Abstract
Cardiomyocytes acquire their primary specialized function (contraction) before exiting the cell cycle. In this regard, proliferation and differentiation must be precisely coordinated for proper cardiac morphogenesis. Here, we have investigated the complex transcriptional mechanisms employed by cardiomyocytes to coordinate antagonistic cell-cycle and differentiation gene programs through the molecular dissection of the core cardiac transcription factor, MEF2. Knockdown of individual MEF2 proteins, MEF2A, -C, and -D, in primary neonatal cardiomyocytes resulted in radically distinct and opposite effects on cellular homeostasis and gene regulation. MEF2A and MEF2D were absolutely required for cardiomyocyte survival, whereas MEF2C, despite its major role in cardiac morphogenesis and direct reprogramming, was dispensable for this process. Inhibition of MEF2A or -D also resulted in the activation of cell-cycle genes and down-regulation of markers of terminal differentiation. In striking contrast, the regulation of cell-cycle and differentiation gene programs by MEF2C was antagonistic to that of MEF2A and -D. Computational analysis of regulatory regions from MEF2 isoform-dependent gene sets identified the Notch and Hedgehog signaling pathways as key determinants in coordinating MEF2 isoform-specific control of antagonistic gene programs. These results reveal that mammalian MEF2 family members have distinct transcriptional functions in cardiomyocytes and suggest that these differences are critical for proper development and maturation of the heart. Analysis of MEF2 isoform-specific function in neonatal cardiomyocytes has yielded insight into an unexpected transcriptional regulatory mechanism by which these specialized cells utilize homologous members of a core cardiac transcription factor to coordinate cell-cycle and differentiation gene programs.
Keywords: cardiomyocyte, differentiation, gene regulation, transcription factor, transcriptomics, antagonistic
Introduction
Cardiac development involves precise integration of specification, proliferation, and differentiation gene programs in cardiomyocytes and multiple non-muscle cell types (1–3). The gene-regulatory network that drives heart organogenesis is controlled by a core of evolutionarily conserved cardiac transcription factors (TFs)2 (4, 5). MEF2 is a key member of this core group whose activity is essential for cardiac development in numerous animal species (6, 7). Vertebrates have evolved multiple isoforms of MEF2 (MEF2A, -B, -C, and -D), thereby broadening the gene-regulatory potential of this cardiac TF and adding additional layers of complexity to the transcriptional circuitry of cardiac organogenesis. The diverse regulatory roles of MEF2 in the heart are exemplified by the wide array of cardiovascular phenotypes associated with knock-out or inhibition of individual MEF2 genes in vertebrate model systems (8–13). Although there is no doubt that MEF2 is required for differentiation and regulates structural genes in muscle (14, 15), the repertoire of gene programs controlled by this family in cardiomyocytes is largely unknown. Based on the range of cellular processes mediated by MEF2 isoforms in other specialized cell types, such as neurons, the regulatory potential of this core cardiac TF has not been fully realized in cardiomyocytes.
The perinatal period is a critical time for proper cardiac maturation, during which differentiated cardiomyocytes with persisting proliferative capacity are programmed to permanently exit the cell cycle (16–18). Cardiomyocyte quiescence will prevent further growth of the heart through cell division, instead promoting growth by increases in cardiomyocyte size. Perhaps not coincidentally, it is also the time during which the mammalian heart begins to lose its ability to regenerate (19, 20). Neonatal cardiomyocytes also undergo radical switches in metabolic pathways and contractile protein isoforms to fully commit to the mature differentiated phenotype (21, 22). We understand surprisingly little about the mechanisms by which cell-cycle exit and terminal differentiation are coordinated, considering the importance of precisely executing these processes for adult cardiomyocyte homeostasis and function.
We have previously reported that neonatal cardiomyocyte homeostasis is dependent on MEF2. Cytoarchitectural (costamere/muscle focal adhesion) and cell-cycle gene programs in neonatal cardiomyocytes were shown to be regulated by MEF2A and MEF2D protein isoforms, respectively (23, 24). Interestingly, defects in these distinct gene programs both led to severely impaired cardiomyocyte survival. Based on the importance of MEF2 for cardiomyocyte survival, we wanted to explore the global gene programs that mediate this process further by performing an unbiased genome-wide pathway analysis of all MEF2 protein isoforms.
The gene-regulatory function of MEF2 isoforms has typically been investigated through the analysis of a representative member. Here we present a comprehensive morphological and genome-wide transcriptomic analysis on the three major cardiac MEF2 isoforms, MEF2A, -C, and -D, in neonatal cardiomyocytes under identical conditions. MEF2A and -D, but not MEF2C, were found to be essential for cardiomyocyte survival. Consistent with these differential effects on survival, a detailed computational analysis uncovered distinct roles for these protein isoforms in cell-cycle and structural gene regulation. The results of this study reveal an entirely unexpected antagonistic regulatory role among MEF2 isoforms and have provided a mechanistic understanding of the intricate transcriptional relationships among homologous proteins in a core cardiac transcription factor family.
Results
Neonatal cardiomyocyte survival is dependent on MEF2A or -D but not MEF2C
The coordination of cell-cycle and differentiation pathways is essential for neonatal cardiomyocyte homeostasis and maturation. Perturbations in these processes can lead to catastrophic events, including programmed cell death. We have previously reported a requirement for MEF2A and -D in neonatal cardiomyocyte survival through their regulation of the cytoarchitecture (costamere) and cell cycle, respectively (23, 24). The precise relationship between these and other MEF2 isoforms in regulating survival has not been fully explored. We sought to dissect the transcriptional mechanisms of this critical process in neonatal cardiomyocytes.
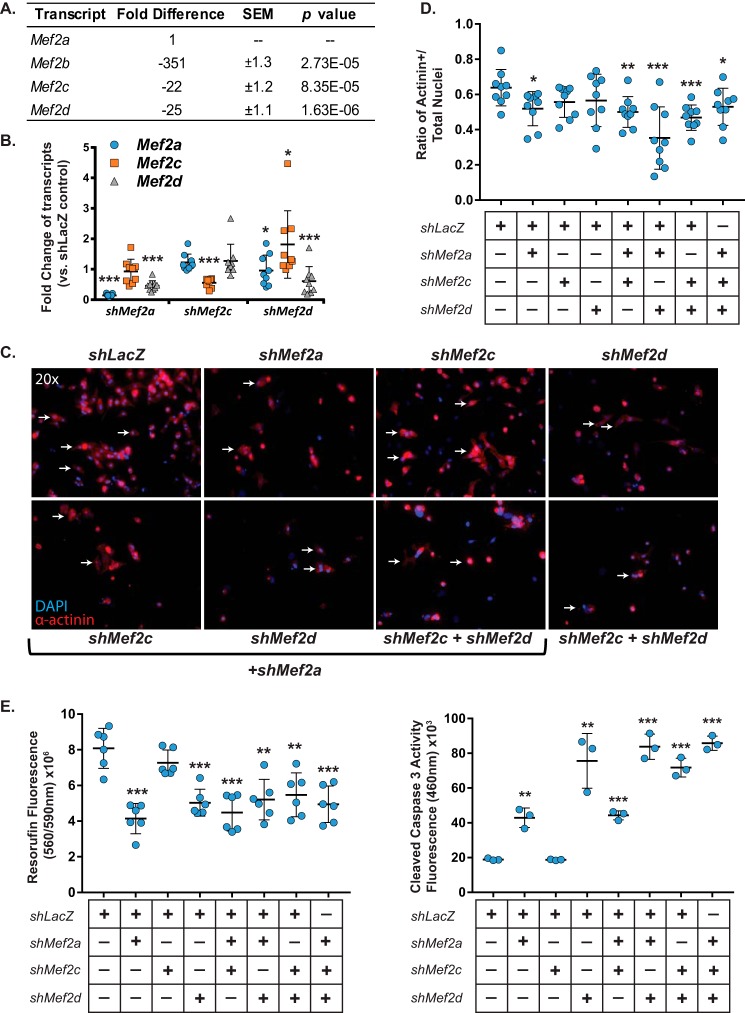
Although expression of Mef2 transcripts has been qualitatively examined in mouse cardiac development (26), the relative expression of the four mammalian Mef2 transcripts has never been quantified specifically in cardiomyocytes. Using quantitative RT-PCR, we found that Mef2a is the most abundant isoform in neonatal rat ventricular myocytes (NRVMs) (Fig. 1A). Mef2c and -d displayed similar expression levels and were 22–25-fold lower than Mef2a, respectively. Mef2b transcripts were largely undetectable in NRVMs, expressed at levels 350-fold lower than that of Mef2a. Based on these results, MEF2A, -C, and -D were deemed to be the predominant MEF2 isoforms expressed in NRVMs, and our analysis is restricted to these three relevant factors.
Figure 1.
Neonatal cardiomyocyte survival is dependent on MEF2A or -D but not MEF2C. A, qRT-PCR analysis of relative expression of Mef2 isoform transcripts in untreated NRVMs shows that Mef2a transcripts are the most abundant, with Mef2b transcripts expressed at a 350-fold lower level and Mef2c and -d expression at ∼20-fold lower levels. B, qRT-PCR analysis of specific shRNA knockdown efficiency shows significant knockdown of targeted Mef2 transcripts in NRVMs. C, immunofluorescent images of NRVMs treated with combinations of Mef2 shRNAs show a decrease in cell numbers in cultures treated with Mef2a and -d shRNA, but not Mef2c shRNA alone. NRVMs are characterized by α-actinin immunoreactivity and counterstained with DAPI. D, quantification of the ratio of α-actinin-positive cells versus the total number of nuclei of n = 9 fields/treatment shows a significant decrease in α-actinin-positive cells when NRVM cultures were treated with Mef2a shRNA or with combinations containing Mef2a or Mef2d shRNA but not with Mef2c shRNA alone. E, NRVMs treated with Mef2a or -d shRNA exhibit significantly decreased viability and significantly increased cleaved-caspase-3 activity (left and right, respectively). Data are means (n = 3) ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Neonatal cardiomyocytes were transduced with MEF2 isoform-specific shRNA adenoviruses and examined 3 days post-transduction. Analysis of MEF2 expression in each of the isoform knockdowns revealed efficient inhibition of the respective MEF2 isoform (Fig. 1B). We have previously demonstrated the isoform specificity of these shRNAs and did not observe cross-reactivity with other MEF2 proteins (23, 25). Interestingly, we observed down-regulation of Mef2d in MEF2A-deficient NRVMs and a reciprocal down-regulation of Mef2a in MEF2D-deficient NRVMs (Fig. 1B). In contrast, MEF2C deficiency did not affect endogenous expression of Mef2a or -d. Finally, with the exception of a modest up-regulation of Mef2c in MEF2D-deficient NRVMs (Fig. 1B, right), there was no compensatory up-regulation of Mef2 transcripts in response to acute depletion of individual MEF2 proteins. Given the previously described isoform specificity of these shRNAs, we conclude that the reciprocal down-regulation of Mef2d and Mef2a transcripts in MEF2A- and MEF2D-deficient NRVMs, respectively, is a biological effect of transcriptional cross-regulation within the MEF2 family in NRVMs.
