Abstract
Hyperphosphorylation and aggregation of the neuronal protein tau are responsible for neurodegenerative diseases called tauopathies. Dysregulation of the alternative splicing of tau exon 10 results in alterations of the ratio of two tau isoforms, 3R-tau and 4R-tau, which have been seen in several tauopathies. Transactive response DNA-binding protein of 43 kDa (TDP-43) is involved in the regulation of RNA processing, including splicing. Cytoplasmic aggregation of TDP-43 has been observed in the brains of individuals with chronic traumatic encephalopathy or Alzheimer's disease, diseases in which neurofibrillary tangles of hyperphosphorylated tau are hallmarks. Here, we investigated the role of TDP-43 in tau exon 10 splicing. We found that TDP-43 promoted tau exon 10 inclusion, which increased production of the 4R-tau isoform. Moreover, TDP-43 could bind to intron 9 of tau pre-mRNA. Deletion of the TDP-43 N or C terminus promoted its cytoplasmic aggregation and abolished or diminished TDP-43-promoted tau exon 10 inclusion. Several TDP-43 mutations associated with amyotrophic lateral sclerosis or frontotemporal lobar degeneration with ubiquitin inclusions promoted tau exon 10 inclusion more effectively than wild-type TDP-43 but did not affect TDP-43 cytoplasmic aggregation in cultured cells. The ratio of 3R-tau/4R-tau was decreased in transgenic mouse brains expressing human TDP-43 and increased in the brains expressing the disease-causing mutation TDP-43M337V, in which cytoplasmic TDP-43 was increased. These findings suggest that TDP-43 promotes tau exon 10 inclusion and 4R-tau expression and that disease-related changes of TDP-43, truncations and mutations, affect its function in tau exon 10 splicing, possibly because of TDP-43 mislocalization to the cytoplasm.
Keywords: alternative splicing, Alzheimer disease, TAR DNA-binding protein 43 (TDP-43) (TARDBP), tau protein (tau), tauopathy
Introduction
Tau is a major neuronal microtubule-associated protein that plays an important role in the assembly and stabilization of microtubules. Hyperphosphorylation of tau leads to its aggregation into neurofibrillary tangles, a neuropathological hallmark of Alzheimer's disease (AD)2 and related neurodegenerative diseases called tauopathies (1–4). Adult human brain expresses six tau isoforms encoded from a single gene by alternative splicing of exons 2, 3, and 10 of its pre-mRNA (5). Exon 10 encodes the second microtubule-binding repeat (6), and its alternative splicing generates tau isoforms with three or four microtubule-binding repeats, named 3R-tau or 4R-tau, which are under developmental and cell type-specific regulation. In normal adult human brain, approximately equal levels of 3R- and 4R-tau are expressed (6, 7). Alteration in the 3R-tau/4R-tau ratio is sufficient to trigger neurodegeneration in frontotemporal dementia and might also play a role in other neurodegenerative disorders such as Pick's disease, corticobasal degeneration, and progressive nuclear palsy (8–11).
Alternative splicing of tau exon 10 can be regulated by many splicing factors and their kinases and phosphatases (12–18). Transactive response DNA-binding protein of 43 kDa (TDP-43) is an RNA- and DNA-binding protein involved in transcriptional repression, RNA splicing, and RNA stability (19). It is the major component of neuronal inclusions in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin inclusions (FTLD-U) (20–22). TDP-43 pathology has now been detected in a number of other neurodegenerative diseases, many of which are associated with tau pathology, including Pick's disease, corticobasal degeneration, chronic traumatic encephalopathy, dementia with Lewy bodies, and AD (23–26). However, whether TDP-43 regulates the alternative splicing of tau exon 10 and TDP-43 pathology is involved in the altered tau exon 10 splicing in any tauopathies remain elusive.
In the present study, we found that TDP-43 promoted tau exon 10 inclusion. Both the N and C termini of TDP-43 were found to be required for the promotion of tau exon 10 inclusion. ALS- and FTLD-U-causing mutations of TDP-43 promoted tau exon 10 inclusion more effectively than the wild-type protein. TDP-43 bound to tau pre-mRNA at six binding sites in intron 9. These results suggested that TDP-43 might be involved in the regulation of tau exon 10 alternative splicing and tau pathogenesis in some tauopathies.
Results
TDP-43 promotes tau exon 10 inclusion
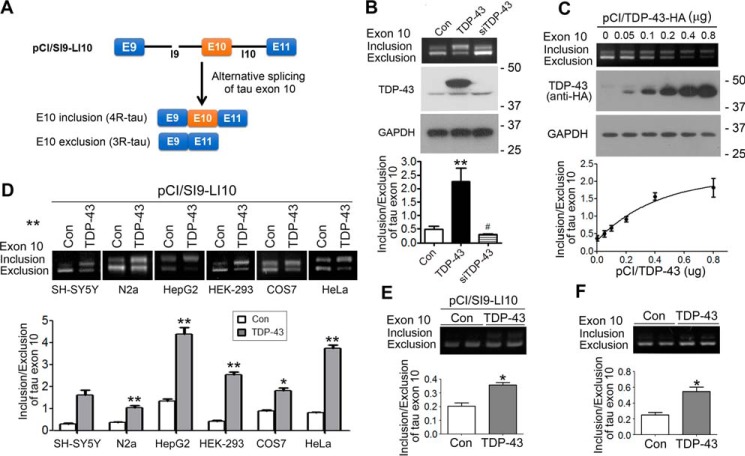
Tau exon 10 encodes the second of the four microtubule-binding repeats. Its alternative splicing generates 4R-tau or 3R-tau. To study the regulation of alternative splicing of tau exon 10 by TDP-43, we used mini-tau gene pCI/SI9-LI10 consisting of tau exons 9, 10, and 11; part of intron 9; and the full length of intron 10 (Fig. 1A) to transfect together with pCI/TDP-43 or siTDP-43 into N2a cells. After transfection for 48 h, we measured the splicing products of tau exon 10 by reverse transcription-PCR (RT-PCR) and found that overexpression of TDP-43 enhanced tau exon 10 inclusion, whereas knockdown of TDP-43 showed the opposite effect (Fig. 1B). To confirm the role of TDP-43 in tau exon 10 splicing, we co-transfected pCI/SI9-LI10 with various amounts of pCI/TDP-43 into HEK-293FT cells. We found that overexpression of TDP-43 promoted tau exon 10 in a dose-dependent manner (Fig. 1C). These results suggest that TDP-43 promotes tau exon 10 inclusion in HEK-293FT cells.
Figure 1.
TDP-43 promotes tau exon 10 inclusion. A, graphic representation of tau mini-gene SI9-LI10. B, TDP-43 was overexpressed or knocked down with siTDP-43 in tau mini-gene (pCI/SI9-LI10)-transfected N2a cells. The splicing products of tau exon 10 were determined by RT-PCR 48 h after transfection, and the level of tau exon 10 inclusion/exclusion was calculated. The expression of TDP-43 was detected by Western blotting. Alteration of TDP-43 affected the alternative splicing of tau exon 10. C, various amounts of pCI/TDP-43·HA were co-transfected with pCI/SI9-LI10 into HEK-293FT cells for 48 h. The alternative splicing products of tau exon 10 were analyzed by RT-PCR. The expression of TDP-43 tagged with HA was detected by Western blotting. The ratio of inclusion/exclusion of tau exon 10 was plotted against the amount of pCI/TDP-43. TDP-43 was found to enhance tau exon 10 inclusion dose-dependently. D, SH-SY5Y, N2a, HepG2, COS7, HEK-293FT, and HeLa cells were co-transfected with pCI/SI9-LI10 and pCI/TDP-43·HA for 48 h. The splicing products of tau exon 10 were determined, and the ratio of tau exon 10 inclusion and exclusion was calculated. TDP-43 was found to promote tau exon 10 inclusion independently of cell types. E and F, primary cortical neurons were co-transfected with pCI/TDP-43 and pCI/SI9-LI10 (E) or transfected with pCI/TDP-43 (F) for 48 h. The splicing products of exogenous (E) and endogenous (F) tau exon 10 were determined by RT-PCR. TDP-43 was found to promote tau exon 10 inclusion in primary cortical neurons. Data are presented as mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; #, p < 0.05; *, versus control; #, versus TDP-43. Con, control.
To examine whether TDP-43 modulates tau exon 10 splicing in different types of cells, we co-transfected pCI/SI9-LI10 with pCI/TDP-43 into SH-SY5Y, N2a, HepG2, HEK-293FT, COS7, and HeLa cells and analyzed the splicing products of tau exon 10 by RT-PCR. We found that overexpression of TDP-43 promoted tau exon 10 inclusion in all types of cells studied (Fig. 1D), confirming that TDP-43 promotes tau exon 10 inclusion in diverse cell types, including neuron-derived cells.
To learn whether TDP-43 regulates splicing of tau exon 10 in neurons, the primary cultured cortical neurons were transfected with pCI/TDP-43 or together with pCI/SI9-LI10 and the alternative splicing products of endogenous or exogenous tau exon 10 were analyzed by RT-PCR. We found that overexpression of TDP-43 promoted both exogenous (Fig. 1E) and endogenous tau exon 10 inclusion significantly (Fig. 1F). These results suggest that TDP-43 regulates tau exon 10 splicing and that overexpression of TDP-43 enhances tau exon 10 inclusion.
