is represented by endogenous respiration and substrate oxidation by heterotrophs and was not reported. The potential anammox activity was measured as the production of nitrogen gas (headspace pressure) according to Dapena‐Mora et al. (2007) with NH4 + and NO2 − at initial concentrations of 70 mg N l−1 each. Nitrate uptake rate measurements were used for measurements of potential heterotrophic denitrifying activity. The tests were performed at anoxic conditions in reject water diluted with distilled water at an initial COD concentration of 200 mg O2 l−1, with NaNO3 at an initial concentration of 100 mg N l−1. Samples for measurement of NO3 −were taken every 12 minutes for 4 hours.
DNA extraction
DNA was separately extracted from three carriers at each sampling occasion. From each carrier, 30 mg of biofilm was used for extraction using the FastDNA spin kit for soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturers' recommendations. The concentration of the extracted DNA was measured using a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific NanoDrop products, Wilmington, DE, USA).
Quantitative PCR
qPCR was used for quantification of autotrophic nitrogen converting bacteria according to Persson et al. (2014). In brief, primers for the 16S rRNA gene were used to target all bacteria, anammox bacteria, Nitrospira and Nitrobacter and primers for the amoA gene were used to target AOB. The qPCR was carried out on an iQ5 (Bio‐Rad Laboratories. Inc., Hercules, CA, USA) thermal cycler using the SYBR green chemistry. Plasmid target gene inserts were used as standards. The results are presented as copy number fractions of the nitrogen converging bacteria to all bacteria, to get relative abundances. Differences in the relative abundances between periods I–VI was tested by one‐factor analysis of variance (ANOVA). Prior to ANOVA, variance homogeneity was confirmed by Levene's test. To assess whether there was a link between the reactor conditions and the relative abundance of the key microbial groups at each sampling occasion (n = 17), the preceding reactor concentrations of nitrogen species, COD and alkalinity was averaged over 10 days. Correlations between these average concentrations and the relative abundances were tested for using a linear model (Pearson's r).
High throughput amplicon sequencing
PCR was carried out using the primers 515F and 806R to amplify partial V4 region sequences of the 16S rRNA gene (Caporaso et al., 2011) with dual indexing of the primers (Kozich et al., 2013). Sequencing was performed on an Illumina MiSeq (Illumina Inc., San Diego, CA, USA) using the MiSeq Reagent Kit v2 with PhiX control library spiked in at 7.5%. For details on PCR, purification, and quality control, see supporting methods. The obtained sequences were processed in Mothur (Schloss et al., 2009) for assembly of contigs, denoising, removal of putative chimera, alignment, classification and construction of operational taxonomic units (OTUs) at 97% taxonomic identity (Kozich et al., 2013). For classification with the Bayesian classifier within Mothur, the Greengenes database v. 13.8.99 (McDonald et al., 2012) was used at 80% confidence threshold. Prior to analysing alpha‐ and beta‐diversity, the OTU dataset was subsampled at 10 000 sequences. Raw sequence reads were deposited at the NCBI Sequence Read Archive, no. SRP059362.
Fluorescence in situ hybridization and confocal laser scanning microscopy
For FISH, samples were taken in the periods II, IV and VI. The carriers were fixed in paraformaldehyde (4% w/v) for 8 h at 4°C. For FISH on biofilm suspension, two carriers form each sampling period were used. The fixed biofilm was brushed off the carriers and homogenized in PBS before storage in PBS‐ethanol (1:1) at −20°C. The biofilm suspensions (2–4 μl) were spotted on diagnostic microscope slides (8 × 6 mm diameter wells; Menzel GmbH, Braunschweig, Germany) for FISH and images were acquired from 10 random fields of view for each carrier. For FISH on biofilm cryosections, the carriers with fixed biofilm were embedded, frozen and cryosectioned in 20–25 μm thick slices which were captured on microscope slides and subjected to FISH. FISH was carried out at 46°C for 2 h for biofilm suspensions and 4 h for biofilm cryosections according to Almstrand et al. (2014). The probes (Table S1) were 5′ labelled with Cy3, Cy5, or Alexa 488. The relative abundances of the anammox bacteria, AOB and NOB was estimated on biofilm suspensions as the ratios of the FISH‐targeted biovolumes of the specific populations to the total bacteria (EUB338 I‐IV probe mix, Table S1) using daime 2.1 (Daims et al., 2006). See supporting methods for details about embedding, cryosectioning, image acquisition and image analysis.