Previous studies have described distinct loss-of-function cardiac phenotypes for mammalian MEF2 family members in vivo (11–13). Because these studies examined the consequences of chronic deficiency of individual MEF2 proteins and in the context of the whole heart, we investigated the effects of acute inhibition of MEF2 family members specifically in isolated cardiomyocytes within a defined temporal window. Inhibition of MEF2A resulted in reduced cardiomyocyte number, decreased viability, and increased cleaved caspase-3 activity, an indicator of programmed cell death (Fig. 1, C–E). MEF2D inhibition also resulted in a significant reduction in cardiomyocyte viability and increase in caspase-3 activity (Fig. 1E), consistent with our previous analysis (24). By contrast, inhibition of MEF2C did not impair NRVM survival (Fig. 1) even at higher multiplicities of infection. These results indicate that either MEF2A or MEF2D is necessary for neonatal cardiomyocyte survival, and MEF2C is not.
To determine whether MEF2 proteins function redundantly in cardiomyocyte survival, we performed double and triple MEF2 isoform knockdowns. As shown in Fig. 1 (C–E), pairwise inhibitions of MEF2 isoforms did not modulate the impaired viability phenotype observed in MEF2A or -D deficiency, and an effect of MEF2 inhibition did not emerge in the MEF2A-deficient, MEF2D-deficient, or MEF2A/D-deficient NRVMs. These results demonstrate that MEF2C is dispensable for cardiomyocyte survival and does not have a role masked by the presence of MEF2A or -D. Whereas MEF2A and -D are both required for cardiomyocyte survival, MEF2A inhibition led to earlier loss of viability with adverse effects observable by 48 h (data not shown).
MEF2 overexpression in MEF2A-deficient cardiomyocytes
To determine whether overexpression of any MEF2 isoform is sufficient to rescue the MEF2A-deficient loss of viability, we overexpressed MEF2A, MEF2C, MEF2D, and MEF2-VP16, a constitutive MEF2 activator, in MEF2A-deficient myocytes. Transient overexpression of the various MEF2 protein isoforms or MEF2-VP16 did not significantly increase the number of actinin-positive cardiomyocytes in MEF2A-deficient NRVMs (Fig. 2, A and B). However, overexpression of MEF2A, MEF2D, or MEF2-VP16 significantly decreased cleaved caspase-3 activity in MEF2A-deficient NRVMs (Fig. 2C). By contrast, overexpression of MEF2C did not significantly reduce cleaved caspase-3 activity in MEF2A-deficient NRVMs (Fig. 2C). The ability of MEF2A, MEF2D, or MEF2-VP16 to reduce the activity of caspase-3 but not increase cell number suggests that decreased activity of this pro-apoptotic factor is not sufficient to prevent loss of cardiomyocytes or that insufficient time has elapsed to see a beneficial effect on cellular viability.
Figure 2.
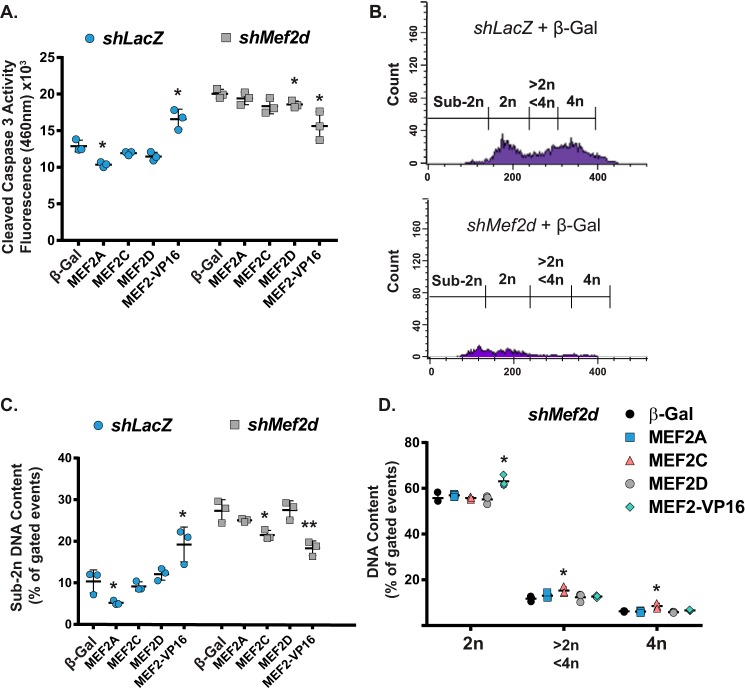
Overexpression of MEF2A, MEF2D, or MEF2-VP16 reduces NRVM apoptosis caused by MEF2A depletion. A, immunofluorescent images of NRVMs treated with Mef2a shRNA and an overexpression construct show that overexpression of MEF2 constructs does not modulate the number of α-actinin-positive cells. B, quantification of the ratio of α-actinin-positive cells versus the total number of nuclei of n = 9 fields/treatment shows no effect of MEF2 overexpression rescue on the viability of NRVMs treated with Mef2a shRNA. C, overexpression of MEF2A, -D, or -VP16 leads to a reduction in pro-apoptotic cleaved-caspase-3 activity in Mef2a shRNA-treated NRVMs. D, propidium iodide-stained flow cytometry shows the emergence of a significant cell population with sub-2n DNA content in Mef2a shRNA-treated NRVMs. E, quantification of flow cytometry cell populations measuring DNA content shows an increase in cells with sub-2n DNA content upon treatment with Mef2a shRNA and a reduction of this group upon overexpression of MEF2 constructs. F, measurement of NRVMs containing 2n, 2n-4n, and 4n DNA. Data are means (n = 3) ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Based on this intriguing isoform-specific difference, we bolstered our analysis by measuring DNA degradation using propidium iodide staining followed by flow cytometry. As shown in Fig. 2D, shLacZ control NRVMs contain a population of cells with diploid (2n) through tetraploid (4n) DNA content portraying the known heterogeneity of mono- and binucleated neonatal myocytes (Fig. 2D, left). Inhibition of MEF2A resulted in a significant redistribution of this DNA content profile, with the emergence of a peak representing, myocytes containing sub-2n DNA content (Fig. 2D, right), consistent with elevated DNA fragmentation observed in apoptosis.
We subsequently evaluated the ability of MEF2 isoforms to reduce the sub-2n fragmented DNA fraction and restore normal DNA content in MEF2A-deficient NRVMs. Overexpression of the various MEF2 isoforms reduced the percentage of the sub-2n population but to varying degrees. MEF2A, MEF2C, or MEF2-VP16 significantly reduced the number of myocytes containing sub-2n DNA content, whereas the modest reduction of sub-2n myocytes observed in MEF2D overexpression did not reach statistical significance (Fig. 2E, right set of bars). These results demonstrate that overexpression of MEF2 isoforms is sufficient to diminish DNA fragmentation in MEF2A-deficient NRVMs. Interestingly, whereas MEF2C overexpression failed to reduce cleaved caspase-3 activity in MEF2A-depleted NRVMs, it had a favorable effect on genomic DNA integrity. These results suggest that MEF2C is distinct from other MEF2 isoforms in that its apparent compensatory activity is restricted to a late stage in the apoptotic pathway, while being unable to modulate earlier steps or ultimately affect survival.
Further analysis of the DNA content profile revealed previously uncharacterized effects of MEF2 isoform overexpression on the cardiomyocyte genome. As shown in Fig. 2F (left graph), MEF2A overexpression in MEF2A-deficient NRVMs resulted in a significant reduction in the sub-2n population and a modest increase in myocytes harboring 2n, 2n-4n, and 4n DNA content. These increases probably represent the myocyte population that was rescued from apoptotic DNA fragmentation by MEF2A overexpression. A similar effect and profile were observed for MEF2-VP16. By contrast, MEF2D overexpression in MEF2A-deficienct NRVMs resulted in significantly more myocytes with 2n compared with MEF2A overexpression and fewer cells containing 2n-4n and 4n DNA content. Curiously, MEF2C overexpression in MEF2A-deficient NRVMs displayed a profile quite distinct from overexpression of MEF2A, MEF2D, or MEF2-VP16 in that same background. This population displayed a significant decrease in diploid (2n) NRVMs and a significant increase in myocytes with 2n-4n and 4n DNA content (Fig. 2F, right graph). By thoroughly analyzing the DNA content profile of MEF2A-deficient NRVMs overexpressing various MEF2 proteins, we have demonstrated that, despite their ability to rescue DNA fragmentation (sub-2n), these isoforms elicited differing effects on the genomic DNA content.
MEF2 overexpression in MEF2D-deficient cardiomyocytes
Previously, we reported a function for MEF2D in cardiomyocyte survival and genomic DNA integrity (24). Based on the distinct effects of MEF2 proteins in these processes in MEF2A-deficient cardiomyocytes, we next wanted to evaluate MEF2 overexpression in MEF2D-deficient NRVMs. Overexpression of MEF2D or MEF2-VP16, but not MEF2A or -C, significantly reduced cleaved caspase-3 activity in MEF2D-deficient NRVMs (Fig. 3A). Regarding DNA content, MEF2D depletion caused a widespread reduction in the NRVM population harboring diploid through tetraploid genomic DNA and an increase in the percentage of sub-2n cells (Fig. 3B), a profile consistent with our previous study (24). Interestingly, overexpression of MEF2C or MEF2-VP16, but not MEF2A or -D, significantly reduced the sub-2n population (Fig. 3C, left graph). Moreover, MEF2C overexpression promoted an increase in the 2n-4n and 4n population similar to its effects in MEF2A-deficient NRVMs (Fig. 3C, right graph). Taken together, the rescue experiments suggest that, despite some compensatory effects, MEF2 isoforms have distinct and separable functions in the regulation of cardiomyocyte genomic DNA integrity and survival.
Figure 3.
