Abstract
Toll-like receptor 2 (TLR2) plays a critical role in host defenses against mycobacterial infections. The R753Q TLR2 polymorphism has been associated with increased incidence of tuberculosis and infections with non-tuberculous mycobacteria in human populations, but the mechanisms by which this polymorphism affects TLR2 signaling are unclear. In this study, we determined the impact of the R753Q TLR2 polymorphism on macrophage sensing of Mycobacterium smegmatis. Upon infection with M. smegmatis, macrophages from knock-in mice harboring R753Q TLR2 expressed lower levels of TNF-α, IL-1β, IL-6, and IL-10 compared with cells from WT mice, but both R753Q TLR2- and WT-derived macrophages exhibited comparable bacterial burdens. The decreased cytokine responses in R753Q TLR2-expressing macrophages were accompanied by impaired phosphorylation of IL-1R-associated kinase 1 (IRAK-1), p38, ERK1/2 MAPKs, and p65 NF-κB, suggesting that the R753Q TLR2 polymorphism alters the functions of the myeloid differentiation primary response protein 88 (MyD88)-IRAK-dependent signaling axis. Supporting this notion, HEK293 cells stably transfected with YFP-tagged R753Q TLR2 displayed reduced recruitment of MyD88 to TLR2, decreased NF-κB activation, and impaired IL-8 expression upon exposure to M. smegmatis. Collectively, our results indicate that the R753Q polymorphism alters TLR2 signaling competence, leading to impaired MyD88-TLR2 assembly, reduced phosphorylation of IRAK-1, diminished activation of MAPKs and NF-κB, and deficient induction of cytokines in macrophages infected with M. smegmatis.
Keywords: innate immunity, macrophage, signal transduction, SNP, Toll-like receptor (TLR)
Introduction
Mycobacterium tuberculosis (Mtb)2 targets over a third of the world population, causing tuberculosis or latent infection that could be reactivated because of co-infection with HIV, diabetes, malnutrition, or immunosuppressive interventions (1–3). Mtb infects alveolar macrophages and results in the formation of granulomas where bacilli reside for the period of latency, whereas during active disease, the granuloma becomes necrotic and caseous, leading to Mtb escape and spreading (3, 4). Macrophages detect mycobacterial lipoproteins, lipoarabinomannan, or DNA via membrane-associated TLRs or by cytosolic nucleotide-binding oligomerization domain-containing protein 2, nucleotide-binding oligomerization domain-like receptor, pyrin domain-containing protein 3, absent in melanoma 2, and cyclic GMP-AMP synthase (2, 5–8).
TLRs are expressed on the cell surface (e.g. TLR5), in endosomes (TLR3 and TLR7–9), or in both compartments (e.g. TLR2 and TLR4) (9) in macrophages, neutrophils, dendritic cells, or epithelial and endothelial cells (10). TLR2 plays a critical role in the detection of and host defense against Mtb and other mycobacteria, including Mycobacterium bovis BCG, Mycobacterium avium, or Mycobacterium smegmatis (11–17). Upon binding of ligands to the ectodomain, TLR2 heterodimerizes with TLR1 or TLR6, leading to Toll-IL-1R (TIR) domain assembly to recruit myeloid differentiation primary response protein 88 (MyD88) adapter-like and MyD88 (18). Subsequently, several molecules of IL-1R-associated kinase 4 (IRAK-4), IRAK-1, and IRAK-2 are recruited and interact with MyD88 via their death domains, activating IRAK activities and initiating recruitment of TNFR-associated factor 6 (TRAF6) (19, 20). TRAF6 interacts with and activates TGF-β-activating kinase 1, which propagates signals downstream, activating transcription factors and inducing inflammatory cytokines (21).
Genome-wide association studies have identified several polymorphisms within TLR2, including R753Q, R677W, and P631H, that have been associated with increased risk of infection with Mtb, M. avium, or Mycobacterium abscessus as well as higher incidences of tuberculosis and leprosy (22–29). Our previous studies in HEK293 transfectants stimulated with TLR2 agonists or irradiated Mtb have shown that R753Q TLR2 undergoes deficient tyrosine phosphorylation, compromised TLR6 assembly, and impaired recruitment of Mal and MyD88 (30). However, the impact of the R753Q polymorphism on the signaling capacities of endogenous TLR2 in macrophages during infection with live mycobacteria has not been studied. The mechanisms by which signaling deficiencies of R753Q TLR2 could underlie increased susceptibility to Mtb infection in human carriers of TLR2 SNPs are unknown.