Conflict of Interest
None declared.
Supporting information
Fig. S1. Influent and effluent concentrations of ammonium, nitrite and nitrate in the MBBR. Dashed vertical lines highlight the different periods in the study.
Fig. S2. Biofilm carrier from the MBBR.
Fig. S3. Biomass wet weight of the biofilm carriers. Dashed lines show the transition between different periods. Average values of eight carriers at each sampling occasion. Error bars show standard deviation.
Fig. S4. Comparison between FISH (using EUBmix probe) and staining of cells with SYTO 62.
Table S1. FISH probes used in the study.
Table S2. Diversity of the biofilm communities by high throughput amplicon sequencing. 1000 resamplings of 10 000 sequences. OTUs clustered at 97% sequence similarity.
Table S3. Potential autotrophic nitrogen removing OTUs clustered at 99% sequence similarity.
Table S4. Percentage of anammox, AOB and NOB during the experiment. Data are range of percentages over periods during the experiment (see Table 1 for time periods). FISH data are biovolume of specific probe targeted guilds in percentages of the total biovolume measured by EUB probe mix (see Table S1 for probes). FISH data are from periods II, IV and VI. High throughput amplicon sequencing data are percentages of OTUs from target group out of the total number of OTUs during periods I, IV, V and VI. qPCR data are percentages of the different target groups of the total bacteria measured by a universal primer pair. qPCR data are from periods I – VI. See Experimental Procedures for details.
Data S1. Supporting methods and references.
Acknowledgements
This study was funded by FORMAS (Contract no. 243‐2010‐2259, 211‐2010‐140, 2015‐1515‐30425‐28 and 245‐2014‐1528), SVU (Contract no. 10‐105) and the foundations of Carl Trygger (CTS 12:374), Adlerbertska forskningsstiftelsen and Åke & Greta Lissheds. The authors acknowledge the Centre for Cellular Imaging and the Genomics core facility at the University of Gothenburg for support and use of their equipment and the staff at the Hammarby Sjöstadsverk research facility, Stockholm, for maintenance of the pilot plant. The authors also acknowledge the anonymous reviewers for valuable comments.
Microb Biotechnol (2017) 10(4), 761–772
References
- Acinas, S. G. , Marcelino, L. A. , Klepac‐Ceraj, V. , and Polz, M. F. (2004) Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen, M. , Karst, S. M. , Ziegler, A. S. , Kirkegaard, R. H. , and Nielsen, P. H. (2015) Back to basics–the influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PLoS ONE 10(7): e0132783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstrand, R. , Persson, F. , Daims, H. , Ekenberg, M. , Christensson, M. , Wilén, B.‐M. , et al (2014) Three‐dimensional stratification of bacterial biofilm populations in a moving bed biofilm reactor for nitritation‐anammox. Int J Mol Sci 15: 2191–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baytshtok, V. , Lu, H. J. , Park, H. , Kim, S. , Yu, R. , and Chandran, K. (2009) Impact of varying electron donors on the molecular microbial ecology and biokinetics of methylotrophic denitrifying bacteria. Biotechnol Bioeng 102: 1527–1536. [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G. , Lauber, C. L. , Walters, W. A. , Berg‐Lyons, D. , Lozupone, C. A. , Turnbaugh, P. J. , et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl. 1): 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S. , Takahashi, Y. , Fujii, N. , Yamada, Y. , Satoh, H. , and Okabe, S. (2010) Nitrogen removal performance and microbial community analysis of an anaerobic up‐flow granular bed anammox reactor. Chemosphere 78: 1129–1135. [DOI] [PubMed] [Google Scholar]
- Chu, Z.‐R. , Wang, K. , Li, X.‐K. , Zhu, M.‐T. , Yang, L. , and Zhang, J. (2015) Microbial characterization of aggregates within a one‐stage nitritation–anammox system using high‐throughput amplicon sequencing. Chem Eng J 262: 41–48. [Google Scholar]