TDP-43 binds intron 9 of tau pre-mRNA
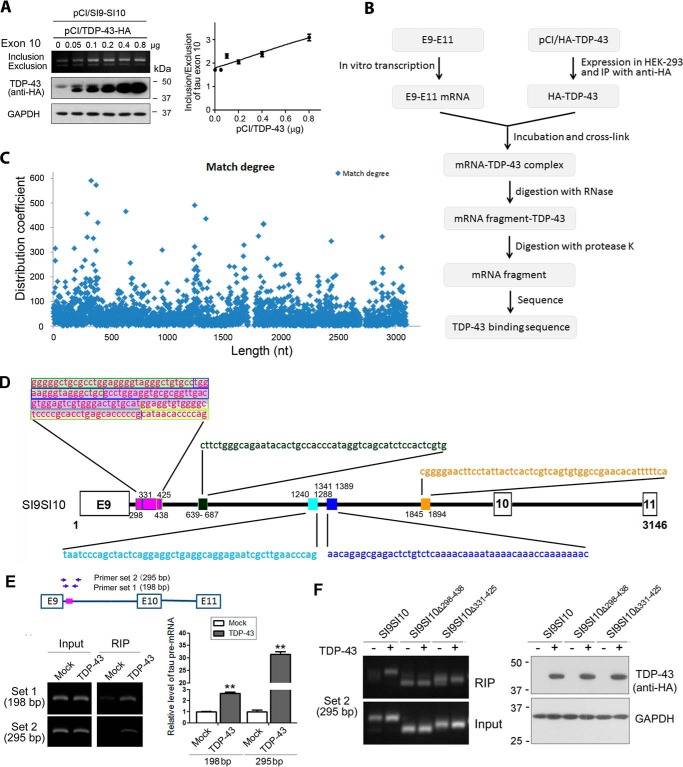
Protein binding could protect DNA or RNA from digestion by DNase or RNase. TDP-43 also promoted tau exon 10 inclusion of mini-tau gene pCI/SI9-SI10 consisting of exons 9, 10, and 11 and partial introns 9 and 10 (Fig. 2A), suggesting that SI9-SI10 contains TDP-43-acting elements. To study the binding of TDP-43 with tau pre-mRNA, we transcribed the linear SI9-SI10 mini-tau gene to pre-mRNA of tau E9–E11 using in vitro transcription system. Meanwhile, we transfected HEK-293FT cells with pCI/TDP-43 and immunoprecipitated TDP-43 with anti-hemagglutinin (HA) but did not elute it from the protein G beads. Then the immunoprecipitated TDP-43 coupled onto protein G beads was incubated with tau E9–E11 pre-mRNA. The binding was cross-linked by UV light. The complex was incubated with RNases and proteinase K sequentially to digest unprotected RNA and proteins, respectively. The pre-mRNA fragments protected by TDP-43 binding from RNase digestion were then subjected to deep sequencing (Fig. 2B). The frequency of pre-mRNA fragments was plotted against the position in the mini-tau gene (Fig. 2C). Using this method, we detected a core binding region located at nucleotides (nt) 298–438. In this region, there were three short binding fragments, nt 298–346, 331–425, and 390–438. Fragment 331–425 was a core binding site, which was flanked by two binding site fragments, nt 298–346 and 390–438 (Fig. 2D). In addition to this core region, we also observed four short binding fragments, nt 639–687, 1240–1288, 1341–1399, and 1845–1894 (Fig. 2D). Interestingly, all these binding sites found here were located in intron 9 (Fig. 2D). These data indicate that TDP-43 may bind intron 9 of tau pre-mRNA, and nt 298–438 may be the core binding region of TDP-43.
Figure 2.
TDP-43 binds intron 9 of tau pre-mRNA. A, various amounts of pCI/TDP-43 were co-transfected with pCI/SI9-SI10 into HEK-293FT cells. The alternative splicing products of tau exon 10 were analyzed by RT-PCR. The expression of TDP-43 tagged with HA was detected by Western blotting. The ratio of inclusion/exclusion of tau exon 10 was plotted against the amount of pCI/TDP-43. B, flow chart of the assay of TDP-43 binding to tau pre-mRNA. C, degree of match of pre-mRNA sequence of SI9-SI10 with TDP-43 in binding assay. SI9-SI10 was transcribed in vitro into pre-mRNA. The pre-mRNA was incubated and UV-cross-linked with immunopurified TDP-43 from HEK-293FT cells. The unbound RNA was then hydrolyzed with RNase, and TDP-43-bound RNA fragments were sequenced. The x axis is the map site value; the y- axis is the distribution coefficient. D, diagram showing the binding sites of the pre-mRNA of tau mini-gene SI9-SI10 with TDP-43 determined from B. A core binding region, nt 298–438, contained a core binding site, nt 331–425, flanked by two binding sites, nt 298–346 and 390–438. There were other binding sites, including nt 639–687, 1240–1288, 1341–1389, and 1845–1894. E, pCI/SI9-LI10 was co-transfected with pCI/TDP-43·HA into HEK-293FT cells. RIP was carried out with anti-HA. Co-immunoprecipitated tau SI9-LI10 pre-mRNA was reverse transcribed into cDNA and amplified with two sets of primers against the core region of tau intron 9 to obtain 198- and 295-bp PCR products. The RT-PCR products were separated by agarose electrophoresis and quantitated by densitometry. The data are presented as mean ± S.D. (error bars) (n = 3). **, p < 0.01. F, pCI/SI9-SI10 or its deletion mutants, pCI/SI9-SI10Δ298–438 and pCI/SI9-SI10Δ331–425, were transfected alone or together with pCI/TDP-43·HA into HEK-293FT cells. RIP was carried out 48 h after transfection. Immunoprecipitated tau SI9-SI10 pre-mRNA was determined by RT-PCR with set 2 primers. The expression of TDP-43 in the input was determined by Western blotting with anti-HA.
To confirm whether TDP-43 binds to tau gene at the core region of intron 9, we performed an RNA immunoprecipitation (RIP) assay. We co-transfected pCI/SI9-LI10 with pCI/TDP-43·HA into HEK-293FT cells, and cells were lysed with RIP lysis buffer on ice for 5 min and then stored at −80 °C. The cell lysates were centrifuged. The TDP-43 was immunoprecipitated with anti-HA. The tau SI9-LI10 pre-mRNA with TDP-43 co-immunoprecipitated by anti-HA was extracted and reverse transcribed with oligo(dT)18 primers. The cDNA was amplified with PCR by two sets of primers against the core region of tau intron 9, resulting in 198- and 295-bp PCR products (Fig. 2E, left panel). We were able to amplify the core binding region of tau pre-mRNA with these two sets of primers from the immunocomplex of TDP-43 (Fig. 2E), suggesting that TDP-43 binds to the core binding region of tau pre-mRNA. Amplification of the core binding region by 295-bp set primers was more specific than by 198-bp set primers (Fig. 2E). These findings suggest that TDP-43 binds to 5′-intron 9 of tau pre-mRNA in cells.
To further confirm the binding of TDP-43 with 5′-intron 9 of tau pre-mRNA, we deleted the TDP-43 core binding region of SI9-SI10 to construct deletion mutants of tau-mini-gene, pCI/SI9-SI10Δ298–438 and pCI/SI9-SI10Δ331–425, and transfected them alone or together with pCI/TDP-43 into HEK-293FT cells. RIP was carried out with anti-HA, and immunoprecipitated pre-mRNA was amplified by RT-PCR with set 2 primers as described above. Consistently, we detected a significant amount of tau pre-mRNA in the immunocomplex with anti-HA from the cells co-transfected with pCI/TDP-43·HA and pCI/SI9-SI10; in contrast, cells transfected with pCI/SI9-SI10 alone showed a minimal level of tau pre-mRNA by RIP with anti-HA, which could be considered as nonspecific RIP (Fig. 2F). Unlike co-transfection of pCI/TDP-43·HA with pCI/SI9-SI10, cells co-transfected with pCI/TDP-43·HA and pC/SI9-SI10Δ298–438 or pCI/SI9-SI10Δ331–425 did not show a detectable increase in the tau pre-mRNA level by RIP with anti-HA when compared with their corresponding control in which cells were transfected with deletion mutations of tau mini-gene alone and in which increased nonspecific RIP was seen (Fig. 2F). Thus, we speculate that deletion of the core region may affect the binding of TDP-43 with the tau SI9-SI10 pre-mRNA.