In this study, we sought to determine the impact of the R753Q TLR2 polymorphism on macrophage responses to live infection with M. smegmatis. Using macrophages from knock-in (KI) mice harboring endogenous R753Q TLR2, we show that the R753Q polymorphism impairs activation of IRAK-1, NF-κB, and MAPKs and induction of inflammatory cytokines during infection. HEK293 transfectants expressing R753Q TLR2 exhibited deficient recruitment of MyD88 to the mutant TLR2 upon exposure to live M. smegmatis. To the best of our knowledge, this is the first demonstration of deficient MyD88-TLR2 assembly and activation of IRAK-1 as the mechanism by which the R753Q polymorphism impairs innate responses of macrophages during infection with live mycobacteria.
Results
The R753Q TLR2 SNP impairs MAPK and NF-κB activation and IL-8 induction upon exposure of HEK293/TLR2 transfectants to M. smegmatis or M. bovis BCG
To examine the impact of the R753Q TLR2 SNP on cell responses to live mycobacteria, we first used HEK293 cells stably expressing YFP-tagged human WT (293/TLR2WT) or R753Q TLR2 (293/TLR2R753Q). Cell activation was judged by phosphorylation of p38 MAPK and p65 NF-κB, activation of the NF-κB-driven luciferase reporter, and induction of IL-8 expression. Exposure of 293/TLR2WT cells to M. smegmatis (multiplicity of infection (m.o.i.) = 50) markedly enhanced phosphorylation of p38 MAPK and p65 NF-κB (Fig. 1A) and led to a 6.1-fold increase in NF-κB reporter activation (Fig. 1B). In contrast, 293/TLR2R753Q cells showed deficient phosphorylation of p38 MAPK and p65 NF-κB and impaired activation of the NF-κB reporter (78% decrease) in response to M. smegmatis and M. bovis BCG (Fig. 1).
Figure 1.
The TLR2 R753Q polymorphism attenuates TLR2-mediated p38 MAPK and NF-κB activation and inhibits IL-8 expression in HEK293 transfectants incubated with live M. bovis BCG or M. smegmatis. A, HEK293 cells stably transfected with the pcDNA3-YFP expression vectors encoding WT or R753Q TLR2s were incubated for the indicated times with M. smegmatis (m.o.i. 50). Cells were lysed, and whole-cell lysates were examined by Western blot analyses using the indicated Abs. The results of a representative experiment (n = 3) are shown. B, 293/TLR2 stable cell lines expressing WT or R753Q TLR2 species were co-transfected with pELAM-Luc and pTK-Renilla-Luc. Cells were treated for 24 h with Pam3CSK4 (1 μg/ml), M. smegmatis, or M. bovis BCG (all at m.o.i. 50), and cell lysates were prepared to determine firefly versus Renilla luciferase activities. The numbers from treated cultures were normalized to medium control values and expressed as -fold induction. C, 293/TLR2 cells stably expressing WT or R753Q TLR2s were treated for 24 h with 1 μg/ml Pam3CSK4, live M. smegmatis, or M. bovis BCG (m.o.i. 50). RNA was isolated, converted to cDNA, and subjected to real-time PCR analysis with the respective primers. Data were processed according to the 2−ΔΔCT method (58). D, cells were stimulated for 24 h with 1 μg/ml Pam3CSK4, M. smegmatis, or M. bovis BCG (m.o.i. 50). IL-8 levels were determined in supernatants by ELISA. The data (mean ± S.D.) of a representative experiment (n = 3) are shown. **, p < 0.01; ***, p < 0.001.
Because MAPKs and NF-κB mediate the expression of inflammatory cytokines (21), we next examined induction of IL-8 in HEK293 cells expressing WT or R753Q TLR2 in response to mycobacteria. Stimulation of 293/TLR2WT cells with Pam3CSK4, M. smegmatis, or M. bovis BCG resulted in ∼10-, ∼43-, and ∼2.5-fold up-regulation in the levels of IL-8 mRNA, whereas 293/TLR2R753Q transfectants showed 74%, 64%, and 58% inhibition in the activation of these responses, respectively (Fig. 1C). Similarly, in contrast to a marked up-regulation of IL-8 secretion observed in 293/TLR2WT cells exposed to Pam3CSK4, M. smegmatis, or M. bovis BCG, a significantly lower induction of IL-8 secretion was observed in 293/TLR2R753Q in response to stimulation with Pam3CSK4 or mycobacteria (Fig. 1D). Immunoblot analyses revealed a similar expression of WT versus R753Q TLR2 in HEK293 stable transfectants (Fig. 1A and data not shown), indicating that the observed differences in cell responses to live M. smegmatis or M. bovis BCG were not due to altered expression of the mutant TLR2. These data indicate that the R753Q SNP attenuates the ability of TLR2 to activate p38 MAPK- and p65 NF-κB-driven pathways in response to M. smegmatis and M. bovis BCG infection, leading to deficient induction of IL-8.