Overexpression of MEF2D or MEF2-VP16 reduces NRVM apoptosis caused by MEF2D depletion. A, overexpression of MEF2D or MEF2-VP16 leads to a reduction in pro-apoptotic cleaved-caspase-3 activity in Mef2a shRNA-treated NRVMs. B, propidium iodide-stained flow cytometry shows the emergence of a significant cell population with sub-2n DNA content in Mef2d shRNA-treated NRVMs. C, quantification of flow cytometry cell populations measuring DNA content shows an increase in cells with sub-2n DNA content upon treatment with Mef2d shRNA and a reduction of this group upon overexpression of MEF2 constructs. D, measurement of NRVMs containing 2n, 2n-4n, and 4n DNA. Data are means (n = 3) ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Genome-wide transcriptomics and comparative analysis of individual MEF2 knockdown in cardiomyocytes
The contrasting effect of MEF2A, -C, and -D on cardiomyocyte survival and DNA content profile suggested distinct regulatory functions of these isoforms despite their largely indistinguishable transcriptional activities in vitro. This hypothesis is additionally supported by previously characterized distinct in vivo loss-of-function phenotypes (11–13). Thus, we performed global gene expression profiling to determine what sets of genes and cellular processes are regulated by individual MEF2 proteins in neonatal cardiomyocytes.
Transcriptomic analysis of NRVMs depleted of individual MEF2 proteins resulted in overlapping but largely distinct dysregulated gene sets. Using stringent statistical criteria (see “Experimental procedures”), the most striking effect on gene expression in NRVMs was observed in MEF2A knockdown. As shown in Fig. 4A, depletion of MEF2A revealed 3,197 significantly dysregulated genes, representing 43% of the total number of dysregulated genes. MEF2C and MEF2D deficiency resulted in only 471 and 786 dysregulated genes, or 6.4 and 10.6% of the total, respectively. In terms of overlapping dysregulated genes, MEF2A and -D gene sets had 20% of the genes in common, which was the highest percentage of all of the MEF2 isoform comparisons (Fig. 4A, right). This was followed by 9% of commonly dysregulated genes between MEF2A and -C. The lowest overlap was observed in the genes shared between MEF2C and -D, representing 2.8% of the total. Finally, only 8.2% of the dysregulated genes were shared in all three MEF2 isoform knockdowns (Fig. 4A, right).
Figure 4.
Comparative analysis of MEF2 isoform-specific transcriptome. A, genes dysregulated in individual Mef2 isoform knockdowns were grouped into statistically similar gene sets. Left, a composite Venn diagram incorporating all overlapping gene sets as determined by Tukey's honest significant difference post hoc test (q < 0.05). Right, summary of total significantly dysregulated (q < 0.05) genes in each Mef2 isoform shRNA knockdown. B, qRT-PCR analysis of a subset of genes preferentially dysregulated in an individual Mef2 shRNA knockdown microarray. Dnm3, dynamin 3; Ccl5, chemokine (C-C motif) ligand 5; Filip1, filamin A interacting protein 1; Lrrc39, leucine-rich repeat-containing 39; Cirbp, cold-inducible RNA-binding protein; Adamtsl2, ADAMTS-like 2; Cdh8, cadherin 8; Mdga2, MAM domain-containing glycosylphosphatidylinosital anchor 2; Kit, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene; Sept4, septin 4; Upk1b, uroplakin 1B; Pten, phosphatase and tensin homolog. Data are means (n = 3) ± S.D. (error bars). *, p < 0.05; ** p < 0.01; *** p < 0.001.
These dysregulated gene expression levels were subsequently validated by qRT-PCR analysis on a subset of the top dysregulated genes (up- and down-regulated) from each individual MEF2 knockdown. As shown in Fig. 4B, the vast majority of genes in each MEF2 isoform gene set examined displayed the expected dysregulation.
Classification of cellular processes in MEF2 knockdown gene sets
To gain insight into the distinct roles of the MEF2 family in NRVMs, the dysregulated, non-overlapping genes in each subgroup were categorized into cellular processes using Ingenuity Pathway Analysis® (IPA).
IPA of MEF2 isoform-sensitive gene sets revealed vastly different cellular processes in the preferentially dysregulated target genes from each MEF2 knockdown (Table 1). Many genes distinctly sensitive to MEF2A play roles in cell-cell junction signaling and cancer pathways. By contrast, genes preferentially regulated by MEF2C are involved in energy production in mitochondria, and genes regulated by MEF2D are involved in cellular growth (HIPPO pathway) and survival (PI3K signaling). Finally, the group of genes sensitive to all three MEF2 isoforms functions in multiple aspects of the cell cycle, including DNA replication and DNA damage checkpoints. Notably, this overlapping category did not have pathways in common with any of the significantly enriched processes in the individual MEF2 gene sets. These results reveal the breadth of cellular processes under MEF2 control and suggest that these gene programs have evolved sophisticated transcriptional mechanisms to differentiate among MEF2 isoforms in cardiomyocytes.
Table 1.
Canonical pathways associated with genes preferentially dysregulated in individual Mef2 shRNA treatments
Ingenuity Pathway Analysis® of canonical pathways preferentially dysregulated by Mef2 shRNA treatment was conducted. The analyzed gene set represents genes that were only statistically dysregulated in knockdown of a single MEF2 factor. The top five most significantly regulated canonical pathways are presented with the number of genes dysregulated in relation to the accepted number of genes associated with each canonical pathway (ratio). TCA, tricarboxylic acid.
| Canonical pathway | p | Ratio |
|---|---|---|
| MEF2A | ||
| Germ cell-Sertoli cell junction signaling | 1.07E−08 | 50:160 (0.312) |
| Molecular mechanisms of cancer | 1.81E−08 | 90:365 (0.247) |
| 14-3-3-mediated signaling | 1.98E−07 | 38:117 (0.325) |
| Protein ubiquitination pathway | 2.13E−07 | 66:255 (0.259) |
| Death receptor pathway | 3.13E−07 | 32:92 (0.348) |
| MEF2C | ||
| Mitochondrial dysfunction | 1.96E−07 | 13:171 (0.076) |
| TCA cycle II (eukaryotic) | 2.17E−05 | 4:23 (0.174) |
| Fatty acid oxidation III (unsaturated, odd number) | 2.40E−05 | 2:3 (0.667) |
| Oxidative phosphorylation | 3.05E−05 | 8:109 (0.073) |
| Methylmalonyl pathway | 3.67E−05 | 2:4 (0.50) |
| MEF2D | ||
| HIPPO signaling | 2.08E−04 | 11:86 (0.128) |
| PI3K signaling in B lymphocytes | 5.78E−04 | 13:128 (0.102) |
| Neuregulin signaling | 1.03E−03 | 10:88 (0.114) |
| Proline degradation | 1.23E−03 | 2:2 (1.00) |
| Fc receptor-mediated phagocytosis in macrophages and monocytes | 1.57E−03 | 10:93 (0.108) |
| All MEF2 | ||
| Cell cycle control of chromosomal replication | 0.000000499 | 8:27 (0.296) |
| Hepatic fibrosis/hepatic stellate cell activation | 4.47E−05 | 17:198 (0.086) |
| Antiproliferative role of TOB in T-cell signaling | 6.70E−05 | 6:26 (0.231) |
| Cell cycle: G1/S checkpoint regulation | 6.94E−05 | 9:64 (0.141) |
| Factors promoting cardiogenesis in vertebrates | 2.52E−04 | 10:92 (0.109) |
Complex and antagonistic patterns of dysregulated gene expression among MEF2 family members
The emergence of a common cell-cycle gene program regulated by all MEF2 isoforms piqued our interest because this could potentially explain the effects on cardiomyocyte survival and genomic DNA content. Because this subset of genes is sensitive to all MEF2 isoforms, we hypothesized that, based on their largely indistinguishable transcriptional activities in vitro, these genes would be similarly regulated by each isoform and provide a logical starting point to mechanistically dissect MEF2-dependent gene regulation in cardiomyocytes.
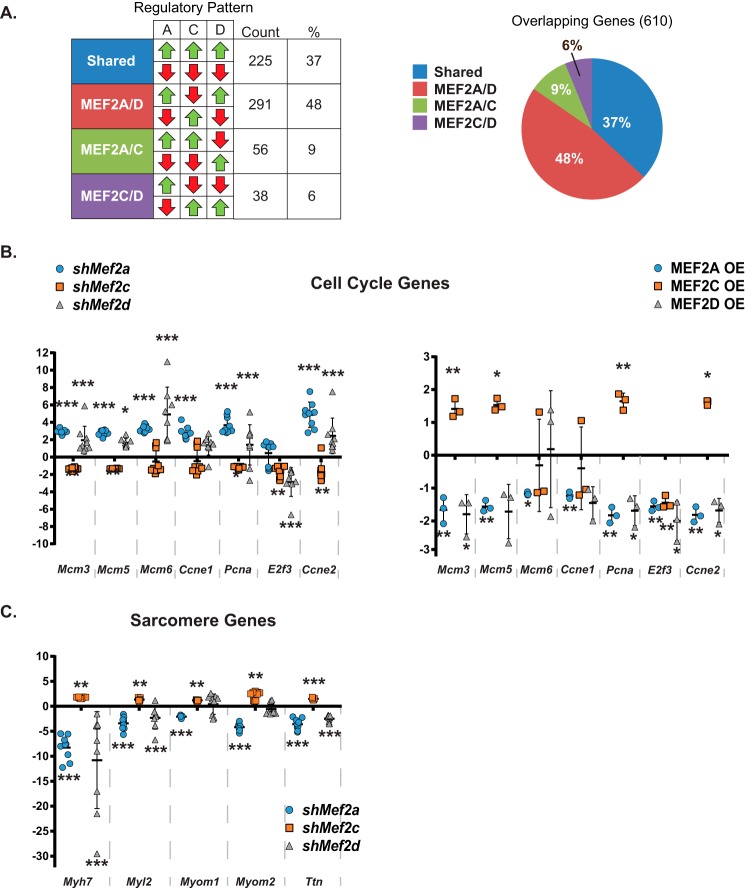
Remarkably, analysis of the dysregulated patterns of the 610 common genes revealed that the vast majority of these genes were not similarly affected by depletion of specific MEF2 isoforms (Fig. 5A, note the red and green arrows). These comparisons revealed four distinct groups based on similarity of dysregulation in all three MEF2 knockdowns or combinations of two MEF2 isoforms (e.g. shared, MEF2A/D, MEF2A/C, and MEF2C/D). Strikingly, only about one-third of the MEF2 isoform-sensitive genes were dysregulated in the same direction (shared), but nearly two-thirds of the genes were differentially affected and showed the opposite dysregulation in at least one MEF2 isoform knockdown (Fig. 5A, right). Of these, the most predominant pattern was the similarity in gene dysregulation between inhibition of MEF2A or -D and the opposite dysregulation by MEF2C depletion as shown in the MEF2A/D row (48%; Fig. 5A, left table). These patterns suggest that a subset of pathways in cardiomyocytes dependent on all MEF2 isoforms are not regulated similarly, extending the notion of isoform-specific regulation to genes functioning in common cellular processes.