Both N and C termini of TDP-43 are required for promotion of tau exon 10 inclusion
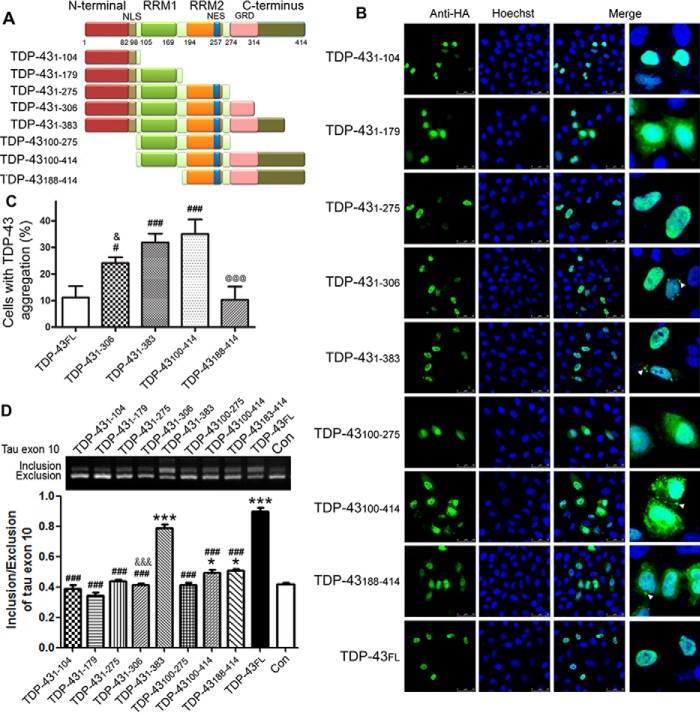
TDP-43 is composed of a nuclear localization signal (NLS), two highly conserved RNA-recognition motifs (RRMs), and a glycine-rich C terminus (Fig. 3A). Its truncation might be associated with its pathogenicity (27). To study the function of different domains in regulating tau exon 10 alternative splicing, we constructed several deletion mutations of TDP-43 (Fig. 3A) and studied their subcellular location in HeLa cells by immunofluorescence staining with anti-HA. We found that TDP-43FL was localized extensively in the nucleus of the cells (Fig. 3B). The N terminus of TDP-43 contains an NLS. Thus, TDP-431–104 was also located in the nucleus (Fig. 3B). Deletion of the first 99 amino acids (aa) in TDP-43100–275 and TDP-43100–414 led to their localization in both the nucleus and cytoplasm (Fig. 3B). Interestingly, we found that TDP-431–179 was localized in both the nucleus and cytoplasm (Fig. 3B), whereas TDP-431–275 and TDP-431–383 were mainly seen in the nucleus. These data suggest that RRM1 and RRM2 may contain a suppressor and an enhancer, respectively, to the nuclear localization of TDP-43.
Figure 3.
The N and C termini of TDP-43 are required for promotion of tau exon 10 inclusion. A, schematic diagram of TDP-43 truncations used. B, the truncation mutants of TDP-43 tagged with HA were expressed in HeLa cells for 48 h and immunostained with monoclonal anti-HA followed by FITC-conjugated goat anti-mouse IgG (green). Hoechst stain (blue) was used to stain nuclei. C, percentage of cells with TDP-43 aggregates in TDP-43-transfected cells. Deletion of the N or C terminus of TDP-43 was found to promote its cytoplasmic aggregation. D, deletion mutants of TDP-43 were co-transfected with pCI/SI9-LI10 into HEK-293FT cells. The splicing products of tau exon 10 were determined. Deletion of either the N or C terminus of TDP-43 was found to abolish its function in promoting tau exon 10 inclusion. The data are presented as mean ± S.D. (error bars) (n = 3). *, p < 0.05; ***, p < 0.001; #, p < 0.05; ###, p < 0.001; &, p < 0.05; &&&, p < 0.001; @@@, p < 0.001; *, versus control; #, versus TDP-43FL; &, versus TDP-431–383; @, versus TDP-43100–414. Scale bar, 50 μm. Con, control; NES, nuclear export signal; GRD, glycine-rich domain.
Aggregates of TDP-43 were identified as the major component of the predominately cytoplasmic inclusions observed in ALS and FTLD-U (1, 2). To study the role of deletion mutations in TDP-43 aggregation, we counted the cells with aggregates resulting from the overexpression of TDP-43 or its deletion mutants. We found that there were aggregates in the cytoplasm in cells with overexpression of TDP-431–306, TDP-431–383, TDP-43100–414, TDP-43188–414, or TDP-43FL but not with TDP-431–104, TDP-431–179, TDP-431–275, and TDP-43100–275 (Fig. 3B). The percentage of cells with TDP-43 aggregates was higher in cells with overexpression of TDP-431–306 or TDP-431–383 than that with TDP-43FL, suggesting that the C terminus of TDP-43 may inhibit self-aggregation. However, we did not find TDP-43 aggregates in the cells expressing TDP-431–275, TDP-431–179, and TDP-431–104 (Fig. 3, B and C). The cells with overexpression of TDP-43100–414 were more affected with aggregates than those with TDP-43FL (Fig. 3, B and C). However, the percentage of TDP-43188–414 cells with aggregates was much less than in TDP-43100–414 cells (Fig. 3, B and C). Thus, aa 100–187 may have a putative aggregation suppression activity. These data suggest that both the N and C termini suppress the aggregation, and removal of the first 99 aa and/or the last 31 aa promotes the aggregation of TDP-43.
To study the role of different domains of TDP-43 in the regulation of tau exon 10 splicing, we co-transfected HEK-293FT cells with pCI/SI9-LI10 and pCI/TDP-43 or its deletion mutants for 48 h and then measured the expression level of splicing products of tau exon 10 by RT-PCR. We found that the full-length TDP-43 promoted tau exon 10 inclusion (Fig. 3D). TDP-431–383 in which the last 31 aa were deleted had similar ability to promote tau exon 10 inclusion as full-length TDP-43. Mutants with further deletion from C terminus, including TDP-431–306, TDP-431–275, TDP-431–179, and TDP-431–104, were unable to promote tau exon 10 inclusion.
N-terminal deletion mutations TDP-43100–414 and TDP-43188–414 promoted tau exon 10 inclusion slightly (Fig. 3D). Compared with TDP-43FL, these two N-terminal deletion mutants showed a very weak effect, but both of them had similar activity in the promotion of tau exon 10 inclusion (Fig. 3D). These findings suggest that the first 99 aa of TDP-43 are also important for the promotion of tau exon 10 inclusion. As expected, deletion of both N- and C-terminal domains in TDP-43100–275 totally abolished its activity in tau exon 10 inclusion (Fig. 3D). These results suggest that the glycine-rich C-terminal 108 aa of TDP-43 are required for its function, and the N terminus containing the NLS enhances the promotion of tau exon 10 inclusion.
ALS and FTLD-U mutations of TDP-43 increase its activity to promote tau exon 10 inclusion
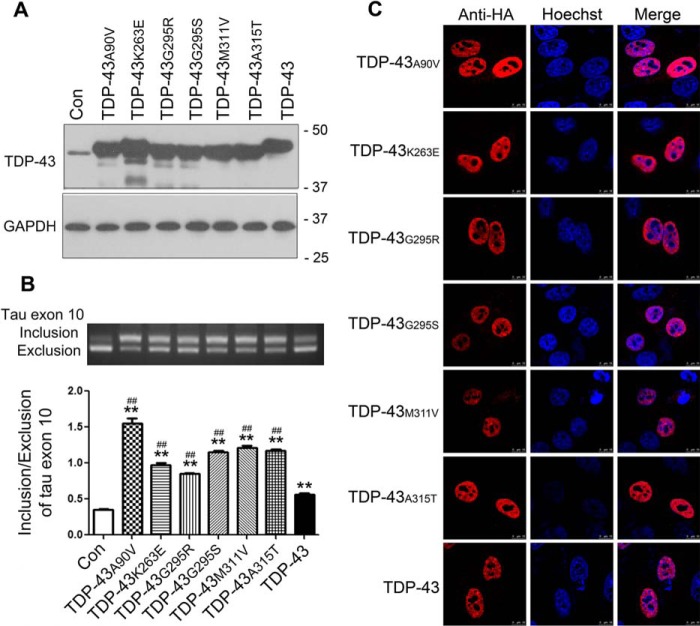
TDP-43 is the major component of neuronal inclusions in ALS and FTLD-U (20), and many disease-causing mutations are located in TDP-43's C-terminal region in individuals with these diseases (22). To study whether the disease-related mutations have any effect on the activity of TDP-43 in the regulation of tau exon 10 splicing, we constructed several ALS/FTLD-U mutations, TDP-43A90V, TDP-43K263E, TDP-43G295R, TDP-43G295S, TDP-43M311V, and TDP-43A315T, and co-expressed them with pCI/SI9-LI10 in HEK-293FT cells. We determined their expressions by Western blotting and measured the splicing products of tau exon 10 by RT-PCR. We found that TDP-43 and these disease mutants all enhanced tau exon 10 inclusion (Fig. 4, A and B). However, the disease-causing mutations promoted tau exon 10 inclusion more effectively than the wild-type TDP-43 (Fig. 4, A and B).
Figure 4.
ALS/FTLD-U mutations of TDP-43 enhance its activity in promotion of tau exon 10 inclusion. A and B, different mutants of pCI/TDP-43·HA were co-transfected with pCI/SI9-LI10 into HEK-293FT for 48 h. The protein levels of TDP-43 and GAPDH were determined by Western blotting (A) and the splicing products of tau exon 10 were detected by RT-PCR (B). ALS/FTLD-U-related TDP-43 mutations were found to enhance tau exon 10 inclusion more effectively. C, ALS/FTLD-U mutants of TDP-43 tagged with HA were overexpressed in HeLa cells for 48 h and immunostained with monoclonal anti-HA followed by TRITC-conjugated goat anti-mouse IgG (red). Hoechst stain (blue) was used to stain nuclei. No cytoplasmic aggregation was found in cells in which ALS/FTLD-U mutations of TDP-43 were overexpressed. The data are represented as mean ± S.D. (error bars). **, p < 0.01 versus control; ##, p < 0.01 versus TDP-43. Scale bar, 50 μm. Con, control.