Macrophages from mice expressing endogenous R753Q TLR2 exhibit impaired activation of IRAK-1, MAPKs, and NF-κB and deficient induction of inflammatory cytokines upon infection with M. smegmatis
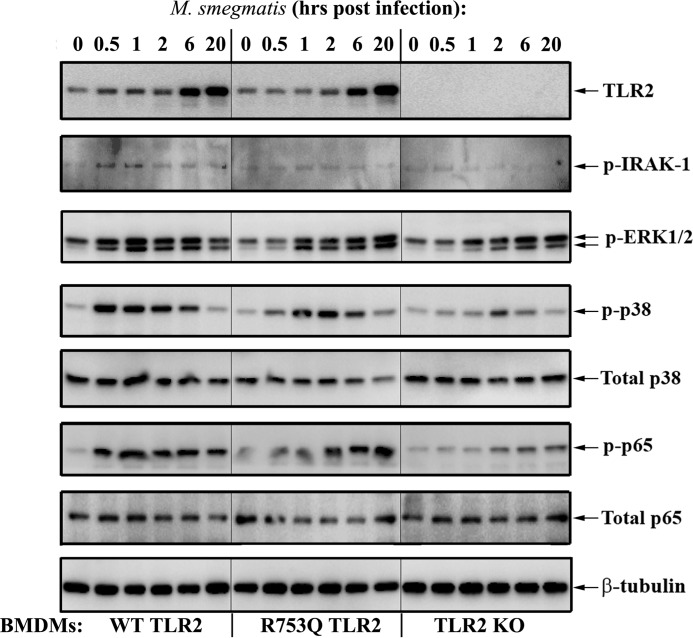
To confirm the results obtained in overexpression systems in macrophages expressing the endogenous TLR2 mutation, we engineered KI mice harboring the R753Q TLR2 SNP (R753Q KI mice) (Fig. 2A). The presence of the R753Q SNP was confirmed by Southern blotting, sequencing of DNA samples, and PCR genotyping (Fig. 2, B and C, and data not shown). BMDMs from WT, TLR2 KO, or R753Q TLR2 KI mice were used to study the effect of the R753Q SNP on TLR signaling and expression of TNFα, IL-1β, IL-6, and IL-10, cytokines involved in macrophage anti-mycobacterial defenses (31–33). Infection of WT BMDMs with M. smegmatis led to robust phosphorylation of IRAK-1, p38, ERK1/2 MAPK, and p65 NF-κB (Fig. 3, left panels). TLR2 KO BMDMs exhibited minimal activation of these signaling intermediates (Fig. 3, right panels), whereas significantly attenuated activation of IRAK-1, MAPKs, and NF-κB were seen in BMDMs from R753Q TLR2 KI mice (Fig. 3, center panels).
Figure 2.
Generation of TLR2 R753Q KI mice. A, the structure of the mouse TLR2 gene, the targeting vector, the targeted allele, and the constitutive allele after Cre recombination. After negative selection for identifying the C57BL/6 embryonic stem cells carrying the R753Q TLR2 mutation, the constitutive R753Q KI allele was obtained after Cre recombination. The stars indicate the sites of mutation. B, the CGC-to-CAG replacement was verified by DNA sequencing. Shown are sequencing data from WT (top panel), heterozygous (HET, center panel), and homozygous KI (bottom panel) mice. C, PCR analysis of DNA samples obtained from tail snips of WT and TLR2 R753Q KI mice. The WT and mutant alleles were identified by their sizes of 191 and 316 bp, respectively.
Figure 3.
Macrophages harboring R753Q TLR2 show impaired activation of IRAK-1, MAPKs, and NF-κB upon mycobacterial infection. BMDMs from WT, TLR2 KO, or R753Q TLR2 KI mice were infected for 2 h with M. smegmatis (m.o.i. 10), washed, incubated in gentamycin-containing medium, and lysed at the indicated times after infection. Whole-cell lysates were examined by Western blot analyses with the indicated Abs. The results of a representative experiment (n = 5) are shown.