Figure 5.
Distinct regulatory patterns of overlapping genes sensitive to all MEF2 isoforms. A, breakdown of genes dysregulated in each individual knockdown based on the pattern of dysregulation across all knockdowns. Nearly 50% of commonly dysregulated genes are dysregulated in the same direction by Mef2a and -d shRNA treatment and in the opposite direction by Mef2c shRNA treatment. B, qRT-PCR analysis of a subset of genes dysregulated in the same direction by Mef2a and -d shRNA treatment and dysregulated in the opposite direction by Mef2c shRNA treatment (n = 9; left) and MEF2A, -C, and -D overexpression (n = 3; right). Mcm3, minichromosome maintenance component 3; Mcm5, minichromosome maintenance component 5; Mcm6, minichromosome maintenance component 6; Ccne1, cyclin e1; Pcna, proliferating cell nuclear antigen; E2f3, E2F transcription factor 3; Ccne2, cyclin e2. C, qRT-PCR analysis of a subset of sarcomere genes dysregulated in the same direction by Mef2a and -d shRNA treatment and in the opposite direction by Mef2c shRNA treatment (n = 9). Myh7, myosin heavy chain 7, cardiac muscle, β; Myl2, myosin light polypeptide 2, regulatory, cardiac, slow; Myom1, myomesin 1; Myom2, myomesin 2; Ttn, titin. Data are means (n = 9) ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Given the unexpected and disparate effects of dysregulation within the common group, we performed IPA on those genes within the various regulatory patterns to obtain a comprehensive analysis of the types of cellular processes for each pattern. As shown in Table 2, the pathways for each regulatory pattern were dramatically different. Genes regulated in the same manner by all three MEF2 isoforms (shared; all up- or all down-regulated) have been shown to function primarily in fibrosis and amino acid biosynthesis. Genes regulated in the same manner by MEF2A and -C (A/C) or MEF2C and -D (C/D) play a role in signal transduction, but the specific ligand-receptor pathways regulated by each of these pairs were distinct. The most enriched pathway identified in the A/C pattern of regulation belonged to ephrin signaling, a critical pathway in cell positioning and guidance in development. The genes present in the C/D regulatory pattern were enriched for cytokine and interferon pathways. The most intriguing result, however, was obtained for the genes exhibiting the MEF2A and D (A/D) regulatory pattern, where genes were similarly affected by MEF2A or -D depletion but displayed the opposite dysregulation by MEF2C depletion. Pathway analysis of this particular cohort, which had the largest collection of genes, revealed the cell cycle as the most significantly enriched pathway. This helps to explain the enrichment of the cell-cycle program in our analysis of the overlapping group of 610 genes indicated in Table 1 (All MEF2). Moreover, these data suggest a previously undescribed role for MEF2A and -C protein isoforms in cell-cycle control in cardiomyocytes and independently confirm our previous observations of MEF2D-dependent regulation of the cell cycle (24).
Table 2.
Pathway analysis of regulatory patterns of genes dysregulated in each MEF2 knockdown
Shown is IPA canonical pathway analysis of genes dysregulated in the same direction in each treatment (shared) and pathways dysregulated in the same direction by Mef2a and -c shRNA treatment or Mef2c and -d shRNA treatment (MEF2A/C or MEF2C/D) or in the same direction by Mef2a and -d shRNA treatment (MEF2A/D).
| Canonical pathway | p | Ratio |
|---|---|---|
| Shared | ||
| Hepatic fibrosis/hepatic stellate cell activation | 5.23E−04 | 8:183 (0.044) |
| Phenylalanine degradation I (aerobic) | 5.80E−04 | 2:4 (0.50) |
| Serine biosynthesis | 9.75E−04 | 2:5 (0.40) |
| Superpathway of serine and glycine biosynthesis I | 2.02E−03 | 2:7 (0.286) |
| Antiproliferative role of TOB in T-cell signaling | 2.16E−03 | 3:26 (0.115) |
| MEF2A/D | ||
| Cell cycle control of chromosomal replication | 1.64E−09 | 8:27 (0.296) |
| Role of CHK proteins in cell cycle checkpoint control | 8.27E−06 | 7:55 (0.127) |
| Factors promoting cardiogenesis in vertebrates | 3.24E−05 | 8:92 (0.087) |
| GADD45 signaling | 1.03E−04 | 4:19 (0.210) |
| DNA damage-induced 14-3-3 signaling | 1.03E−04 | 4:19 (0.210) |
| MEF2A/C | ||
| Ephrin receptor signaling | 9.52E−03 | 3:174 (0.017) |
| Glycerol-3-phosphate shuttle | 1.00E−02 | 1:4 (0.250) |
| Melatonin degradation II | 1.00E−02 | 1:4 (0.250) |
| Ephrin B signaling | 1.45E−02 | 2:73 (0.027) |
| Glycerol degradation I | 1.50E−02 | 1:6 (0.167) |
| MEF2C/D | ||
| Type I diabetes mellitus signaling | 1.01E−03 | 3:110 (0.027) |
| Role of JAK2 in hormone-like signaling | 1.70E−03 | 2:34 (0.059) |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 1.91E−03 | 3:137 (0.022) |
| Interferon signaling | 1.91E−03 | 2:36 (0.056) |
| Activation of IRF by cystolic pattern recognition receptors | 5.57E−03 | 2:62 (0.032) |
Based on this interesting finding, we next validated the expression of select cell-cycle genes in the individual MEF2 isoform knockdowns. These genes have been shown to function in DNA replication and checkpoint control. As shown in Fig. 5B (left), inhibition of MEF2A or -D resulted in the up-regulation of the cell-cycle genes Mcm3, -5, -6, Ccne1, -2, and Pcna. Up-regulation of the cell-cycle program did not lead to increased proliferation but rather cell-cycle reentry followed by programmed cell death and is entirely consistent with our previous characterization of MEF2D-deficient NRVMs (24). By contrast, inhibition of MEF2C resulted in down-regulation of cell-cycle gene expression (Fig. 5B, left). To reinforce the observed antagonistic regulatory effect on the cell cycle within the MEF2 family, we overexpressed these proteins in NRVMs. As predicted, overexpression of MEF2A or -D repressed whereas MEF2C induced cell-cycle gene expression (Fig. 5B, right).
We complemented the cell-cycle analysis by examining the expression of representative sarcomere genes, which are established markers of differentiated cardiomyocytes. Expression of the sarcomere genes myosin heavy chain 7 (Myh7), myosin light chain 2 (Myl2), myomesin 1 and 2 (Myom1 and -2), and titin (Ttn), in MEF2A- and MEF2D-deficient NRVMs was significantly down-regulated, whereas these genes were up-regulated in MEF2C-deficient cells (Fig. 5C). These data reveal a previously unappreciated mechanism of MEF2-dependent gene regulation in cardiomyocytes, one in which MEF2 protein isoforms antagonistically regulate gene programs despite their similar transcriptional activities in vitro. Specifically, MEF2A and -D are required for repression of cell-cycle genes and activation of a subset of sarcomeric markers, and MEF2C plays an antagonistic role by activating cell-cycle genes and repressing sarcomeric markers of differentiation.
Identification of distinct transcription factor modules associated with MEF2-dependent cell-cycle and sarcomere genes
Given the lack of evidence demonstrating differences in DNA consensus sequence preferences of MEF2 isoforms, we hypothesized that the antagonistic regulatory patterns resulted from specific interactions with transcriptional coregulators. Therefore, upstream regulatory regions were computationally analyzed for discrete TF modules harboring a MEF2-binding site within 50 base pairs of a predicted TF-binding site. To identify these TF modules enriched in cell-cycle genes, we performed module-based motif enrichment analysis (Genomatix). Modules were considered significantly enriched in a set of promoters if the Z-score was > 2. As shown in Table 3, although similar TF modules were found to be overrepresented in both cell-cycle and sarcomere genes, the vast majority were unique to each gene set. Regarding the similar TF modules between the two groups, the basic helix-loop-helix (bHLH) proteins (or E-box-binding factors), such as HAND, Twist, and Mesp, play prominent and diverse roles in cardiogenesis, such as specification, proliferation, and differentiation (27–29); thus, it is not surprising to find them overrepresented in both gene categories. Similarly, the MADS box factor SRF and the steroid receptor RXR, which have pleiotropic gene-regulatory functions (30, 31), were enriched in TF modules within the regulatory regions of both cell-cycle and sarcomere genes. Analysis of the largely unique modules revealed transcriptional regulators that function downstream of Notch and Hedgehog signaling. These evolutionarily conserved signal transduction pathways emerged as the most interesting candidates because of their important role not only in developmental gene regulation but also in cardiogenesis (32, 33). These upstream signals could be the lynchpin to establish isoform selectivity in the MEF2-dependent regulation of cell-cycle and sarcomere gene programs in cardiomyocytes in cardiac development.
Table 3.