To learn the subcellular localization of these disease-causing, TDP-43 mutants, we overexpressed them in HeLa cells and then stained cells by immunofluorescence. We found that, like wild-type TDP-43, all these disease-causing mutant proteins were located in the nucleus (Fig. 4C). Different from the several truncations of TDP-43 described above, we did not observe significant differences in the cytoplasmic aggregation of TDP-43 in the cells transfected with wild-type TDP-43 (TDP-43WT) or with the disease-causing mutations (Fig. 4C). These results suggest that the disease-related TDP-43 mutations detected here do not affect subcellular location of TDP-43 in cultured cells but enhance its activity in the promotion of tau exon 10 inclusion.
Increased 3R-tau/4R-tau ratio in TDP-43M337V mouse brains
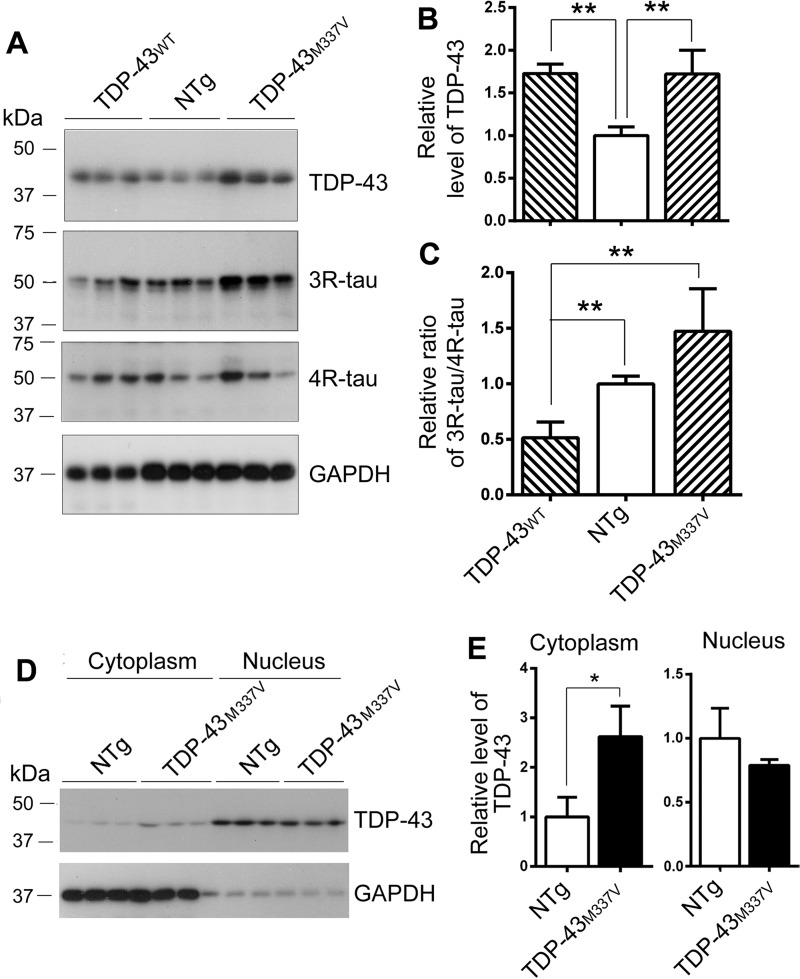
Alternative splicing of tau exon 10 generates 3R-tau and 4R-tau. To study the role of TDP-43 in tau exon 10 splicing in vivo, we measured the levels of 3R-tau and 4R-tau in the brains of non-transgenic mice (NTg) and transgenic mice overexpressing TDP-43WT and an ALS-causing mutation of TDP-43 (TDP-43M337V) by Western blotting. We found that transgenic TDP-43WT and TDP-43M337V mice expressed similar levels of TDP-43 (Fig. 5, A and B). However, compared with NTg mice, the ratio of 3R-tau and 4R-tau was decreased in the brains of TDP-43WT mice and showed a trend of increase in the TDP-43M337V mice. The ratio of 3R-tau and 4R-tau in TDP-43M337V mouse brains was higher than that in the brains of TDP-43WT mice.
Figure 5.
The ratio of 3R-tau and 4R-tau is decreased in TDP-43 and increased in TDP-43M337V transgenic mouse brains. A–C, the levels of TDP-43 and tau in the brain homogenates from hemizygous human TDP-43WT and mutated human TDP-43M337V transgenic mice and the NTg mice were analyzed by Western blotting. Blots were developed with anti-TDP-43 (A260), anti-3R-tau (RD3), anti-4R-tau (RD4), and anti-GAPDH (A). The relative level of TDP-43 was calculated after normalization with GAPDH (B). The relative ratio of 3R-tau and 4R-tau was calculated after normalization with the corresponding loading controls (C). D and E, the brains from TDP-43M337V transgenic mice and NTg mice were homogenized. The cytoplasm and nucleus were separated by centrifugation. TDP-43 and GAPDH were analyzed by Western blotting (D). The levels of TDP-43 in the cytoplasmic and nuclear fractions were normalized with GAPDH (E). The data are represented as mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
In the cultured cells, we found that all ALS/FTLD-U-causing mutations enhanced tau exon 10 inclusion (Fig. 4B). However, we found that the 3R-tau/4R-tau ratio was increased in TDP-43M337V mice. Alternative splicing of pre-mRNA is carried out at the nucleus. It is well known that pathological TDP-43 is dislocated into the cytoplasm from the nucleus (20, 28). Thus, we separated nuclear and cytoplasmic fractions by centrifugation and then analyzed the levels of TDP-43 and GAPDH by Western blotting. We found, as expected, that TDP-43 existed in the nuclei, and GAPDH was in the cytoplasm predominantly (Fig. 5, D and E). The level of TDP-43 in cytoplasm in the TDP-43M337V mice was 2.5-fold higher than that in the NTg mice, and its level in nuclei was apparently lower than that in the NTg mice (Fig. 5E). These results suggest that TDP-43M337V may suppress tau exon 10 inclusion and enhance 3R-tau expression via cytoplasmic localization in vivo.
Discussion
TDP-43 is the major protein identified in the intracellular ubiquitinated inclusions in ALS and FTLD-U (20). In the present study, we found that TDP-43 bound intron 9 of TDP-43 pre-mRNA and promoted tau exon 10 inclusion, leading to an increase of 4R-tau expression in cultured cells. Deletion of the last 108 aa or the first 99 aa significantly decreased or abolished the ability of TDP-43 in promoting tau exon 10 inclusion. Thus, both the N and C termini were required, and the N terminus was beneficial for its function in the regulation of tau exon 10 splicing. ALS/FTLD-U mutations showed stronger activity than wild-type TDP-43 in enhancing tau exon 10 inclusion. The 3R-tau/4R-tau ratio was decreased in the TDP-43WT mouse brains and trended to increase in the TDP-43M337V mouse brains with accompanying dislocation of TDP-43 from the nucleus to the cytoplasm. Thus, disease-associated abnormalities, including truncations and mutations, of TDP-43 promote its cytoplasmic aggregation and depletion from the nucleus, leading to enhancement of tau exon 10 exclusion and 3R-tau expression (Fig. 6).
Figure 6.
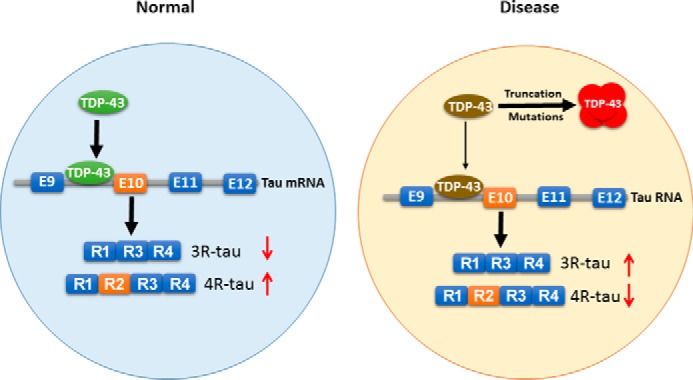
The proposed mechanism by which TDP-43 regulates tau expression. TDP-43 acts on intron 9 and promotes tau exon 10 inclusion in physiological condition. In the disease condition, truncations or mutations lead to its cytoplasmic aggregation and nuclear depletion. Loss of function of TDP-43 (as a result of the nuclear depletion) leads to suppression of tau exon 10 inclusion and 4R-tau expression, which consequently may contribute to the neurofibrillary pathology.