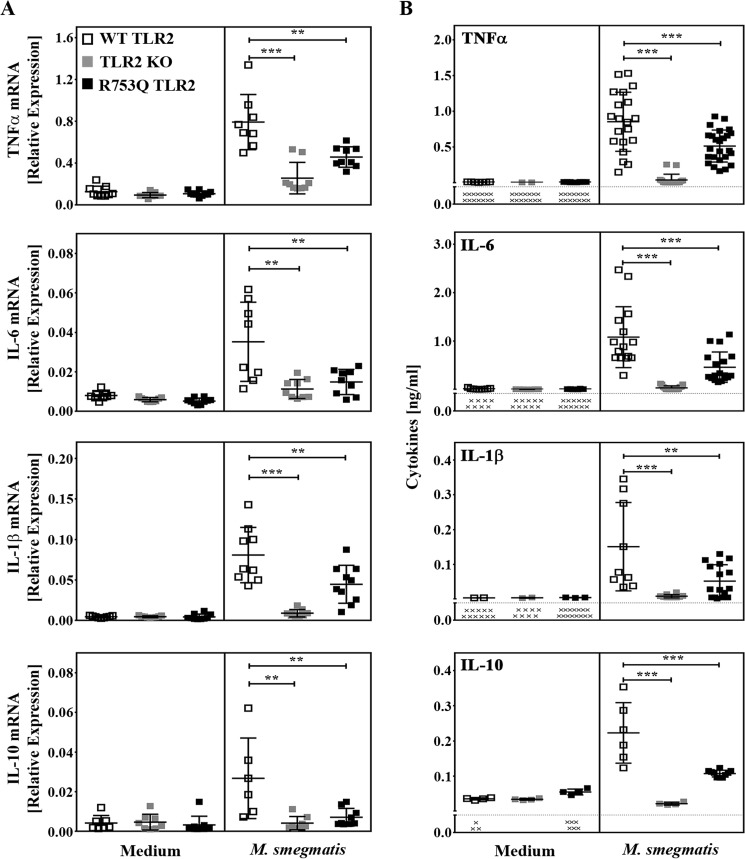
Because MAPKs and NF-κB govern the expression of inflammatory cytokines that regulate host immune responses against pathogens (10, 21), we next determined the impact of the R753Q TLR2 SNP on the expression of TNF-α, IL-1β, IL-6, and IL-10. BMDMs from WT mice responded to infection with M. smegmatis by 4- to 18-fold up-regulation of TNF-α, IL-1β, IL-6, and IL-10 mRNA expression as well as a marked increase in the secreted levels of these cytokines (Fig. 4, A and B). In contrast, there was a lack of production of these cytokines in TLR2 KO BMDMs, whereas BMDMs from R753Q KI mice expressed significantly lower levels of cytokine mRNA and protein. These results indicate that the R753Q SNP impairs TLR2-mediated activation of IRAK1, MAPKs, NF-κB, and induction of inflammatory cytokines in macrophages infected with M. smegmatis.
Figure 4.
Macrophages from R753Q TLR2 KI mice exhibit attenuated cytokine production during infection with M. smegmatis. BMDMs from WT, TLR2 KO, or R753Q TLR2 KI mice were infected for 2 h with M. smegmatis (m.o.i. 10), and the cells were washed and incubated in gentamycin-containing medium. A, RNA was isolated, converted to cDNA, and subjected to real-time PCR analysis with the respective gene primers. The data depict TNF-α mRNA levels (2 h after infection), IL-6 and IL-1β mRNA (24 h after infection), and IL-10 mRNA (6 h after infection). B, following M. smegmatis infection, TNF-α, IL-6, IL-10 (24 h after infection), and IL-1β (48 h after infection) levels were analyzed in cell-free supernatants by ELISA. Summary data (mean ± S.D., n = 3) are depicted. **, p < 0.01; ***, p < 0.001.
The R753Q polymorphism does not affect TLR2 expression and mycobacterial uptake
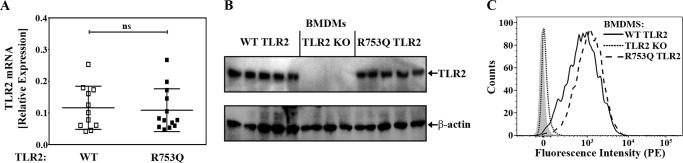
TLR2 traffics between the plasma membrane and endosomes and has been implicated in bacterial uptake and phagocytosis (34–39). Thus, the R753Q SNP could impair macrophage responses to M. smegmatis because of altered TLR2 expression or uptake of bacilli. To address these questions, we first compared the expression of WT and mutant TLR2 in BMDMs derived from WT versus R753Q TLR2 KI mice. Real-time PCR and Western blot analyses of BMDM samples obtained from the respective mice revealed comparable mRNA and protein levels of total WT and R753Q TLR2 variants (Fig. 5, A and B). Furthermore, FACS analyses showed similar levels of cell surface TLR2 species in BMDMs derived from WT or R753Q TLR2 KI mice (Fig. 5C).