Enriched transcription factor-binding site analysis
Binding site enrichment analysis was performed using the Genomatix software suite to determine enriched motifs adjacent (within 50 bp) of MEF2 consensus binding sites in the regulatory region 5 kb upstream of the putative transcriptional start site of antagonistically regulated genes (differentiation genes at the top, cell cycle/proliferation genes at the bottom). The table provides a description of each candidate co-regulator for each enriched motif with a Z-score (Z-scores > 2.0 were considered significantly enriched). The binding factors and their relevant binding domains are included when known. Finally, a brief summary of relevant regulatory function is provided.
| TF module | Module description | Z-Score | Known factor(s) | Binding domain | Function |
|---|---|---|---|---|---|
| Differentiation | |||||
| V$HAND V$MEF2 | Twist subfamily of class B bHLH transcription factors | 4.33 | Hand1, Hand2, Lyl1, Mesp1, Mesp2 Nhlh1, Nhlh2, Scx, Tal1, Tal2, Tcf3, Tcf4, Tcf12, Tcf15, Twist1, Twist2 | bHLH | Positive regulator of striated muscle differentiation, secondary heart field specification |
| V$EBOX V$MEF2 | E-box-binding factors | 4.22 | Atf6, Atf6b, Max, Mga, Mlx, Mlxip, Mlxipl, Mnt, Mxd3, Mxd4, Mxi1, Myc, Mycl, Mycn, Tcfl5, Usf1, Usf2 | bHLH-ZIP, bZIP | Wnt signaling pathway, positive regulator of apoptosis |
| V$MEF2 V$RXRF | RXR heterodimer-binding sites | 3.53 | Nr1h2, Nr1h3, Nr1i2, Nr1i3, Rara, Rarb, Rarg, Rxra, Rxrb, Rxrg, Thra, Thrb, Vdr | C4 zinc-finger domain | Cardiac muscle proliferation, canonical Wnt signaling, ventricular cardiac muscle differentiation |
| V$EGRF V$MEF2 | EGR/nerve growth factor-induced protein C and related factors | 3.41 | Egr1–4, Wt1 | C2H2 zinc-finger domain | Heart development, BMP signaling, positive regulation of cell proliferation |
| V$MEF2 V$RBPF | RBPJ-κ | 2.85 | Rbpj, Rbpjl | RHR | Notch-binding transcriptional regulator, cardiac differentiation |
| V$MEF2 V$SRFF | Serum-response element-binding factor | 2.84 | Srf | MADS box | Positive regulator of cell differentiation |
| V$HESF V$MEF2 | Vertebrate homologues of enhancer of split complex | 2.73 | Bhlhe40, Bhlhe41, Helt, Hes1, Hes2, Hes3, Hes5, Hes6, Hes7, Hey1, Hey2, Heyl, Itgb3bp, Sohlh1, Sohlh2 | bHLH | Notch signaling in cardiac development, cardiac hypertrophy in response to stress |
| V$HOXH V$MEF2 | HOX-MEIS1 heterodimers | 2.72 | Hoxa10, Hoxa11, Hoxa13, Hoxa9, Hoxa9l, Hoxb13, Hoxb9, Hoxc10, Hoxc12, Hoxd10, Hoxd12, Hoxd13, Hoxd9, Meis1–3, | TALE class homeodomain, homeodomain | Embryonic patterning, skeletal muscle tissue development |
| V$MEF2 V$PBXC | PBX-MEIS complexes | 2.14 | Meis1–3, Pbx1–4, Pknox1, Pknox2 | TALE class homeodomain | Heart development |
| Cell cycle | |||||
| V$HAND V$MEF2 | Twist subfamily of class B bHLH transcription factors | 4.33 | Hand1, Hand2, Lyl1, Mesp1, Mesp2 Nhlh1, Nhlh2, Scx, Tal1, Tal2, Tcf3, Tcf4, Tcf12, Tcf15, Twist1, Twist2 | bHLH | Positive regulator of striated muscle differentiation, secondary heart field specification |
| V$FKHD V$MEF2 | Forkhead domain factors | 4.1 | Foxa1–3, Foxb2, Foxc1-2, Foxd1–4, Foxe1, Foxe3, Foxf1, Foxg1, Foxj1-3, Foxk1-2, Foxl1-2, Foxm1, Foxn2-3, Foxo1, Fox3-4, Foxo6, Foxp1–5, Foxq1, Foxs1 | Forkhead domain (winged helix) | G2/M transition, anatomical structure morphogenesis, cardiac muscle proliferation, ventricular cardiac muscle morphogenesis |
| V$MEF2 V$RXRF | RXR heterodimer-binding sites | 3.53 | Nr1h2, Nr1h3, Nr1i2, Nr1i3, Rara, Rarb, Rarg, Rxra, Rxrb, Rxrg, Thra, Thrb, Vdr | C4 zinc-finger domain | Cardiac muscle proliferation, canonical Wnt signaling, ventricular cardiac muscle |
| V$AP1R V$MEF2 | MAF- and AP1-related factors | 3.06 | Bach1-2, Maf, Mafa, Mafb, Maff, Mafg, Mafk, Nfe2, Nfe2l1–3, Nrl, RGD1562865 | BTB-POZ bZIP, bZIP | Regulator of anatomical structure morphogenesis, embryonic development |
| V$MEF2 V$SRFF | Serum-response element-binding factor | 2.84 | Srf | MADS box | Positive regulator of cell differentiation |
| V$IRXF V$MEF2 | Iroquois homeobox factors | 2.63 | Irx1–6, Mkx | Homeodomain | Heart development, negative regulator of myoblast differentiation |
| V$MEF2 V$P53F | p53 tumor supressor | 2.47 | Tp53, Tp63, Tp73 | p53 domain | Cell-cycle arrest, DNA-damage response, regulator of mitotic cell cycle |
| V$GLIF V$MEF2 | GLI zinc finger family | 2.05 | Gli1–3, Glis1–3, Zic1–5 | C2H2 zinc-finger domain | Heart development/looping, cell proliferation, Hedgehog signaling |
Notch and Hedgehog signaling coordinate MEF2 isoform-specific regulation of cell-cycle and differentiation programs
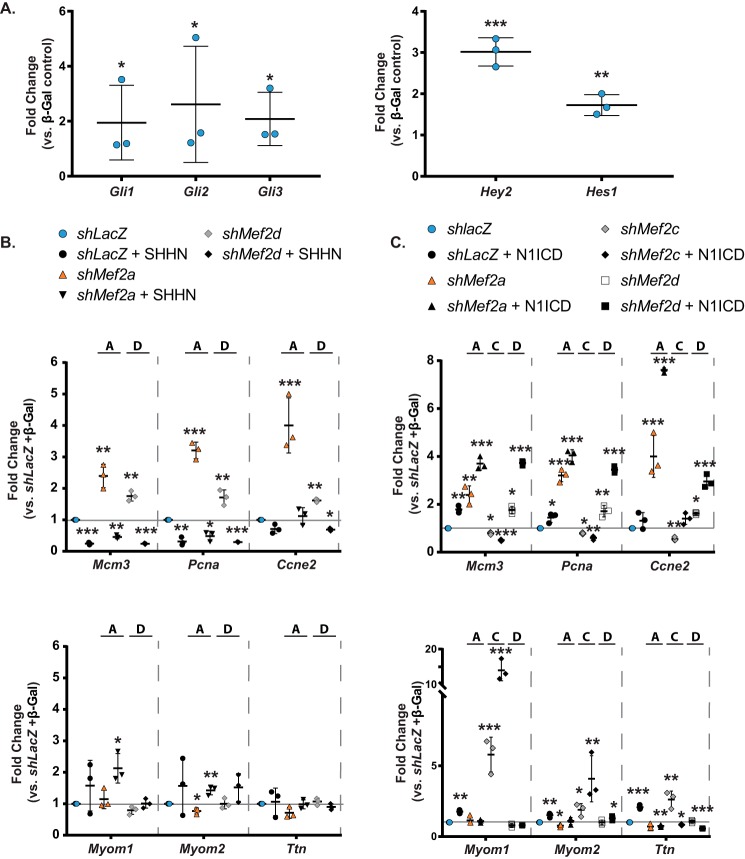
To determine whether Notch and/or Hedgehog signaling modulate MEF2-dependent gene regulation in a program-specific fashion, we initially evaluated the ability of NRVMs to respond to these pathways by examining the expression of their known downstream target genes, some of which are mediators of these signals. As shown in Fig. 6A, transduction of NRVMs with adenoviruses overexpressing constitutively active Sonic hedgehog (SHH-N) and the Notch intracellular domain (N1ICD) induced expression of the GLI (Gli1–3) and vertebrate enhancer of split (Hey2 and Hes1) TFs, respectively.
Figure 6.
Modulation of MEF2A- and -D-dependent cell-cycle and sarcomere gene regulation by Notch and Hedgehog signaling. A, transduction with constructs overexpressing the N-terminal SHH protein (SHH-N; left) or the Notch 1-intracellular domain (N1ICD; right) leads to efficient induction of downstream transcripts. Gli1, GLI family zinc finger 1; Gli2, GLI family zinc finger 2; Gli3, GLI family zinc finger 3; Hey2, hairy/enhancer-of-split related with YRPW motif 2; Hes1, hairy and enhancer of split 1. B, qRT-PCR analysis of a subset of cell-cycle (top) and differentiation (bottom) transcripts in combinatorial transductions with Mef2a or Mef2d shRNA and SHHN overexpression. C, qRT-PCR analysis of a subset of cell-cycle (top) and differentiation (bottom) transcripts in combinatorial transductions of Mef2a, -c, or -d shRNA and N1ICD overexpression. Data are means (n = 3) ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus shLacZ + β-gal double control.
Overexpression of SHH-N was found to diminish the up-regulation of cell-cycle gene expression in MEF2A- or MEF2D-deficient NRVMs (Fig. 6B, top). SHH-N alone also had a repressive effect on cell-cycle gene expression. In contrast, SHH-N had no significant effect on the expression of sarcomere genes either in the presence or absence of MEF2A or -D (Fig. 6B, bottom). Finally, inhibition of MEF2C in NRVMs overexpressing SHH-N displayed variable and inconsistent expression patterns on both cell-cycle and sarcomere gene programs (data not shown).
Conversely, constitutive Notch activity further up-regulated cell-cycle gene expression in both MEF2A- and MEF2D-deficient NRVMs, whereas MEF2C inhibition had the opposite effect (Fig. 6C, top). Notch alone had a modest effect on the expression of cell-cycle genes. These data suggest that the presence of MEF2A or -D, but not MEF2C, represses the ability of Notch signaling to activate cell-cycle gene expression. In a reciprocal fashion, Notch-mediated activation of sarcomere genes required the presence of MEF2A or -D, but not MEF2C (Fig. 6C, bottom). Interestingly, α-actinin immunofluorescence revealed that activation of Hedgehog or Notch in MEF2A- or D-deficient NRVMs was unable to rescue the abnormal morphology and number of cardiomyocytes (Fig. 7, A and B).
Figure 7.
Hedgehog or Notch signaling does not alter the morphology of Mef2-depleted NRVMs. A, immunofluorescent images of NRVMs treated with Mef2a shRNA and an overexpression construct show that overexpression of constitutively active N1ICD and SHHN does not modulate the ratio of α-actinin-positive cells to total nuclei in negative control (shLacZ) and shMef2a-treated cultures, but overexpression of N1ICD does reduce the number of α-actinin-positive cells in shMef2c-treated cultures, and SHHN does reduce the number of α-actinin-positive cells in shMef2d-treated cultures. B, quantification of α-actinin-positive cells to total nuclei in n = 3 fields/treatment group. C, model of MEF2A and -D regulation of Notch and Hedgehog signaling in NRVMs. Data are means (n = 3) ± S.D. (error bars) versus shRNA + β-gal negative control.