Alternative splicing of tau exon 10 generates 3R-tau and 4R-tau, which are equally expressed in adult human brain (6, 7). Several frontotemporal dementia and parkinsonism linked to chromosome 17 with tau pathology (FTDP-T)-causing mutations do not change the primary structure of the protein but alter the ratio of 3R-tau/4R-tau, suggesting that dysregulation of tau exon 10 splicing is enough to cause a neurodegenerative disease (29–31). Alternative splicing of tau exon 10 is regulated by splicing factors acting on the cis-elements located mainly on exon 10 and intron 10 (13, 32). These splicing factors include serine- and arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins as well as tissue-specific factors (33). We recently found that SR proteins SRp55, 9G8, SC35, and ASF/SF2 regulate the alternative splicing of tau exon 10 (18, 32, 34, 35). In the present study, we found that TDP-43 also regulated tau exon 10 splicing. Overexpression of TDP-43 in cultured cells or in transgenic mice enhanced 4R-tau expression. Importantly, we found that some of the TDP-43 mutations that occur in ALS and FTLD-U made this protein more active in promoting tau exon 10 inclusion in cultured cells.
TDP-43 is a DNA- and RNA-binding protein that regulates RNA metabolism, including transcription, alternative splicing, and stability of RNA. It was previously reported that TDP-43 inhibits the splicing of cystic fibrosis transmembrane conductance regulator exon 9 (36), apolipoprotein A-II exon 3 (37), and eukaryotic translation termination factor 1 (38) and enhances the splicing of breast cancer gene 1 and DNA polymerase-interacting protein 3 (38, 39). The present study adds tau as one more target gene whose alternative splicing is regulated by TDP-43. Considering that alteration of the tau 3R-tau/4R-tau balance can cause neurodegeneration (40, 41), the role of TDP-43 in regulating tau exon 10 splicing found in the present study is highly significant.
TDP-43 contains two RNA-recognition motifs, the function of which is involved in RNA processing and stabilization (42). The glycine-rich C terminus may drive a toxic gain of function (42). Using various N-terminal and/or C-terminal deletion mutants of TDP-43, we found that deletion of the first 99 aa significantly reduced whereas deletion of the last 108 aa totally abolished its effect in the promotion of tau exon 10 inclusion. Thus, both the N and C termini of TDP-43 are required for its activity to enhance tau exon 10 inclusion. The N-terminal domain of TDP-43 contains an NLS, aa 82–98, and the RNA-splicing machinery is located in the nucleus. Therefore, deletion of the first 99 aa promoted TDP-43 cytoplasmic translocation, leading to nuclear depletion and a decrease in its activity. Further deletion of 88 aa in TDP-43188–414 did not affect its function. However, deletion of N-terminal domains plus two RRMs abolished the function. Thus, RRMs are required for TDP-43 to enhance tau exon 10 inclusion. Interestingly, deletion of the last 31 aa did not affect TDP-43-promoted tau exon 10 inclusion, suggesting that the last 31 aa may not be important in the regulation of tau exon 10 splicing. However, TDP-431–306 in which the last 108 aa were deleted lost its function in the regulation of tau exon 10 splicing.
In addition to truncation, pathological TDP-43 is ubiquitinated, hyperphosphorylated, oxidized, and/or acetylated (43–46). Cysteine oxidation and lysine acetylation of TDP-43 at RRM domains impair its RNA-binding and -splicing function in cystic fibrosis transmembrane conductance regulator (43, 44). Here, we found that either N-terminal or C-terminal truncation of TDP-43 impaired its function in tau exon 10 inclusion. The effect of the above posttranslational modifications on the function of TDP-43 in tau exon 10 splicing, however, remains unclear.
TDP-43 pathology has been observed in 57% of AD cases and 87% of chronic traumatic encephalopathy cases. We recently found that the TDP-43 level was reduced in AD brain (58). However, no correlation of TDP-43 expression with the dysregulated tau exon 10 splicing was observed in AD brain (47). Tau exon 10 splicing is regulated by multiple factors and their phosphorylation (48). Up-regulation of Dyrk1A resulting from overexpression in Down syndrome or truncation by activated calpain I in AD brain leads to increase of phosphorylation of splicing factors, such as ASF, SC35, and SRp55 and suppresses tau exon 10 inclusion (17, 32, 34). TDP-43 is a phosphoprotein. Its function and subcellular location are regulated by phosphorylation. Mutation, truncation, and phosphorylation promote TDP-43 cytoplasmic aggregation, resulting in its depletion from the nucleus. CK1 phosphorylates TDP-43 and affects its function (49). In AD brain, CK1 is overexpressed (50). However, whether the overexpression of CK1 may affect the function of TDP-43 in tau exon 10 splicing and consequently contribute to tau pathology in AD brain remains to be determined.
TDP-43 is the major component of ubiquitinated inclusions in most cases of ALS and FTLD-U (20, 21), and multiple mutations in TARDBP have been discovered (51, 52). It is interesting to note that most of the TDP-43 mutations found in ALS and FTLD-U cases are located in the glycine-rich domain (20, 21). A previous study found that ALS/FTLD-U-causing mutations D169G, G294A, Q331K, M337V, Q343R, N390D, and N390S significantly increase cell numbers with TDP-43162–414 aggregation in SH-SY5Y cells. Other ALS/FTLD-U-causing mutations G287S, G290A, G298S, A315T, G348C, R361S, and A382T do not significantly affect TDP-43162–414 aggregation (53). In the present study, we did not find a significant difference in the intracellular aggregation between wild-type TDP-43 and the ALS/FTLD-U-causing TDP-43 mutations, which may be due to different cell types, different mutations, and different variants of TDP-43. They used TDP-43162–414, but we used TDP-431–414.
Through expression of the disease-causing mutants in mice and genome-wide RNA splicing analyses, mutant TDP-43Q331K has been shown to retain normal or enhanced activity for facilitating splicing of some RNA targets but “loss of function” for others in cortices (54). All the mutations tested in the present study enhanced the activity of TDP-43 to promote tau exon 10 inclusion in cultured cells. Moreover, we did not observe any significant cytosolic aggregation in these TDP-43 mutants. In the present study, we found a decrease of 3R-tau/4R-tau ratio in the TDP-43WT transgenic mouse brains, suggesting that TDP-43 promotes tau exon 10 inclusion. However, we found that the 3R-tau/4R-tau ratio was increased in the TDP-43M337V mouse brains, and this was accompanied by an increase of cytoplasmic TDP-43 and a decrease of nuclear TDP-43. Unfortunately, we did not determine the role of TDP-43M337V in tau exon 10 splicing in cultured cells. However, all TDP-43 mutants detected in the present study enhanced tau exon 10 inclusion without a significantly cytoplasmic aggregation. Thus, we speculate that overexpression of TDP-43M337V may also enhance tau exon 10 inclusion due to lack of cytoplasmic aggregation, which remains to be determined. TDP-43M337V, an ALS mutation of TDP-43, may suppress tau exon 10 inclusion and promote 3R-tau expression through depletion of TDP-43 from the nucleus in vivo. Therefore, these mutations might contribute to neurodegeneration through disturbing the normal 1:1 ratio of 3R-tau/4R-tau in these ALS and FTLD-U cases. However, to date, no such study has shown that tau exon 10 is dysregulated in the individuals with TDP-43 mutation in vivo.
In addition, we found cytoplasmic TDP-43 aggregation in cells with overexpression of some TDP-43 deletion mutants. TDP-43100–414 showed the highest cytoplasmic aggregation rates, suggesting that the N terminus may contain antiaggregation activity. It has been reported that pathological truncations of TDP-43, 35-kDa TDP-4391–414 and 25-kDa TDP-43220–414, promote its self-aggregation (55). Cytoplasmic aggregation of TDP-43 increases dramatically in SH-SY5Y cells overexpressing TDP-43162–414, TDP-43218–414, or TDP-431–161 but not TDP-43274–414, TDP-431–217, TDP-431–273, or TDP-431–314 (53). In the present study, we found that aggregation of TDP-43 was dramatically increased in the cells overexpressing TDP-43100–414 but not TDP-43188–414, suggesting that RRM1 may contain a potent aggregation domain. Similarly, deletion of the last 31 aa at the C terminus increased TDP-43 self-aggregation, and deletion of a further 77 aa reduced its self-aggregation. Thus, the last 31 aa serve as the strong antiaggregation domain, and aa 307–383 may be a potent aggregation domain.
In conclusion, we found that TDP-43 regulated tau exon 10 splicing by promoting its inclusion. TDP-43 could bind intron 9 of tau pre-mRNA. Both the N and C termini of TDP-43 were required for its function in promotion of tau exon 10 inclusion. Several ALS/FTLD-U-causing mutations of TDP-43 promoted tau exon 10 inclusion effectively. Thus, nuclear depletion of TDP-43 resulting from cytoplasmic aggregation in diseases dysregulates the alternative splicing of tau exon 10 and consequently alters the ratio of 3R-tau/4R-tau. These findings provide a mechanistic linkage between TDP-43 pathology and tau pathology that co-exist in AD and some other tauopathies and shed new light on how TDP-43 abnormality may cause neurodegeneration.