Figure 5.
The R753Q SNP does not affect TLR2 expression. A, RNA was isolated from BMDMs obtained from WT or R753Q TLR2 KI mice, converted to cDNA, and analyzed by real-time PCR. Data (mean ± S.D.) of the indicated number of mice from three experiments are shown. ns, not significant. B, whole-cell lysates were examined by Western blot analyses using Abs against total mouse TLR2 and β-tubulin. The results of five (WT and R753Q TLR2) and four (TLR2 KO) individual mice for each genotype are depicted. C, BMDMs from WT, KO, and R753Q TLR2 KI mice were stained with PE-conjugated anti-mouse CD282 (TLR2) Ab or isotype control IgG-PE, followed by FACS analyses of PE fluorescence. Isotype control staining profiles for WT BMDMs mice are shown on the left by a gray shadow (isotype control staining for the TLR2 KO and R753Q TLR2 BMDMs is not shown and exhibited a pattern identical to that of WT TLR2 BMDMs). The data of a representative experiment (n = 5) are shown.
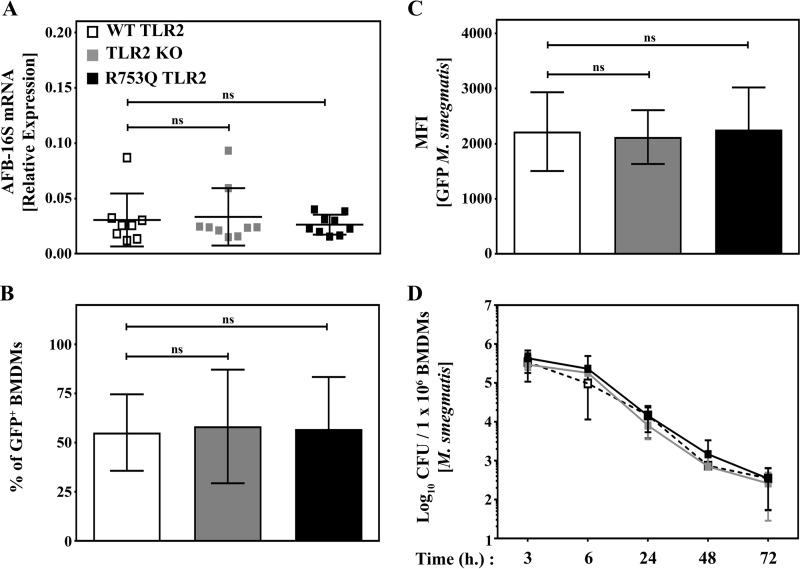
Second, we assessed whether the presence of the R753Q SNP affects bacterial uptake by macrophages. Real-time PCR analyses showed that BMDMs from WT, R753Q TLR2 KI, or TLR2 KO mice exhibit comparable levels of mycobacterial 16S rRNA (40) 2 h after infection with M. smegmatis (m.o.i. 10) (Fig. 6A). Similarly, FACS demonstrated similar percentages and mean fluorescent intensity (MFI) of GFP-positive WT, R753Q TLR2 KI, and TLR2 KO bone marrow-derived macrophages (BMDMs) after infection with GFP-expressing M. smegmatis (Fig. 6, B and C). Furthermore, we also found similar bacterial cfu in cell lysates from M. smegmatis-infected BMDMs isolated from WT versus R753Q TLR2 mice or mice lacking TLR2 (Fig. 6D). Taken together, these results indicate that the R753Q SNP does not affect TLR2 expression or uptake and survival of M. smegmatis in macrophages.
Figure 6.
Macrophages harboring WT or R753Q TLR2 show similar bacterial burdens after infection with M. smegmatis. BMDMs from WT, TLR2 KO, or R753Q TLR2 KI mice were infected for 2 h with M. smegmatis (m.o.i. 10). A, RNA was isolated, converted to cDNA, and examined by real-time PCR with the respective gene-specific primers to assess mycobacterium 16S rRNA expression. Data (mean ± S.D.) of the indicated numbers of individual mice from three experiments are depicted. ns, not significant. B and C, macrophages were incubated for 2 h with GFP-expressing M. smegmatis (m.o.i. 10), washed, and fixed, and bacterial uptake was assessed by FACS analyses judged by percentages of GFP-positive BMDMs (B) or by MFI values of GFP fluorescence (C). Summary results (mean ± S.D.) of three experiments are shown. D, cells were processed as in B and C and incubated for the indicated times in gentamycin-containing cDMEM. Cells were lysed, and serial dilutions of cell lysates were plated on bacterial agar dishes to determine cfu. The total number of cfu per 1 × 106 cells was calculated, and the data (mean ± S.D.) of 8 mice/group are shown.