- Costa, M. C. , Carvalho, L. , Leal, C. D. , Dias, M. F. , Martins, K. L. , Garcia, G. B. , et al (2014) Impact of inocula and operating conditions on the microbial community structure of two anammox reactors. Environ Technol 35: 1811–1822. [DOI] [PubMed] [Google Scholar]
- Daims, H. , Lucker, S. , and Wagner, M. (2006) Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8: 200–213. [DOI] [PubMed] [Google Scholar]
- Dapena‐Mora, A. , Fernandez, I. , Campos, J. L. , Mosquera‐Corral, A. , Mendez, R. , and Jetten, M. S. M. (2007) Evaluation of activity and inhibition effects on Anammox process by batch tests based on the nitrogen gas production. Enzyme Microb Tech 40: 859–865. [Google Scholar]
- De Clippeleir, H. , Vlaeminck, S. E. , De Wilde, F. , Daeninck, K. , Mosquera, M. , Boeckx, P. , et al (2013) One‐stage partial nitritation/anammox at 15 degrees C on pretreated sewage: feasibility demonstration at lab‐scale. Appl Microbiol Biotechnol 97: 10199–10210. [DOI] [PubMed] [Google Scholar]
- Dosta, J. , Fernandez, I. , Vazquez‐Padin, J. R. , Mosquera‐Corral, A. , Campos, J. L. , Mata‐Alvarez, J. , and Mendez, R. (2008) Short‐ and long‐term effects of temperature on the Anammox process. J Hazard Mater 154: 688–693. [DOI] [PubMed] [Google Scholar]
- Gilbert, E. M. , Agrawal, S. , Karst, S. M. , Horn, H. , Nielsen, P. H. , and Lackner, S. (2014) Low temperature partial nitritation/anammox in a moving bed biofilm reactor treating low strength wastewater. Environ Sci Technol 48: 8784–8792. [DOI] [PubMed] [Google Scholar]
- Gilbert, E. M. , Agrawal, S. , Schwartz, T. , Horn, H. , and Lackner, S. (2015) Comparing different reactor configurations for Partial Nitritation/Anammox at low temperatures. Water Res 81: 92–100. [DOI] [PubMed] [Google Scholar]
- Gruber‐Dorninger, C. , Pester, M. , Kitzinger, K. , Savio, D. F. , Loy, A. , Rattei, T. , et al (2014) Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson, D.J. , Persson, F. and Jansen, J.L. (2014) Manammox – mainstream anammox at Sjölunda WWTP. IWA World Water Congress and Exhibition, September 21‐26, 2014. Lisbon, Portugal: IWA; [Google Scholar]
- Hendrickx, T. L. , Kampman, C. , Zeeman, G. , Temmink, H. , Hu, Z. , Kartal, B. , and Buisman, C. J. (2014) High specific activity for anammox bacteria enriched from activated sludge at 10 degrees C. Bioresour Technol 163: 214–221. [DOI] [PubMed] [Google Scholar]
- Hibbing, M. E. , Fuqua, C. , Parsek, M. R. , and Peterson, S. B. (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. Y. , Lotti, T. , de Kreuk, M. , Kleerebezem, R. , van Loosdrecht, M. , Kruit, J. , et al (2013) Nitrogen removal by a nitritation‐anammox bioreactor at low temperature. Appl Environ Microbiol 79: 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isanta, E. , Reino, C. , Carrera, J. , and Perez, J. (2015) Stable partial nitritation for low‐strength wastewater at low temperature in an aerobic granular reactor. Water Res 80: 149–158. [DOI] [PubMed] [Google Scholar]
- Kartal, B. , Kuenen, J. G. , and van Loosdrecht, M. C. M. (2010) Sewage treatment with Anammox. Science 328: 702–703. [DOI] [PubMed] [Google Scholar]
- Kindaichi, T. , Yuri, S. , Ozaki, N. , and Ohashi, A. (2012) Ecophysiological role and function of uncultured Chloroflexi in an anammox reactor. Water Sci Technol 66(12): 2556–2561. [DOI] [PubMed] [Google Scholar]
- Kozich, J. J. , Westcott, S. L. , Baxter, N. T. , Highlander, S. K. , and Schloss, P. D. (2013) Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. , and Lin, J. G. (2010) Co‐existence of anammox and denitrification for simultaneous nitrogen and carbon removal‐strategies and issues. J Hazard Mater 178: 1–9. [DOI] [PubMed] [Google Scholar]
- Lackner, S. , Gilbert, E. M. , Vlaeminck, S. E. , Joss, A. , Horn, H. , and van Loosdrecht, M. C. (2014) Full‐scale partial nitritation/anammox experiences ‐ An application survey. Water Res 55: 292–303. [DOI] [PubMed] [Google Scholar]