Experimental procedures
Plasmids, antibodies, and other reagents
pCI/TDP-43 tagged with HA was generated by PCR amplification from pCAG/TDP-43 (Origene). ALS/FTLD-U mutations of TDP-43, including TDP-43A90V, TDP-43K263E, TDP-43G295R, TDP-43G295S, TDP-43M311V, and TDP-43A315T, were generated using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). The deletion mutations of TDP-43, TDP-431–104, TDP-431–179, TDP-431–275, TDP-431–306, TDP-431–383, TDP-43100–275, TDP-43100–414, and TDP-43188–414, tagged with HA were amplified using pCI/TDP-43 as a template and constructed into pCI-neo. All constructs were confirmed by DNA sequence analysis. pCI/SI9-LI10 containing a tau minigene, SI9-LI10, comprising tau exons 9, 10, and 11; part of intron 9; and the full length of intron 10, and pCI/SI9-SI10 containing tau exons 9, 10, and 11 and part of introns 9 and 10 were a gift from Dr. Jianhua Zhou of the University of Massachusetts Medical School (34, 35, 56). The deletion mutants of tau mini-gene, pCI/SI9-SI10Δ298–438 and pCI/SI9-SI10Δ331–425, were constructed by using pCI/SI9-SI10 as template and verified by DNA sequencing. Monoclonal anti-HA (1:20,000) was obtained from Sigma (catalog number H9658, lot number 112M4841). Polyclonal anti-TDP-43 (1:1000) (A260, catalog number 3449S, lot number 1) was from Cell Signaling Technology. siRNA of mouse TDP-43 (a pool of three siRNAs, including sc-154072A (sense, GUGUUUGUUGGACGUUGUAtt; antisense, UACAACGUCCAACAAACACtt), sc-154072B (sense, CCUAAUGCCCUUACCUAAAtt; antisense, UUUAGGUAAGGGCAUUAGGtt), and sc-154072C (sense, GUUAGUUACUUGCCGUAAAtt; antisense, UUUACGGCAAGUAACUAACtt), and anti-GAPDH (1:2000) (catalog number sc-25778, lot number D3015) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-3R-tau (1:2000) (RD3, catalog number 05-803, lot number JBC1863429) and mouse monoclonal anti-4R-tau (1:500) (RD4, catalog number 05-804, lot number 2073108) were purchased from Millipore (Temecula, CA). Peroxidase-conjugated anti-mouse and anti-rabbit IgGs were obtained from Jackson ImmunoResearch Laboratories (1:5000) (West Grove, PA). Fluorescein isothiocyanate (FITC)- or tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse IgG and Hoechst 33342 were from Invitrogen. The ECL kit was from Thermo Fisher Scientific (Rockford, IL).
Animals
Non-transgenic C57BL/6 mice (NTg mice), C57BL/6-Tg(PrnpTARDBP)3cPtrc/J (hTDP-43 transgenic mice, stock number 016608), and C57BL/6-Tg(Prnp-TARDBP*M337V)4Ptrc/J (hTDP-43M337V transgenic mice, stock number 017604) were purchased from The Jackson Laboratory and maintained at Case Western Reserve University. All mice were weaned on postnatal day 30 and genotyped by PCR analysis of DNA extracted from a punched ear. The animals were housed in cages at 24 ± 1 °C at 50–60% humidity.
Cell culture and transfection
HepG2, COS7, HEK-293FT, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) at 37 °C (5% CO2). N2a and SH-SY5Y cells were maintained in DMEM/F-12 medium with 10% FBS. All transfections were performed with Lipofectamine 2000 (Invitrogen), Lipofectamine LTX (Invitrogen), or FuGENE HD (Roche Diagnostics) according to the manufacturers' instructions.
Primary culture of cortical neurons and transfection
Isolation and culture of mouse cerebral cortical neurons were conducted in accordance with a previous method (57). The brains were obtained aseptically from fetal mice after craniotomy. A median sagittal incision was made, and then the brain was placed in a Petri dish with precooled Dulbecco's Hanks' buffer. The Petri dish was placed on ice. After removal of the cerebellum and subcortical tissue, meninges and blood vessels were dissociated, and the cerebral cortex was cut into pieces, digested in 0.25% trypsin at 37 °C for 15 min, and agitated into a cell suspension. Following filtration and centrifugation at 1000 rpm for 5 min, the pellet was resuspended with Neurobasal medium supplemented with 2% B27 (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were adjusted to about 1 × 106/ml and seeded in a 24-well plate precoated with poly-d-lysine (Sigma) at 37 °C at 5% CO2. On the 2nd day, the cells were treated with 10 μm Ara-C for 24 h to inhibit non-neuronal cells. The medium was replaced once every 2 days. Transfections were performed with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions.
Knockdown of TDP-43 with RNA interference
For inhibition of TDP-43 expression, N2a cells were transfected with short interfering RNAs of mouse TDP-43 (siTDP-43) (Santa Cruz Biotechnology) using Lipofectamine 2000. After 48-h transfection, the alternative splicing products of tau exon 10 and TDP-43 expression were analyzed. The same amount of scrambled siRNA was used as a control.
Quantitation of tau exon 10 splicing by RT-PCR
Total cellular RNA was extracted from cultured cells using the RNeasy® mini kit (Qiagen, Valencia, CA) according to the manufacturer's instruction. One microgram of total RNA was used for first-strand cDNA synthesis with oligo(dT)18 using the Omniscript reverse transcription kit (Invitrogen). PCR was performed using Prime STARTM HS DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) with forward primer 5′-GGTGTCCACTCCCAGTTCAA-3′ and reverse primer 5′-CCTGGTTTATGATGGATGTTGCCTAATGAG-3′ to measure alternative splicing of tau exon 10 of mini-tau gene and with forward primer 5′-AACACCGCCACCCGGGAG-3′ and reverse primer 5′-GTCTGTCTTGGCTTTGGCATTCTC-3′ for the measurement of the alternative splicing of mouse tau exon 10. The PCR conditions were: 98 °C for 5 min, 30 cycles of 98 °C for 10 s and 68 °C for 40 s, and then 68 °C for 10 min for extension. The PCR products were resolved on 1.5% agarose gels, visualized by ethidium bromide staining, and quantitated using the Molecular Imager system (Bio-Rad).
Preparation of cytoplasmic and nuclear fractionations
Mouse brain tissue was homogenized in 9 volumes of cold buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10 mm β-mercaptoethanol, 2.0 mm EDTA, 1.0 mm Na3VO4, 50 mm NaF, 1.0 mm AEBSF, 10 μg/ml each of aprotinin, leupeptin, and pepstatin). The brain homogenates were centrifuged at 900 × g for 5 min, and the pellets were homogenized and centrifuged again as described above. The two supernatants were pooled together as the cytoplasm fraction, and the pellet was the nuclear fraction.
Western blotting
Cultured cells were lysed in 1× Laemmli sample buffer (125 mm Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate (SDS), 10% glycerol, 2% 2-mercaptoethanol, 0.004% bromphenol blue) and boiled for 5 min. The protein concentration of cell lysates was measured using the PierceTM 660nm Protein Assay kit (Thermo Fisher Scientific). Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrically blotted onto PVDF membrane. The membrane was blocked with 5% fat-free milk in TBS (50 mm Tris-HCl, pH 7.4, 150 mm NaCl) for 30 min and then incubated with primary antibody in 5% fat-free milk and 0.5% NaN3 in TBS overnight at room temperature. After washing three times with TBST (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.05% Tween 20), the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. Following three washes with TBST, the membrane was incubated with ECL for 1 min and then exposed to HyBlot CL® autoradiography film (Denville Scientific, Inc., Holliston, MA).
Immunofluorescence staining
HeLa cells were plated onto coverslips in 24-well plates 1 day prior to transfection. Cell transfection with pCI/TDP-43 or its mutants tagged with HA at the N terminus was carried out as described above. Two days after transfection, the cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature; blocked with 10% goat serum in 0.2% Triton X-100, PBS for 2 h at 37 °C; and incubated with monoclonal anti-HA (1:1000) overnight at 4 °C. After incubation with secondary antibodies (FITC- or TRITC-conjugated goat anti-mouse IgG; 1:500), the cells were incubated with 5 μg/ml Hoechst 33342 for 15 min at room temperature and mounted with Fluoromount-G. The immunofluorescence was revealed with a Leica TCS SP2 laser-scanning confocal microscope.
Detection of the binding region of tau mRNA with TDP-43
For TDP-43 immunopurification, HEK-293FT cells were transfected with pCI/TDP-43·HA using FuGENE HD for 48 h and lysed by sonication in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.2% sodium deoxycholate, 50 mm NaF, 1 mm Na3VO4, 2 mm EDTA, 1 mm AEBSF, 10 μg/ml each of aprotinin, leupeptin, and pepstatin). The cell lysate was centrifuged at 15,000 × g for 5 min at 4 °C. The extract was incubated with protein G beads precoupled with monoclonal anti-HA overnight at 4 °C. The beads were extensively washed with TBS and suspended in RNase-free TBS.
For DNA template preparation, we first cleaned the RNase from pCI/SI9-SI10 plasmid by digestion with proteinase K (100 μg/ml; Sigma) in 50 mm Tris-Cl, pH 7.5, 5 mm CaCl2, 0.5% SDS for 1 h at 37 °C. The plasmid DNA was extracted with Tris-EDTA-saturated, pH 8.0, phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated with ethanol. The pCI/SI9-SI10 plasmid was linearized with restriction endonuclease NdeI (20 units/μl; New England Biolabs) overnight at 37 °C and extracted with Wizard SV Gel and PCR Clean-Up System (Promega). The linearized pCI/SI9-SI10 was used as the template for pre-mRNA synthesis in vitro.