R753Q TLR2 exhibits impaired recruitment of MyD88 upon M. smegmatis infection
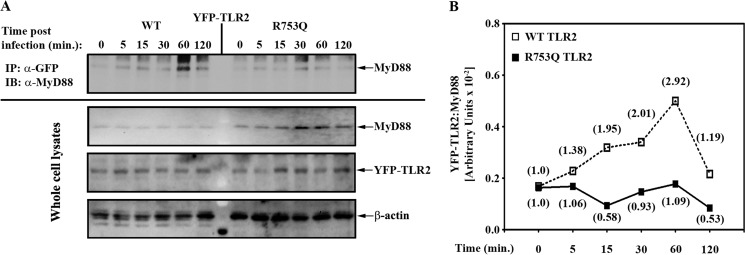
To gain insights into the mechanisms by which the R753Q SNP alters TLR2 signaling, we analyzed the recruitment of endogenous MyD88 to transfected YFP-tagged WT or mutant TLR2 species in HEK293 cell transfectants exposed to live M. smegmatis. Co-immunoprecipitation analyses revealed that exposure of 293/TLR2WT to M. smegmatis resulted in association of endogenous MyD88 with WT TLR2 as early as 5 min, with a steady accumulation of TLR2-associated MyD88 up to 60 min after infection, followed by a decline at 120 min (Fig. 7). In contrast, we noted drastically impaired MyD88 recruitment to the mutant TLR2 species in 293/TLR2R753Q cells incubated with M. smegmatis (Fig. 7). Comparable levels of total WT, R753Q TLR2, and MyD88 proteins were observed in whole cell lysates, indicating that differences in M. smegmatis-inducible MyD88-TLR2 associations cannot be accounted for by variations in total levels of the interacting proteins. Thus, the R753Q polymorphism impairs the ability of TLR2 to recruit MyD88 after infection with live M. smegmatis, leading to deficient activation of IRAK-1, NF-κB, and MAPKs and induction of inflammatory cytokines in macrophages infected with live M. smegmatis.
Figure 7.
The impact of the R753Q polymorphism on MyD88 recruitment to TLR2 in response to live M. smegmatis. HEK293 cells stably expressing YFP-tagged human WT or R753Q TLR2 species were incubated for the indicated times with M. smegmatis (m.o.i. 50). A, YFP-TLR2 proteins were immunoprecipitated (IP) from whole-cell lysates with anti-GFP Ab, and MyD88 recruitment was analyzed by immunoblotting (IB) with anti-MyD88 Ab. Whole-cell lysates were examined by Western blot analyses using Abs against GFP, MyD88, and β-actin. The results of a representative experiment (n = 3) are depicted. B, quantifications of the Western blot data shown in A. The data plotted are arbitrary values obtained by normalizing the band intensity for the YFP-TLR2-associated MyD88 to the band intensities of total YFP-TLR2 and then normalizing this to the band intensity of β-actin. The numbers in the brackets represent -fold induction values calculated by dividing arbitrary values for the intensities of bands in mycobacteria-infected cells by those measured in uninfected cells.
Discussion
Genome-wide association studies have demonstrated the association of a microsatellite polymorphism in the intron of TLR2 and the R753Q TLR2 polymorphism with increased incidence of tuberculosis in several cohorts of patients (22–25, 41). Furthermore, the presence of TLR2 SNPs has been linked to infections with other mycobacteria, e.g. M. bovis, M. avium, and M. abscessus (26–29). Studies with PBMCs obtained from patients expressing the R753Q SNP showed attenuated NF-κB activation and cytokine release in response to stimulation with Borrelia burgdorferi lysates (42). We reported that the R753Q SNP alters the electrostatic potential of the TIR2 domain and impairs the ability of TLR2 to dimerize with TLR6 and recruit MyD88 in cells stimulated with TLR2 agonists or irradiated Mtb, leading to decreased cytokine expression (30). However, it has been unknown how the endogenous R753Q SNP affects TLR2 responses during infection of macrophages with live mycobacteria.