- Laureni, M. , Weissbrodt, D.G. , Szivak, I. , Robin, O. , Nielsen, J.L. , Morgenroth, E. and Joss, A. (2015) Activity and growth of anammox biomass on aerobically pre‐treated municipal wastewater. Water Res 80: 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureni, M. , Falås, P. , Robin, O. , Wick, A. , Weissbrodt, D. G. , Nielsen, J. L. , et al (2016) Mainstream partial nitritation and anammox: long‐term process stability and effluent quality at low temperatures. Water Res 101: 628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchman, E. , Edwards, K. F. , and Klausmeier, C. A. (2015) Microbial resource utilization traits and trade‐offs: implications for community structure, functioning, and biogeochemical impacts at present and in the future. Front Microbiol 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Li, D. , Zeng, H. , Li, X. , Zeng, T. , Chang, X. , et al (2012) Biodiversity and quantification of functional bacteria in completely autotrophic nitrogen‐removal over nitrite (CANON) process. Bioresource Technol 118: 399–406. [DOI] [PubMed] [Google Scholar]
- Lotti, T. , Kleerebezem, R. , Hu, Z. , Kartal, B. , Jetten, M. S. , and van Loosdrecht, M. C. (2014a) Simultaneous partial nitritation and anammox at low temperature with granular sludge. Water Res 66C: 111–121. [DOI] [PubMed] [Google Scholar]
- Lotti, T. , Kleerebezem, R. , van Erp Taalman Kip, C. , Hendrickx, T.L. , Kruit, J. , Hoekstra, M. and van Loosdrecht, M.C. (2014b) Anammox growth on pretreated municipal wastewater. Environ Sci Technol 48: 7874–7880. [DOI] [PubMed] [Google Scholar]
- Lotti, T. , Kleerebezem, R. , and van Loosdrecht, M. C. (2015) Effect of temperature change on anammox activity. Biotechnol Bioeng 112: 98–103. [DOI] [PubMed] [Google Scholar]
- Lücker, S. , Schwarz, J. , Gruber‐Dorninger, C. , Spieck, E. , Wagner, M. , and Daims, H. (2015) Nitrotoga‐like bacteria are previously unrecognized key nitrite oxidizers in full‐scale wastewater treatment plants. ISME J 9: 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Bao, P. , Wei, Y. , Zhu, G. , Yuan, Z. , and Peng, Y. (2015) Suppressing nitrite‐oxidizing bacteria growth to achieve nitrogen removal from domestic wastewater via anammox using intermittent aeration with low dissolved oxygen. Sci Rep 5: 13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D. , Price, M. N. , Goodrich, J. , Nawrocki, E. P. , DeSantis, T. Z. , Probst, A. , et al (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy, S. J. , Starnawska, A. , Starnawski, P. , Saunders, A. M. , Nierychlo, M. , Nielsen, P. H. , and Nielsen, J. L. (2016) Identification of active denitrifiers in full‐scale nutrient removal wastewater treatment systems. Environ Microbiol 18: 50–64. [DOI] [PubMed] [Google Scholar]
- Morgenroth, E. , Obermayer, A. , Arnold, E. , Bruhl, A. , Wagner, M. , and Wilderer, P. A. (2000) Effect of long‐term idle periods on the performance of sequencing batch reactors. Water Sci Technol 41(1): 105–113. [Google Scholar]
- Ni, B. J. , Ruscalleda, M. , and Smets, B. F. (2012) Evaluation on the microbial interactions of anaerobic ammonium oxidizers and heterotrophs in Anammox biofilm. Water Res 46: 4645–4652. [DOI] [PubMed] [Google Scholar]
- Okabe, S. , Kindaichi, T. , and Ito, T. (2005) Fate of 14C‐labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl Environ Microbiol 71: 3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiki, M. , Shimokawa, M. , Fujii, N. , Satoh, H. , and Okabe, S. (2011) Physiological characteristics of the anaerobic ammonium‐oxidizing bacterium ‘Candidatus Brocadia sinica’. Microbiology 157: 1706–1713. [DOI] [PubMed] [Google Scholar]
- Park, H. , Rosenthal, A. , Jezek, R. , Ramalingam, K. , Fillos, J. , and Chandran, K. (2010) Impact of inocula and growth mode on the molecular microbial ecology of anaerobic ammonia oxidation (anammox) bioreactor communities. Water Res 44: 5005–5013. [DOI] [PubMed] [Google Scholar]
- Pellicer‐Nàcher, C. , Franck, S. , Gülay, A. , Ruscalleda, M. , Terada, A. , Al‐Soud, W. A. , et al (2014) Sequentially aerated membrane biofilm reactors for autotrophic nitrogen removal: microbial community composition and dynamics. Microb Biotechnol 7: 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, J. , Lotti, T. , Kleerebezem, R. , Picioreanu, C. , and van Loosdrecht, M. C. (2014) Outcompeting nitrite‐oxidizing bacteria in single‐stage nitrogen removal in sewage treatment plants: a model‐based study. Water Res 66C: 208–218. [DOI] [PubMed] [Google Scholar]