As depicted in the model in Fig. 7C, the ability of the Hedgehog signaling pathway to modulate MEF2A- and MEF2D-dependent gene regulation was restricted to the cell-cycle program. By contrast, Notch modulated MEF2-dependent regulation of both cell-cycle and differentiation programs. Specifically, this signaling pathway exacerbated the effect of MEF2A and -D, but not MEF2C, knockdown on cell-cycle programs in NRVMs. Conversely, the effect of Notch on sarcomere gene expression was enhanced by MEF2C, but not MEF2A or -D, inhibition.
Discussion
The mechanisms of coordinating cell-cycle and differentiation programs in cardiomyocytes remain poorly understood. Here, we have used acute isoform-specific knockdown of the mammalian MEF2 proteins to demonstrate that protein isoforms of this evolutionarily conserved, core cardiac transcription factor have distinct regulatory roles in neonatal cardiomyocytes. Interestingly, we have also uncovered a previously uncharacterized antagonistic regulatory role between individual members of the MEF2 family in the regulation of cell-cycle and differentiation gene programs. These disparate roles are most evident in cardiomyocyte survival, where a differential requirement of specific MEF2 isoforms is observed for this process. These results reveal a mechanism whereby antagonistic regulatory pathways can function simultaneously to promote proper development and growth of the heart.
To our knowledge, MEF2 represents the first transcription factor to have a dual and antagonistic function in the regulation of both cell-cycle and differentiation (sarcomere) gene programs in cardiomyocytes. The forkhead box transcription factor family was previously demonstrated to have antagonistic roles in neonatal cardiomyocyte cell-cycle withdrawal and proliferation (34). However, the opposing function of two family members, FoxO and FoxM1, was restricted to the cell-cycle program. These TFs were shown to coordinate the regulation of cell-cycle activators and inhibitors depending on nutrient availability or growth factor levels. FoxO was demonstrated to prevent neonatal cardiomyocyte proliferation through direct negative regulation of IGF1, whereas FoxM1 positively regulated this growth factor gene. In addition, these differential regulatory effects were mediated upstream by the activation status of AMPK. Because differentiation genes were not examined in that study, it is unknown whether the forkhead TFs regulate this gene program in an antagonistic fashion.
Another example of antagonistic transcriptional regulation is found in the relationship between Irx3 and Irx5. These two transcription factors share redundant functions during the development of the mammalian heart but appear to play divergent roles in adult cardiac electrophysiology (35). Single knockouts of each Irx factor lead to a loss in appropriate depolarization gradient, but double knockouts restore the appropriate polarization gradient. These results suggest that Irx5 is required to repress the function of Irx3 in specific regions of the adult heart and provides an additional example of homologous cardiac transcription factors playing an antagonistic relationship that is required for normal cardiac homeostasis.
We have shown that MEF2A and MEF2D are necessary for neonatal cardiomyocyte survival and that these factors were able to diminish programmed cell death, as determined by cleaved caspase-3 activity, in the context of MEF2A deficiency. Interestingly, MEF2C overexpression in MEF2A-deficient NRVMs failed to rescue cleaved caspase-3 activation but was capable of reducing apoptotic DNA degradation. Ultimately, MEF2C overexpression had no effect on survival, suggesting that MEF2C activity is not functionally redundant with and cannot compensate for MEF2A in the more proximal events of programmed cell death.
We have previously described canonical pathways in the common (or co-regulated) set of dysregulated genes in individual MEF2 knockdowns in C2C12 skeletal myoblasts (25). Because striated muscle cell types share many gene programs, we compared the co-regulated MEF2 pathways between C2C12 cells and NRVMs. Overall, there was no obvious overlap in the pathways regulated by all MEF2 isoforms between skeletal and cardiac myocytes. The co-regulated MEF2 pathways in C2C12 cells were dominated by signal transduction cascades, whereas the co-regulated pathways in NRVMs were predominantly associated with cell-cycle control. This difference may reflect the contexts in which the analyses were performed, such that the co-regulated MEF2-dependent cell signaling is more important for the differentiation process (myoblast-to-myotube), whereas precise cell-cycle control is necessary for permanent cell-cycle withdrawal and maturation of differentiated neonatal myocytes.
Comparison of pathways preferentially sensitive to inhibition of individual MEF2 proteins in skeletal myoblast with our present data in cardiomyocytes shows distinct roles for MEF2 isoforms in these related striated muscle cell types. Specifically, comparison of gene sets preferentially sensitive to MEF2A in cardiomyocytes and skeletal myoblasts revealed that only two pathways, molecular mechanisms of cancer and germ cell-Sertoli cell junction signaling, were shared in both muscle cell types. The remaining pathways in NRVMs and skeletal myoblasts dependent on MEF2A did not have any obvious functional relationships to each other. A recently published study investigated the overlap of MEF2A binding in skeletal myoblasts and cardiomyocytes using ChIP-exo (36). Whereas many of their enriched gene ontology terms were distinct from our canonical pathway analysis, induction of apoptosis and cell death were among the enriched pathways in cardiomyocytes. These results support our observations that MEF2A is required for cardiomyocyte survival.
Interestingly, several cell-cycle pathways (G1/S checkpoint regulation and estrogen-mediated S-phase entry) were shown to be significantly enriched in MEF2C-deficient skeletal myoblasts (25). This is in stark contrast to the enrichment of this gene program in MEF2A- or MEF2D-deficient cardiomyocytes. The most significantly enriched canonical pathways preferentially dysregulated by MEF2C inhibition in NRVMs are associated with energy metabolism, suggesting a distinct role for MEF2C in cardiomyocyte metabolism that is not shared in skeletal muscle. Because NRVMs were cultured in medium containing minimal fatty acids, it would be interesting to evaluate the requirement of MEF2C in myocyte survival under different metabolic conditions. Despite these differences, MEF2C had a significant regulatory effect on cell-cycle gene expression in cardiomyocytes. The observation that MEF2C positively regulates cell-cycle genes may reflect its role in cardiac reprogramming (37), such that its activity is required to drive early steps in differentiation and coordinate proliferation of immature cardiomyocytes.
Inhibition of MEF2D displayed the most disparate effect on pathway enrichment in both striated muscle cell types. There was very little overlap between specific pathways preferentially dysregulated in skeletal and cardiac myocytes. For this MEF2 isoform, cytokine, hypoxia, and AMPK signaling were enriched in skeletal myoblasts, whereas HIPPO, PI3K, and neuregulin pathways were sensitive to MEF2D in NRVMs. The majority of pathways preferentially dysregulated by MEF2D inhibition in both models are generally associated with signal transduction, suggesting a distinct role for MEF2D in integrating cell signaling in both muscle cell types.
The presence of canonical MEF2-binding sites in both cell-cycle and sarcomere gene sets suggested that differential regulation might be mediated by program-exclusive co-regulators. Indeed, we observed minimal overlap in overrepresented transcription factor-binding sites between the cell-cycle and sarcomere genes. Among the unique transcription factor-binding sites enriched in cell-cycle and sarcomere genes were those belonging to TFs that mediate Hedgehog and Notch signaling, respectively. The Hedgehog signaling pathway has an early role in patterning of the developing heart (33). Curiously, the Hedgehog signaling pathway has been shown to regulate Mef2c expression to promote cardiomyocyte differentiation in P19 embryocarcinoma cells (38). Our present report shows that Hedgehog also functions in differentiated cardiomyocytes, and in these postnatal myocytes, it modulates MEF2A- and MEF2D-dependent regulation of the cell cycle but not the sarcomere gene program.
Notch has been shown to play an important role in cardiac morphogenesis in multiple cell lineages in the heart, including cardiomyocytes (32). In addition, previous studies have shown that the Notch signaling pathway modulates MEF2 activity in mammalian skeletal myoblasts (39, 40). Notch also synergizes with MEF2 to promote proliferation in fly development (41). Although a specific function for Notch has not been ascribed to sarcomere gene regulation, it has been shown to promote cell-cycle reentry of neonatal cardiomyocytes (42). Our present data reveal that Notch induction of proliferative markers is repressed by MEF2A and -D, introducing a potential mechanism for the loss of proliferative capacity observed in those studies. Recently, inhibition of Notch was shown to increase MEF2C binding on structural gene promoters in the context of cardiac reprogramming (43). We show that activation of Notch and inhibition of MEF2C together in NRVMs further up-regulated Notch-induced structural gene expression, indicating a differential, context-specific utilization of this pathway in the regulation of cardiac structural genes. Taken together, our results reinforce the relevance of a Notch-MEF2 transcriptional pathway in cardiomyocytes and that modulation of MEF2 isoform-specific activity by this upstream signal is a key mechanism coordinating cell-cycle control and differentiation in cardiomyocytes.
We have demonstrated that protein isoforms of the mammalian MEF2 transcription factor family antagonistically regulate cell-cycle and differentiation gene programs in neonatal cardiomyocytes. Moreover, the ability of MEF2A, -C, and -D to regulate disparate gene programs is modulated by Hedgehog and Notch signaling. The results of this study point to an intricate regulatory mechanism involving the differential utilization of homologous members of a core cardiac transcription factor that determines the differentiation state of cardiomyocytes. Relating to this point, the coexistence of active and repressive MEF2 complexes in tumors was recently described (44). Altogether, functional differences in MEF2-dependent gene regulation may arise from their relative expression levels, distinct dimeric combinations, interaction with transcriptional and chromatin cofactors, and/or modulation of their activity by key developmental signaling pathways. In the future, it would be interesting to determine whether the TFs that function downstream of Notch and Hedgehog physically and genetically interact with MEF2 protein isoforms to coordinate the antagonistic regulation of cell-cycle and differentiation programs in cardiac development and reprogramming.
Experimental procedures
Cell culture
NRVMs were isolated from 1–2-day-old SASCO Sprague-Dawley neonatal rats (Charles River Laboratories). Briefly, neonatal rats were sacrificed by decapitation, and whole hearts were removed. Ventricles were then isolated and transferred to 1× Hanks' buffered saline solution with 0.06% trypsin and incubated overnight at 4 °C. The following day, 10 mg/ml collagenase II (Worthington) in 1× Hanks' buffered saline solution was added to isolate individual cardiomyocytes. Resulting suspensions were preplated to remove contaminating fibroblasts and plated at a density of 1 × 106 cells/well in 6-well dishes in DMEM (Invitrogen) containing 10% fetal bovine serum. All tissue culture plates that were seeded with NRVMs were pretreated with 0.1% gelatin for at least 1 h before seeding. After 24 h in culture, NRVMs were washed with 1× PBS, and cells were placed in DMEM with 0.5× Nutridoma-SP (Roche Applied Sciences).