This paper demonstrates that exposure of HEK293 cells expressing transfected R753Q TLR2 to live M. smegmatis or M. bovis BCG resulted in deficient activation of p38 MAPK and NF-κB and impaired induction of IL-8. To confirm our results in macrophages expressing endogenous mutant TLR2, we engineered a new KI mouse strain harboring the mutant R753Q TLR2. Infection of BMDMs from R753Q TLR2 KI mice with M. smegmatis led to impaired activation of IRAK-1, MAPKs, and NF-κB and decreased induction of TNF-α, IL-1β, IL-6, and IL-10 compared with the responses elicited by cells from WT mice, whereas TLR2−/− BMDMs exhibited the most pronounced deficiencies. Of note, we observed comparable levels of total and cell surface-associated WT and R753Q TLR2 proteins in macrophages, indicating that the effects of the R753Q SNP are not due to lower expression of the mutant TLR2. These results are consistent with conclusions obtained while studying the effect of the R753Q TLR2 SNP on manifestations of amyloidosis in patients with familial Mediterranean fever (43) and with our previous findings in HEK293 transfectants (30). However, there is a possibility that the R753Q SNP could affect dynamic trafficking of TLR2 between the plasma membrane, Golgi, and endosomes or its engagement of the co-receptors CD14, TLR1, TLR6, CD36, or integrins (9, 14, 44), requiring further studies.
Because TLR2 has been reported to regulate bacterial uptake and phagocytosis (34–39), we determined whether the R753Q SNP alters M. smegmatis uptake in the macrophage. Our results revealed a similar uptake of GFP-expressing M. smegmatis in BMDMs from WT, TLR2 KO, or R753Q TLR2 KI mice, as judged by comparable expression of 16S bacterial RNA, numbers and percentages of GFP-positive macrophages, and levels of bacterial cfu. These data are consistent with previous reports showing comparable uptake of H37Rv M. tuberculosis and M. bovis BCG and similar percentages of bacilli in the cytoplasm of WT and TLR2 KO BMDMs (45). Thus, differences between WT versus R753Q TLR2 in eliciting signaling and cytokine expression during M. smegmatis infection are not due to altered levels of the mutant TLR2 or changes in bacterial uptake.
In response to stimulation with defined TLR2 agonists or during mycobacterial infection, TLR2 expression in macrophages is increased (46, 47). Interestingly, we demonstrate comparable increases in TLR2 levels in macrophages from WT or TLR2 R753Q KI mice infected with M. smegmatis. In addition to NF-κB, there are other transcription factors that bind to the promoter region of TLR2 and drive its expression, including Sp1, STAT5, and the Ets family members Sp1, Sp3, and, possibly, PU.1 (48–50). Thus, preserved induction of such transcription factors could account for the comparable up-regulation of TLR2 expression seen in WT versus R753Q KI macrophages. Furthermore, the TLR2 R753Q SNP is likely to represent not just a “loss-of-function” mutation but could uniquely affect different aspects of receptor signaling. Indeed, such a signaling mode is reported for another TLR2 SNP, R677W, whereby R677W TLR2-expressing monocytes respond to exposure to Mycobacterium leprae with impaired induction of TNF-α and IL-12 p40 but increased expression of IL-10 compared with cells expressing WT TLR2 (27). Notably, our preliminary results suggest that the R753Q TLR2 is likely to act by “reprograming” and altering signaling, whereby the SNP inhibits phosphorylation of IRAK1, MAPK, and p65 NF-kB, without affecting PI3K-dependent phosphorylation of Akt (data not shown).
TLR2 engagement by pathogens involves its heterodimerization with TLR1 or TLR6 (51–53), leading to recruitment of the adapters Mal and MyD88 and myddosome assembly (20, 21). Efficient myddosome assembly is critical for optimal activation of IRAK-1, MAPKs, and NF-κB and induction of antibacterial immune responses. We reported that impaired responses of cells transfected with R753Q TLR2 to defined agonists or irradiated Mtb were due to the altered electrostatic potential of the mutant TIR2 domain, deficient TLR2-TLR6 assembly, and impaired recruitment of Mal and MyD88 (30). Using the HEK293 overexpression system, we demonstrate here that exposure of HEK293 cells expressing R753Q TLR2 to live M. smegmatis resulted in diminished ability of the mutant TLR2 to recruit MyD88 and activate IRAK-1. To the best of our knowledge, this is the first report uncovering impaired MyD88 recruitment to R753Q TLR2 and deficient IRAK-1 activation as the mechanistic basis of reduced cell activation in response to mycobacterial infection.