- Persson, F. , Sultana, R. , Suarez, M. , Hermansson, M. , Plaza, E. , and Wilen, B.‐M. (2014) Structure and composition of biofilm communities in a moving bed biofilm reactor for nitritation‐anammox at low temperatures. Bioresource Technol 154: 267–273. [DOI] [PubMed] [Google Scholar]
- Schloss, P. D. , Westcott, S. L. , Ryabin, T. , Hall, J. R. , Hartmann, M. , Hollister, E. B. , et al (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist, H. , Salzgeber, D. , Eugster, J. , and Joss, A. (2008) Anammox brings WWTP closer to energy autarky due to increased biogas production and reduced aeration energy for N‐removal. Water Sci Technol 57(3): 383–388. [DOI] [PubMed] [Google Scholar]
- van der Star, W. R. , Miclea, A. I. , van Dongen, U. G. , Muyzer, G. , Picioreanu, C. , and van Loosdrecht, M. C. (2008) The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotechnol Bioeng 101: 286–294. [DOI] [PubMed] [Google Scholar]
- Surmacz‐Górska, J. , Gernaey, K. , Demuynck, C. , Vanrolleghem, P. and Verstreate, W. (1996) Nitrification monitoring in activated sludge by oxygen uptake rate (OUR) measurements. Water Res 30: 1228–1236. [Google Scholar]
- Vlaeminck, S. E. , Terada, A. , Smets, B. F. , De Clippeleir, H. , Schaubroeck, T. , Bolca, S. , et al (2010) Aggregate size and architecture determine microbial activity balance for one‐stage partial nitritation and anammox. Appl Environ Microbiol 76: 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wett, B. , Omari, A. , Podmirseg, S.M. , Han, M. , Akintayo, O. and Gómez Brandón, M. , et al (2013) Going for mainstream deammonification from bench‐ to full‐scale for maximized resource efficiency. Water Sci Technol 68: 283–289. [DOI] [PubMed] [Google Scholar]
- Xia, Y. , Kong, Y. H. , Thomsen, T. R. , and Nielsen, P. H. (2008) Identification and ecophysiological characterization of epiphytic protein‐hydrolyzing Saprospiraceae (“Candidatus epiflobacter” spp.) in activated sludge. Appl Environ Microbiol 74: 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.Z. , He, Z.L. , Yang, Y.F. , Deng, Y. , Tringe, S.G. and Alvarez‐Cohen, L. (2015) High‐throughput metagenomic technologies for complex microbial community analysis: open and closed formats. MBio 6(1): e02288–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Influent and effluent concentrations of ammonium, nitrite and nitrate in the MBBR. Dashed vertical lines highlight the different periods in the study.
Fig. S2. Biofilm carrier from the MBBR.
Fig. S3. Biomass wet weight of the biofilm carriers. Dashed lines show the transition between different periods. Average values of eight carriers at each sampling occasion. Error bars show standard deviation.
Fig. S4. Comparison between FISH (using EUBmix probe) and staining of cells with SYTO 62.
Table S1. FISH probes used in the study.
Table S2. Diversity of the biofilm communities by high throughput amplicon sequencing. 1000 resamplings of 10 000 sequences. OTUs clustered at 97% sequence similarity.
Table S3. Potential autotrophic nitrogen removing OTUs clustered at 99% sequence similarity.
Table S4. Percentage of anammox, AOB and NOB during the experiment. Data are range of percentages over periods during the experiment (see Table 1 for time periods). FISH data are biovolume of specific probe targeted guilds in percentages of the total biovolume measured by EUB probe mix (see Table S1 for probes). FISH data are from periods II, IV and VI. High throughput amplicon sequencing data are percentages of OTUs from target group out of the total number of OTUs during periods I, IV, V and VI. qPCR data are percentages of the different target groups of the total bacteria measured by a universal primer pair. qPCR data are from periods I – VI. See Experimental Procedures for details.
Data S1. Supporting methods and references.