Adenoviral transduction
Adenoviral vectors bearing shRNA constructs were generated as described previously (25). Mef2 shRNA adenoviruses were used at a multiplicity of infection (MOI) of 50 for all RNA collection and combinatorial knockdown experiments. An MOI of 25 was used in the MEF2 overexpression rescue experiments. β-gal, MEF2A, MEF2C, MEF2D, and MEF2-VP16 overexpression adenoviruses were a generous gift from Jeff Molkentin (Children's Hospital, Cincinnati, OH). For Mef2a knockdown rescue experiments, MEF2A overexpression adenovirus was used at an MOI of 1.25, and other overexpression adenoviruses were used at an MOI of 7.5. For Mef2d knockdown rescue experiments, MEF2A overexpression adenovirus was used at an MOI of 2.5, and other overexpression adenoviruses were used at an MOI of 15. SHH-N overexpression adenovirus was a kind gift of Ronald G. Crystal (Weill Cornell Medicine) and was used at an MOI of 25. N1ICD overexpression adenovirus was a kind gift of Igor Prudovsky (Maine Medical Center Research Institute) and was used at an MOI of 2.5.
Comparative microarray analysis
Forty-eight hours after transduction, total RNA from shLacZ (n = 3)-, shMef2a (n = 3)-, shMef2c (n = 3)-, and shMef2d (n = 3)-treated NRVMs was prepared via TRIzol® isolation (Invitrogen). RNA quality was evaluated using a Bioanalyzer (Agilent), and samples were hybridized to a Rat Gene 2.0 ST at the Boston University Microarray Core Facility (n = 3 per shRNA, 12 arrays total). Microarray data are available in GEO (NCBI) with accession number GSE92861.
Quantitative RT-PCR
Total RNA from NRVM Mef2 knockdown experiments was isolated using TRIzol® isolation and was used to synthesize cDNA using reverse transcriptase with random hexamers (Promega). Quantitative RT-PCR was performed in triplicate wells using SYBR® Green Master Mix (Applied Biosystems) with the 7900HT sequence detection system (Applied Biosystems). The primers used can be found in Table 4. qRT-PCR data were analyzed using the ΔΔCT method.
Table 4.
Rat qRT-PCR primers used in this study
| Gene | Forward | Reverse |
|---|---|---|
| Gapdh | 5′-TGGCAAGTGGAGATTGTTGCC | 5′-AAGATGGTGATGGGCTTCCCG |
| Mef2a | 5′-GAACTCAGTGTGCTCTGTGACTGTGAG | 5′-GCCAGTGCTTGGTGGTCTCT |
| Mef2b | 5′-GAAAGAAAGCCGCTCTGCACAG | 5′-ACCTTCTGGCCCCTCCTCCATA |
| Mef2c | 5′-CAGGGACGAGAGAGAGAAGAAAC | 5′-CAATCTTTGCCTGCTGATCATTAG |
| Mef2d | 5′-CTTTCCTCTCTGGCACTAAGGAC | 5′-CCAGTCTATAACTCTGCATCATC |
| Dnm3 | 5′-TAACCACATCCGTGAGCGAG | 5′-GCGGAAGATTGGTTCCCTGA |
| Ccl5 | 5′-GCTTTGCCTACCTCTCCCTC | 5′-TCCTTCGAGTGACAAAGACGA |
| Filip1 | 5′-GCAGGAGCGAGAGAGGTTGA | 5′-CATCAGGGCGAAGGACTTGA |
| Lrrc39 | 5′-CACGGAGAACAGAAGACCAAG | 5′-CATGATGTCTTCCACAAGCAAA |
| Cirbp | 5′-AGACTACTATGCCAGCCGGA | 5′GGACGCAGAGGGCTTTTACT |
| Adamtsl2 | 5′-CTCACAAGGCAAGGACCAGAC | 5′-CCACTTCAACGCCATCGTAG |
| Cdh8 | 5′-GGAGCCCGACCTGAGAAAT | 5′-TCTAAGCAGCTTTTCCAAGACCA |
| Mdga2 | 5′-CTCATCGTGCAGTATCCCCC | 5′-ACGCCATTCGTAAGTCAGCA |
| Kit | 5′TTTAAAGGTAACAGCAAAGAGCAA | 5′-GTGACCACGAAGCCAATGAG |
| Sept4 | 5′-GGACTGAAGCTGGGGATGAC | 5′-CCGATCCCGGTACAAGTCAG |
| Upk1b | 5′-CAGCCAGTCCAGTGGGAAAT | 5′-GGCGATCCCACACATACCAA |
| Pten | 5′-ACTGCAGAGTTGCACAGTATCCTT | 5′-GCCTCTGACTGGGAATAGTTACTCC |
| Mcm3 | 5′-AACCCGTTCCAAGGATGTCTTTGAG | 5′-GGTTTCCTGTCTGTGGTGACG |
| Mcm5 | 5′-GGACATGATGCTGGCCAAACATGT | 5′-GGCTGCAGTTTCATCTTGCTGAGG |
| Mcm6 | 5′-GACTTCCTGGAAGAGTTCCAGGG | 5′-CGATCCTGGAGGAAGTGAGCTC |
| Pcna | 5′-CGTGAACCTCACCAGCATGTCC | 5′-CCAAGTTGCTCAACGTCTAAGTCCA |
| Ccne1 | 5′-CCAGGATAGCAGTCAGCCTTGG | 5′-TGCTCTCATCCTCGCCTGC |
| Ccne2 | 5′-AATTGTTGGCCACCTGTACTGTCTG | 5′-ACTTCACAGACCTCTAAAAGCCAGTCT |
| E2f3 | 5′-AGGAGCGAGAGATGAGAAAGG | 5′-GTGGTGAGGATCTGGATGTACG |
| Myh7 | 5′-GGAAGAACCTACTGCGACTGCAGGACC | 5′-TGTTTCAAAGGCTCCAGGTCTCAGGGC |
| Myl2 | 5′-GAAGGCCGACTATGTCCGGG | 5′-TGGGGATGGAGAACAGGCTA |
| Myom1 | 5′-GAGAAAAATCGGGCTCGGGT | 5′-GCAGGTGAGATTGAGTGCCT |
| Myom2 | 5′-AAGCCTCTTTGTCTCCCGAA | 5′-TCCCAGAAAGATGAGGAGTACC |
| Ttn | 5′-CACCACCAGTCCCAGAAGTT | 5′-AGACTGCTTCCTTCCGTTCA |
| Ptch1 | 5′-AAGTGTTGCCCCAAACTCCA | 5′-AACAGGCGTAGGCAAGCATC |
| Gli1 | 5′-CTGGTCTGCCCTTTTGCCAC | 5′-GAAAGAGTGACCCCTCAGTGCAG |
| Gli2 | 5′-TCACCATCCATAAGCGGAGC | 5′-GTTGCTCCTGTGTCAGTCCA |
| Gli3 | 5′-TTCCTCCATTACACGTGCCT | 5′-GTGCAAGGAGCGGATGTAGT |
| Hey2 | 5′-AGGGTGTCCGTAGCTCTTCT | 5′-ACTGTGCCCCGGAGTAATTGT |
| Hes1 | 5′-AGCACAGAAAGTCATCAAAGCC | 5′-CTTGGAATGCCGGGAGCTAT |
Immunocytochemistry
Glass coverslips were acid-etched by incubating in a 1 n HCl solution at 65 °C overnight. Coverslips were then rinsed in deionized H2O six times and then rinsed once in 70% ethanol and allowed to dry in a tissue culture hood under UV light irradiation for at least 1 h. Prepared coverslips were then transferred to 6-well plates, and gelatinized and NRVMs were seeded as described above.
NRVMs were transduced and incubated in standard cell culture conditions for 72 h. Cells were then fixed by washing once in 1× PBS and then fixed for 15 min in freshly prepared 4% formaldehyde at room temperature. Fixative was then removed, and coverslips were washed three times in 1× PBS and then stored at 4 °C for up to 2 weeks before immunostaining.
Before staining, coverslips were blocked with Mouse on Mouse (Vector Laboratories) for 1 h at room temperature. During incubation, antibody dilution buffer (0.1% BSA, 0.3% Triton X-100 in 1× PBS) was prepared fresh, and a working primary α-actinin (1:500; A7811, Sigma-Aldrich) antibody solution was prepared. Mouse on Mouse was aspirated, and coverslips were incubated in primary antibody solution overnight at 4 °C with gentle shaking. Primary antibody was aspirated and coverslips were washed three times for 5 min with 1× PBS, and then coverslips were incubated in secondary antibody solution (1:500 AlexaFluor® 586 donkey α-mouse IgG; A10037, Invitrogen) for 1 h at room temperature in the dark. Secondary antibody solution was aspirated, coverslips were rinsed three times for 6 min with 1× PBS, and then coverslips were mounted onto slides with Vectashield with DAPI (Vector) and sealed. Slides were imaged within 1 week of staining using an Olympus DSU spinning disk confocal microscope.
Cell viability assays
NRVMs were cultured in 24-well plates at a cell density of 8 × 104 cells/well and transduced with adenovirus. NRVMs were cultured for 48 h, and then 20 μl of CellTiter-Blue® reagent (Promega) was added to each well. Plates were incubated for 16 h at 37 °C in a tissue-culture incubator, and fluorescence of the medium was measured at 560/590 nm using a Victor3 plate reader (PerkinElmer Life Sciences). Data were normalized to untreated control wells lacking NRVMs.
NRVMs were cultured at a cell density of 1 × 106 cells/well, and total protein lysate was collected. Protein lysates were combined with a fluorogenic caspase-3 substrate, Ac-DEVD-7-amido-4-methylcoumarin (BD Biosciences) to a final concentration of 50 μm. Lysates were then incubated for 1 h at 37 °C. Fluorescence was measured at 440/460 nm using a Victor3 plate reader (PerkinElmer Life Sciences). Data were normalized to total protein levels.
Hypodiploid DNA content was measured using propidium iodide staining and flow cytometry. Briefly, NRVMs were plated at a cell density of 1 × 106 cells/well, transduced with adenovirus, and cultured for a further 72 h. NRVMs were then scraped into the existing medium to retain any cells that had detached from the plate and centrifuged at 1,000 × g for 5 min at 4 °C. Pellets were resuspended in 3.33× PBS, and ice-cold 100% ethanol was added to resuspended pellet to a final concentration of 1× PBS. Cells were stored up to 2 weeks in ethanol solution at 4 °C. On the day of flow cytometry, NRVMs were pelleted as described above and washed once with 1× PBS. The NRVMs were resuspended in propidium iodide staining solution (50 μg/ml propidium iodide (Sigma-Aldrich), 2× PBS, 10 μg/ml RNase A, prepared fresh) for 30 min at room temperature in the dark. Post-staining, cells were analyzed using a FACSCalibur flow cytometer (488-nm excitation/585-nm emission).