In the pCI-neo vector, T7 and T3 RNA polymerase promoters flank the multiple cloning region. Therefore, the pre-mRNA of tau SI9-SI10 was synthesized in vitro with the RiboMAXTM Large Scale RNA Production System-T7 transcription kit (Promega) in a total volume of 100 μl (20 μl of T7 transcription 5× buffer, 30 μl of rNTPs, 30 μl of linear pCI/SI9-SI10 DNA template, and 10 μl of T7 enzyme mixture containing RNA polymerase, recombinant RNasin® ribonuclease inhibitor, and recombinant inorganic pyrophosphatase) for 4 h at 37 °C. RQ1 RNase-free DNase (1 unit/μl; Promega) was added to remove DNA template. The pre-mRNA was extracted withtk;1 Tris-EDTA-saturated, pH 8.0, phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated with ethanol.
The pre-mRNA of tau SI9-SI10 was added into the suspension of TDP-43-anti-HA beads and incubated for 2 h at room temperature. Then the complex of TDP-43 and the pre-mRNA was UV-cross-linked with the Strata-linker 2400 (Stratagene) at 400 mJ/cm2 for 30 min. After washing with TBS, RNase T1 (1000 units/μl; Thermo Scientific) and RNase A (10 μg/μl; Thermo Scientific) were added and incubated for 15 min at 37 °C to remove the RNA unprotected by TDP-43. After washing with TBS, proteinase K (100 μg/ml, Sigma) in 50 mm Tris-Cl, pH 7.5, 5 mm CaCl2, 0.5% SDS was added and incubated for 45 min at 37 °C to digest the TDP-43. The pre-mRNA complex was purified with MicroSpin G-25 columns (GE Healthcare), lyophilized, and sequenced by the Beijing Genomics Institute (Beijing, China).
RNA immunoprecipitation
The RNA immunoprecipitation experiment was performed with the Magna RIPTM kit (Millipore). Briefly, HEK-293FT cells were co-transfected with pCI/TDP-43·HA and pCI/SI9-SI10 or pCI/SI9-SI10 and its deletion mutants for 48 h. The cells were lysed in complete RIP lysis buffer (RIP lysis buffer, protease inhibitor mixture, 0.1 unit/μl RNase inhibitor) on ice for 5 min and frozen at −80 °C. The cell lysates were thawed quickly and centrifuged at 14,000 rpm for 10 min at 4 °C.
The magnetic beads were washed with 0.5 ml of RIP wash buffer, suspended in 100 μl of RIP wash buffer, and incubated with monoclonal anti-HA for 4 h at 4 °C. The beads were washed three times with RIP wash buffer. The anti-HA-binding beads were resuspended in 900 μl of RIP immunoprecipitation buffer (860 μl of RIP wash buffer, 35 μl of 0.5 m EDTA, 5 μl of RNase inhibitor), and 100 μl of the cell extract was added. An aliquot of the extract was saved as “input.” After overnight incubation at 4 °C, the beads were washed six times with ice-cold RIP wash buffer and digested with proteinase K at 55 °C for 30 min in 150 μl of proteinase K buffer (117 μl of RIP wash buffer, 15 μl of 10% SDS, 18 μl of 10 mg/ml proteinase K). The proteins in the input sample were digested in parallel.
The RNA in supernatants was extracted with RNeasy® Plus mini kit (Qiagen) according to the manufacturer's instruction. The same amount of RNA was subjected to first-strand cDNA synthesis using the Omniscript reverse transcription kit (Invitrogen) with oligo(dT)18 primers. The cDNA was amplified by PrimeSTARTM HS DNA polymerase (Takara Bio Inc.) with two sets of primers against tau intron 9: set 1 (forward, 5′-GAGAGTGGCTGGCTGCGCGTGGAGGTGTGG-3′; reverse, 5′-ACTTCCTTGAAGAGGGTCCAGAGGGGACTGG-3′) and set 2 (forward, 5′-ATGTCAAGTCCAAGATCGGCTCCACTGAGAA-3′; reverse, 5′-CACTCACAGTGTAGTGGAGAGCCCAATAAAG-3′). The PCR conditions were: 98 °C for 5 min, 98 °C for 10 s and 68 °C for 45 s for 30 cycles, and then 68 °C for 10 min for extension. The PCR products were resolved in 2% agarose gels and visualized by ethidium bromide staining.
Statistical analysis
The data were presented as the mean ± S.D. Data points were compared by the unpaired two-tailed Student's t test for two-group comparison and by one-way analysis of variance for multiple-group comparison. A value of p < 0.05 was considered statistically significantly.
Author contributions
J. G. performed experiments, analyzed results, and wrote the first draft of the manuscript and figures. F. C. performed experiments. X. W. provided the brain samples of TDP-43 transgenic mice. C.-X. G., K. I., and X. W. provided the reagents, discussed results, and edited manuscript. F. L. designed experiments, analyzed and interpreted results, and wrote the manuscript.
This work was supported by Nantong University, the New York State Office for People With Developmental Disabilities, National Natural Science Foundation of China Grants 81030059 and 31671046, Natural Science Foundation from Jiangsu Education Department Grant 16KJB310013, a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD), and United States Alzheimer's Association Grant DSAD-15-363172. The authors declare that they have no conflicts of interest with the contents of this article.
- AD
- Alzheimer's disease
- TDP-43
- transactive response DNA-binding protein of 43 kDa
- ALS
- amyotrophic lateral sclerosis
- FTLD-U
- frontotemporal lobar degeneration with ubiquitin inclusions
- E
- exon
- I
- intron
- nt
- nucleotides
- RIP
- RNA immunoprecipitation
- NLS
- nuclear localization signal
- RRM
- RNA-recognition motif
- aa
- amino acids
- FL
- full length
- NTg
- non-transgenic mice
- SR
- serine- and arginine-rich
- ASF
- alternative splicing factor
- TARDBP
- TAR DNA-binding protein
- TRITC
- tetramethylrhodamine isothiocyanate
- Ara-C
- arabinocytidine
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride.
References
- 1. Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., and Wisniewski H. M. (1986) Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089 [PubMed] [Google Scholar]
- 2. Ballatore C., Lee V. M., and Trojanowski J. Q. (2007) Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 [DOI] [PubMed] [Google Scholar]
- 3. Montejo de Garcini E., Díez J. C., and Avila J. (1986) Quantitation and characterization of tau factor in porcine tissues. Biochim. Biophys. Acta 881, 456–461 [DOI] [PubMed] [Google Scholar]
- 4. Alonso A., Zaidi T., Novak M., Grundke-Iqbal I., and Iqbal K. (2001) Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andreadis A., Brown W. M., and Kosik K. S. (1992) Structure and novel exons of the human tau gene. Biochemistry 31, 10626–10633 [DOI] [PubMed] [Google Scholar]
- 6. Goedert M., Spillantini M. G., Jakes R., Rutherford D., and Crowther R. A. (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3, 519–526 [DOI] [PubMed] [Google Scholar]
- 7. Kosik K. S., Kowall N. W., and McKee A. (1989) Along the way to a neurofibrillary tangle: a look at the structure of tau. Ann. Med. 21, 109–112 [DOI] [PubMed] [Google Scholar]
- 8. Bronner I. F., ter Meulen B. C., Azmani A., Severijnen L. A., Willemsen R., Kamphorst W., Ravid R., Heutink P., and van Swieten J. C. (2005) Hereditary Pick's disease with the G272V tau mutation shows predominant three-repeat tau pathology. Brain 128, 2645–2653 [DOI] [PubMed] [Google Scholar]
- 9. Neumann M., Schulz-Schaeffer W., Crowther R. A., Smith M. J., Spillantini M. G., Goedert M., and Kretzschmar H. A. (2001) Pick's disease associated with the novel tau gene mutation K369I. Ann. Neurol. 50, 503–513 [DOI] [PubMed] [Google Scholar]
- 10. Yoshida M. (2006) Cellular tau pathology and immunohistochemical study of tau isoforms in sporadic tauopathies. Neuropathology 26, 457–470 [DOI] [PubMed] [Google Scholar]
- 11. Pickering-Brown S., Baker M., Yen S. H., Liu W. K., Hasegawa M., Cairns N., Lantos P. L., Rossor M., Iwatsubo T., Davies Y., Allsop D., Furlong R., Owen F., Hardy J., Mann D., et al. (2000) Pick's disease is associated with mutations in the tau gene. Ann. Neurol. 48, 859–867 [PubMed] [Google Scholar]
- 12. Gao L., Wang J., Wang Y., and Andreadis A. (2007) SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations. Mol. Cell. Neurosci. 34, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartmann A. M., Rujescu D., Giannakouros T., Nikolakaki E., Goedert M., Mandelkow E. M., Gao Q. S., Andreadis A., and Stamm S. (2001) Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol. Cell. Neurosci. 18, 80–90 [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Gao Q. S., Wang Y., Lafyatis R., Stamm S., and Andreadis A. (2004) Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J. Neurochem. 88, 1078–1090 [DOI] [PubMed] [Google Scholar]
- 15. Wang Y., Wang J., Gao L., Lafyatis R., Stamm S., and Andreadis A. (2005) Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c·SRp55 complex that either recruits or antagonizes htra2β1. J. Biol. Chem. 280, 14230–14239 [DOI] [PubMed] [Google Scholar]
- 16. D'Souza I., and Schellenberg G. D. (2006) Arginine/serine-rich protein interaction domain-dependent modulation of a tau exon 10 splicing enhancer: altered interactions and mechanisms for functionally antagonistic FTDP-17 mutations Δ280K and N279K. J. Biol. Chem. 281, 2460–2469 [DOI] [PubMed] [Google Scholar]
- 17. Yin X., Jin N., Gu J., Shi J., Zhou J., Gong C. X., Iqbal K., Grundke-Iqbal I., and Liu F. (2012) Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 287, 30497–30506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu J., Shi J., Wu S., Jin N., Qian W., Zhou J., Iqbal I. G., Iqbal K., Gong C. X., and Liu F. (2012) Cyclic AMP-dependent protein kinase regulates 9G8-mediated alternative splicing of tau exon 10. FEBS Lett. 586, 2239–2244 [DOI] [PubMed] [Google Scholar]
- 19. Buratti E., and Baralle F. E. (2012) TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 37, 237–247 [DOI] [PubMed] [Google Scholar]
- 20. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 21. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., and Oda T. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 22. Lagier-Tourenne C., Polymenidou M., and Cleveland D. W. (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higashi S., Iseki E., Yamamoto R., Minegishi M., Hino H., Fujisawa K., Togo T., Katsuse O., Uchikado H., Furukawa Y., Kosaka K., and Arai H. (2007) Concurrence of TDP-43, tau and α-synuclein pathology in brains of Alzheimer's disease and dementia with Lewy bodies. Brain Res. 1184, 284–294 [DOI] [PubMed] [Google Scholar]
- 24. Uryu K., Nakashima-Yasuda H., Forman M. S., Kwong L. K., Clark C. M., Grossman M., Miller B. L., Kretzschmar H. A., Lee V. M., Trojanowski J. Q., and Neumann M. (2008) Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol. 67, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Josephs K. A., Murray M. E., Whitwell J. L., Parisi J. E., Petrucelli L., Jack C. R., Petersen R. C., and Dickson D. W. (2014) Staging TDP-43 pathology in Alzheimer's disease. Acta Neuropathol. 127, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McKee A. C., Stern R. A., Nowinski C. J., Stein T. D., Alvarez V. E., Daneshvar D. H., Lee H. S., Wojtowicz S. M., Hall G., Baugh C. M., Riley D. O., Kubilus C. A., Cormier K. A., Jacobs M. A., Martin B. R., et al. (2013) The spectrum of disease in chronic traumatic encephalopathy. Brain 136, 43–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregory J. M., Barros T. P., Meehan S., Dobson C. M., and Luheshi L. M. (2012) The aggregation and neurotoxicity of TDP-43 and its ALS-associated 25 kDa fragment are differentially affected by molecular chaperones in Drosophila. PLoS One 7, e31899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cairns N. J., Neumann M., Bigio E. H., Holm I. E., Troost D., Hatanpaa K. J., Foong C., White C. L. 3rd, Schneider J. A., Kretzschmar H. A., Carter D., Taylor-Reinwald L., Paulsmeyer K., Strider J., Gitcho M., et al. (2007) TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am. J. Pathol. 171, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Umeda T., Yamashita T., Kimura T., Ohnishi K., Takuma H., Ozeki T., Takashima A., Tomiyama T., and Mori H. (2013) Neurodegenerative disorder FTDP-17-related tau intron 10 +16C→T mutation increases tau exon 10 splicing and causes tauopathy in transgenic mice. Am. J. Pathol. 183, 211–225 [DOI] [PubMed] [Google Scholar]
- 30. Yen S. H., Hutton M., DeTure M., Ko L. W., and Nacharaju P. (1999) Fibrillogenesis of tau: insights from tau missense mutations in FTDP-17. Brain Pathol. 9, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ingram E. M., and Spillantini M. G. (2002) Tau gene mutations: dissecting the pathogenesis of FTDP-17. Trends Mol. Med. 8, 555–562 [DOI] [PubMed] [Google Scholar]
- 32. Qian W., Liang H., Shi J., Jin N., Grundke-Iqbal I., Iqbal K., Gong C. X., and Liu F. (2011) Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic Acids Res. 39, 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manley J. L., and Krainer A. R. (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi J., Qian W., Yin X., Iqbal K., Grundke-Iqbal I., Gu X., Ding F., Gong C. X., and Liu F. (2011) Cyclic AMP-dependent protein kinase regulates the alternative splicing of tau exon 10: a mechanism involved in tau pathology of Alzheimer disease. J. Biol. Chem. 286, 14639–14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding S., Shi J., Qian W., Iqbal K., Grundke-Iqbal I., Gong C. X., and Liu F. (2012) Regulation of alternative splicing of tau exon 10 by 9G8 and Dyrk1A. Neurobiol. Aging 33, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., and Baralle F. E. (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mercado P. A., Ayala Y. M., Romano M., Buratti E., and Baralle F. E. (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Passoni M., De Conti L., Baralle M., and Buratti E. (2010) UG repeats/TDP-43 interactions near 5′ splice sites exert unpredictable effects on splicing modulation. J. Mol. Biol. 415, 46–60 [DOI] [PubMed] [Google Scholar]
- 39. Fiesel F. C., Weber S. S., Supper J., Zell A., and Kahle P. J. (2012) TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 40, 2668–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gendron T. F., and Petrucelli L. (2009) The role of tau in neurodegeneration. Mol. Neurodegener. 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Panda D., Samuel J. C., Massie M., Feinstein S. C., and Wilson L. (2003) Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc. Natl. Acad. Sci. U.S.A. 100, 9548–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buratti E., and Baralle F. E. (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 43. Cohen T. J., Hwang A. W., Unger T., Trojanowski J. Q., and Lee V. M. (2012) Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 31, 1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen T. J., Hwang A. W., Restrepo C. R., Yuan C. X., Trojanowski J. Q., and Lee V. M. (2015) An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., and Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., and Lee V. M. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niblock M., Hortobágyi T., Troakes C., Al-Sarraj S., Spickett C., Jones R., Shaw C. E., and Gallo J. M. (2016) Lack of association between TDP-43 pathology and tau mis-splicing in Alzheimer's disease. Neurobiol. Aging 37, 45–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qian W., and Liu F. (2014) Regulation of alternative splicing of tau exon 10. Neurosci. Bull. 30, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kametani F., Nonaka T., Suzuki T., Arai T., Dohmae N., Akiyama H., and Hasegawa M. (2009) Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem. Biophys. Res. Commun. 382, 405–409 [DOI] [PubMed] [Google Scholar]
- 50. Ghoshal N., Smiley J. F., DeMaggio A. J., Hoekstra M. F., Cochran E. J., Binder L. I., and Kuret J. (1999) A new molecular link between the fibrillar and granulovacuolar lesions of Alzheimer's disease. Am. J. Pathol. 155, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borroni B., Archetti S., Del Bo R., Papetti A., Buratti E., Bonvicini C., Agosti C., Cosseddu M., Turla M., Di Lorenzo D., Pietro Comi G., Gennarelli M., and Padovani A. (2010) TARDBP mutations in frontotemporal lobar degeneration: frequency, clinical features, and disease course. Rejuvenation Res. 13, 509–517 [DOI] [PubMed] [Google Scholar]
- 52. Kirby J., Goodall E. F., Smith W., Highley J. R., Masanzu R., Hartley J. A., Hibberd R., Hollinger H. C., Wharton S. B., Morrison K. E., Ince P. G., McDermott C. J., and Shaw P. J. (2010) Broad clinical phenotypes associated with TAR-DNA binding protein (TARDBP) mutations in amyotrophic lateral sclerosis. Neurogenetics 11, 217–225 [DOI] [PubMed] [Google Scholar]
- 53. Nonaka T., Kametani F., Arai T., Akiyama H., and Hasegawa M. (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 54. Arnold E. S., Ling S. C., Huelga S. C., Lagier-Tourenne C., Polymenidou M., Ditsworth D., Kordasiewicz H. B., McAlonis-Downes M., Platoshyn O., Parone P. A., Da Cruz S., Clutario K. M., Swing D., Tessarollo L., Marsala M., et al. (2013) ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. U.S.A. 110, E736–E745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Y. J., Caulfield T., Xu Y. F., Gendron T. F., Hubbard J., Stetler C., Sasaguri H., Whitelaw E. C., Cai S., Lee W. C., and Petrucelli L. (2013) The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 22, 3112–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu Q., Guo J., and Zhou J. (2004) A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J. Neurochem. 90, 164–172 [DOI] [PubMed] [Google Scholar]
- 57. Choi D. W., Maulucci-Gedde M., and Kriegstein A. R. (1987) Glutamate neurotoxicity in cortical cell culture. J. Neurosci. 7, 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gu J., Wu F., Xu W., Shi J., Hu W., Jin N., Qian W., Wang X., Iqbal K., Gong C. X., and Liu F. (2017) TDP-43 suppresses tau expression via promoting its mRNA instability. Nucleic Acids Res. 10.1093/nar/gkx175 [DOI] [PMC free article] [PubMed] [Google Scholar]