Computational analysis
Microarray data were annotated with Entrez ID numbers. Statistically significant dysregulation of each gene was evaluated in the individual Mef2-deficient microarrays. Significantly dysregulated genes were sorted into groups based on shared dysregulation between the various treatment groups. IPA (Ingenuity Systems) was used to determine canonical cellular pathways associated with the same gene sets.
Co-regulatory factor enrichment analysis was performed on the above-mentioned proximal promoter region of genes co-regulated by the MEF2 factors using MatInspector from the Genomatix software suite. Analysis was constricted to regions within 50 bp of putative MEF2-binding sites to enrich for potential interactions. The analytical background was composed of a cross-section of genomic promoter sequences to discriminate between enriched motifs and generic promoter regions. Resulting enriched motifs were sorted by Z-score, with a Z-score > 2.0 being considered significantly enriched. Additional data about enriched motifs were extracted from the Genomatix MatBase and NCBI databases.
Statistical analyses
All numerical quantification is representative of the mean ± S.E. of at least three independently performed experimental replicates. Statistically significant differences between two populations of data were determined using Student's t test. p values ≤ 0.05 were considered statistically significant. In cases of qRT-PCR analysis where the number of independent replicates exceeded three, a quartile outlier test was applied to determine significant outlier data. These points were removed, and the expressed quantification does not include these values.
The technical quality of the microarrays was determined using relative log expressions and normalized unscaled S.E. Relative log expressions and normalized unscaled error values greater than 0.1 and 1.05, respectively, are considered out of normal limits. All arrays had median values within the normal limits of these tests. Microarray data were normalized using the robust multiarray average algorithm and were log2-transformed by default. Knockdown efficiency of MEF2 shRNA was determined by calculating the -fold change for each MEF2 isoform knockdown relative to the shLacZ negative control. Significant dysregulation of gene expression was determined using a one-way analysis of variance, and the Benjamini-Hochberg false-discovery rate correction was applied to obtain corrected q values, and a q threshold of < 0.05 was used to determine significant dysregulation. Tukey's honest significant difference post hoc test was performed to identify significantly dysregulated genes and correct for multiple testing error across all intergroup comparisons. A corrected q value of < 0.05 was used to determine statistically significant gene dysregulation among gene groups.
Author contributions
C. A. D. and F. J. N. designed the study and wrote the paper. C. A. D. performed and analyzed all of the experiments in this manuscript. Both authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Naya laboratory for critical reading of the manuscript. We are grateful to Jeff Molkentin (Cincinnati Children's Hospital) for providing the MEF2 overexpression adenoviruses, Ronald Crystal (Weill Cornell Medicine) for the SHH-N adenovirus, and Igor Prudovsky (Maine Medical Center Research Institute) for the N1ICD adenovirus. The Boston University Microarray Core CTSI was instrumental in the analysis of the gene expression data sets.
This work was supported by National Institutes of Health Grant HL73304 (to F. J. N.) and Boston University Bioinformatics CTSI Grant U54-TR001012. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51971).
- TF
- transcription factor
- NRVM
- neonatal rat ventricular myocyte
- qRT-PCR
- quantitative RT-PCR
- IPA
- Ingenuity Pathway Analysis®
- MOI
- multiplicity of infection
- bHLH
- basic helix-loop-helix.
References
- 1. Vincent S. D., and Buckingham M. E. (2010) How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1–41 [DOI] [PubMed] [Google Scholar]
- 2. Srivastava D. (2006) Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–1048 [DOI] [PubMed] [Google Scholar]
- 3. Paige S. L., Plonowska K., Xu A., and Wu S. M. (2015) Molecular regulation of cardiomyocyte differentiation. Circ. Res. 116, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olson E. N. (2006) Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waardenberg A. J., Ramialison M., Bouveret R., and Harvey R. P. (2014) Genetic networks governing heart development. Cold Spring Harb. Perspect. Med. 4, a013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Potthoff M. J., and Olson E. N. (2007) MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 7. Desjardins C. A., and Naya F. J. (2016) The function of the MEF2 family of transcription factors in cardiac development, cardiogenomics, and direct reprogramming. J. Cardiovasc. Dev. Dis. 10.3390/jcdd3030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hinits Y., Pan L., Walker C., Dowd J., Moens C. B., and Hughes S. M. (2012) Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 369, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y. X., Qian L. X., Yu Z., Jiang Q., Dong Y. X., Liu X. F., Yang X. Y., Zhong T. P., and Song H. Y. (2005) Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 579, 4843–4850 [DOI] [PubMed] [Google Scholar]
- 10. Guo Y., Kühl S. J., Pfister A. S., Cizelsky W., Denk S., Beer-Molz L., and Kühl M. (2014) Comparative analysis reveals distinct and overlapping functions of Mef2c and Mef2d during cardiogenesis in Xenopus laevis. PLoS One 9, e87294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Q., Schwarz J., Bucana C., and Olson E. N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Naya F. J., Black B. L., Wu H., Bassel-Duby R., Richardson J. A., Hill J. A., and Olson E. N. (2002) Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 13. Kim Y., Phan D., van Rooij E., Wang D. Z., McAnally J., Qi X., Richardson J. A., Hill J. A., Bassel-Duby R., and Olson E. N. (2008) The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J. Clin. Invest. 118, 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Black B. L., and Olson E. N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 15. Estrella N. L., and Naya F. J. (2014) Transcriptional networks regulating the costamere, sarcomere, and other cytoskeletal structures in striated muscle. Cell Mol. Life Sci. 71, 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahuja P., Sdek P., and MacLellan W. R. (2007) Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol. Rev. 87, 521–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pasumarthi K. B., and Field L. J. (2002) Cardiomyocyte cell cycle regulation. Circ. Res. 90, 1044–1054 [DOI] [PubMed] [Google Scholar]
- 18. Zacchigna S., and Giacca M. (2014) Extra- and intracellular factors regulating cardiomyocyte proliferation in postnatal life. Cardiovasc. Res. 102, 312–320 [DOI] [PubMed] [Google Scholar]
- 19. Foglia M. J., and Poss K. D. (2016) Building and re-building the heart by cardiomyocyte proliferation. Development 143, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N., and Sadek H. A. (2011) Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray T. V., Ahmad A., and Brewer A. C. (2014) Reactive oxygen at the heart of metabolism. Trends Cardiovasc. Med. 24, 113–120 [DOI] [PubMed] [Google Scholar]
- 22. Murphy A. M. (1996) Contractile protein phenotypic variation during development. Cardiovasc. Res. 31, E25–E33 [PubMed] [Google Scholar]
- 23. Ewen E. P., Snyder C. M., Wilson M., Desjardins D., and Naya F. J. (2011) The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J. Biol. Chem. 286, 29644–29653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Estrella N. L., Clark A. L., Desjardins C. A., Nocco S. E., and Naya F. J. (2015) MEF2D deficiency in neonatal cardiomyocytes triggers cell cycle re-entry and programmed Cell death in vitro. J. Biol. Chem. 290, 24367–24380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Estrella N. L., Desjardins C. A., Nocco S. E., Clark A. L., Maksimenko Y., and Naya F. J. (2015) MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J. Biol. Chem. 290, 1256–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edmondson D. G., Lyons G. E., Martin J. F., and Olson E. N. (1994) Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 27. Conway S. J., Firulli B., and Firulli A. B. (2010) A bHLH code for cardiac morphogenesis. Pediatr. Cardiol. 31, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. VanDusen N. J., and Firulli A. B. (2012) Twist factor regulation of non-cardiomyocyte cell lineages in the developing heart. Differentiation 84, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bondue A., and Blanpain C. (2010) Mesp1: a key regulator of cardiovascular lineage commitment. Circ. Res. 107, 1414–1427 [DOI] [PubMed] [Google Scholar]
- 30. Niu Z., Li A., Zhang S. X., and Schwartz R. J. (2007) Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 19, 618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evans R. M., and Mangelsdorf D. J. (2014) Nuclear receptors, RXR, and the big bang. Cell 157, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de la Pompa J. L., and Epstein J. A. (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22, 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rochais F., Mesbah K., and Kelly R. G. (2009) Signaling pathways controlling second heart field development. Circ. Res. 104, 933–942 [DOI] [PubMed] [Google Scholar]
- 34. Sengupta A., Kalinichenko V. V., and Yutzey K. E. (2013) FoxO1 and FoxM1 transcription factors have antagonistic functions in neonatal cardiomyocyte cell-cycle withdrawal and IGF1 gene regulation. Circ. Res. 112, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gaborit N., Sakuma R., Wylie J. N., Kim K. H., Zhang S. S., Hui C. C., and Bruneau B. G. (2012) Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development 139, 4007–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wales S., Hashemi S., Blais A., and McDermott J. C. (2014) Global MEF2 target gene analysis in cardiac and skeletal muscle reveals novel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res. 42, 11349–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., and Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Voronova A., Al Madhoun A., Fischer A., Shelton M., Karamboulas C., and Skerjanc I. S. (2012) Gli2 and MEF2C activate each other's expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res. 40, 3329–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilson-Rawls J., Molkentin J. D., Black B. L., and Olson E. N. (1999) Activated notch inhibits myogenic activity of the MADS-box transcription factor myocyte enhancer factor 2C. Mol. Cell Biol. 19, 2853–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen H., McElhinny A. S., Cao Y., Gao P., Liu J., Bronson R., Griffin J. D., and Wu L. (2006) The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20, 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pallavi S. K., Ho D. M., Hicks C., Miele L., and Artavanis-Tsakonas S. (2012) Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO J. 31, 2895–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Campa V. M., Gutiérrez-Lanza R., Cerignoli F., Díaz-Trelles R., Nelson B., Tsuji T., Barcova M., Jiang W., and Mercola M. (2008) Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J. Cell Biol. 183, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abad M., Hashimoto H., Zhou H., Morales M. G., Chen B., Bassel-Duby R., and Olson E. N. (2017) Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports 8, 548–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Di Giorgio E., Franforte E., Cefalù S., Rossi S., Dei Tos A. P., Brenca M., Polano M., Maestro R., Paluvai H., Picco R., and Brancolini C. (2017) The co-existence of transcriptional activator and transcriptional repressor MEF2 complexes influences tumor aggressiveness. PLoS Genet. 13, e1006752. [DOI] [PMC free article] [PubMed] [Google Scholar]