TLR2 undergoes tyrosine phosphorylation in its cytoplasmic domain (54), and two tyrosines, Tyr-616 and Tyr-761, play a critical role in TLR2-dependent NF-κB activation and cytokine expression (54, 55). It is currently unknown whether Tyr-616 or Tyr-761 are directly phosphorylated and which kinases could be involved in this process. It is noteworthy that the R753Q TLR2 SNP is in a close proximity to Tyr-761, suggesting that, during infection of macrophages with mycobacteria, this SNP may alter TLR2 tyrosine phosphorylation, which has been implicated in enabling efficient TLR2-TLR1 or TLR2-TLR6 signalosome assembly (30, 54). Supporting this notion, we previously reported deficient tyrosine phosphorylation of transfected R753Q human TLR2 in HEK293 cells stimulated with TLR2 agonists or irradiated Mtb, which correlated with impaired TLR2-TLR6 assembly (30). It is plausible that the R753Q SNP alters the TIR2 electrostatic potential and docking interface for protein tyrosine kinases and impairs TLR2 tyrosine phosphorylation. These processes could lead to deficient TLR2 assemblies with TLR6 or TLR1 upon infection of macrophages with live mycobacteria, translating into impaired recruitment of MyD88 and altered signaling. Experiments are ongoing to address this hypothesis. Our initial studies to address the functional significance of the TLR2 R753Q SNP in vivo revealed that mice harboring the TLR2 R753Q SNP i.p. infected with M. smegmatis exhibited increased bacterial burdens in the spleen and liver compared with WT animals (data not shown). These findings may suggest impaired killing of M. smegmatis in vivo in R753Q TLR2-expressing mice. Experiments are in progress to carry out comprehensive analyses of the impact of the R753Q TLR2 SNP on immune responses to mycobacterial species and infection morbidity and mortality in vivo.
Recently, Herrtwich et al. (56) have shown a novel role for TLR2 signaling in regulating the generation of polyploid macrophages, giant cells, and granuloma formation upon infection with Mycobacterium species. Chronic stimulation of TLR2 was found to promote macrophage polyploidy and suppress genomic instability by regulating Myc-induced DNA repair and activating unique gene expression signatures for metabolic and extracellular matrix-remodeling molecules (56). These events could promote a long-lived granuloma-resident macrophage differentiation program that regulates granulomatous tissue remodeling, which is critical in controlling Mtb infection (3, 4). It is an intriguing possibility that altered signaling by R753Q TLR2 could confer unique regulation of Myc-dependent DNA repair and expression profiles of genes involved in the regulation of metabolism and extracellular matrix remodeling, instructing the fate of granulomas in the lung. It is tempting to speculate that impaired TLR2-TLR6 heterodimerization leading to attenuated TLR2-mediated signaling in response to mycobacteria could be responsible for the increased incidence of TB in certain cohorts of patients carrying the TLR2 R753Q SNP. Interventions directed at corrections of impaired myddosome assembly exhibited by the mutant R753Q TLR2 species via application of agonistic peptides or small chemical agonists promoting TLR2-MyD88 complex formation could potentially provide new approaches for the treatment of patients that carry the R753Q SNP.
Experimental procedures
Abs, reagents, and cell lines
The following Abs were used in this study: anti-GFP (Thermo Fisher Scientific, Waltham, MA); anti-FLAG (Sigma-Aldrich, St. Louis, MO); anti-MyD88, anti-p65 NF-κB, anti-TLR2, and anti-β-actin (Santa Cruz Biotechnology Inc., Santa Cruz, CA); and anti-p-p38, anti-p-p65, anti-p-ERK1/2, anti-ERK1/2, anti-p38, anti-p-Akt, anti-Akt, and anti-β-tubulin (Cell Signaling Technology, Inc., Danvers, MA.). S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH (Pam3CSK4) was obtained from InvivoGen (San Diego, CA). Zombie® Aqua cell viability dye, PE-labeled anti-mouse CD282 (TLR2) Ab, and PE-labeled rat IgG2a κ isotype control Ab were from BioLegend (San Diego, CA). HEK293T and HEK293 cells were obtained from the ATCC and maintained in complete DMEM supplemented with 10% FBS (HyClone, Logan, UT), 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Thermo Fisher Scientific) (cDMEM).
Generation of R753Q TLR2 KI mice
R753Q TLR2 KI mice were generated at Cyagen Biosciences (Santa Clara, CA) using C57BL/6J (B6) embryonic stem cells by standard gene-targeting techniques (Fig. 2A). In brief, to engineer the targeting vector, the 5′ homology arm and 3′ homology arm were amplified from bacterial artificial chromosome clone RP23-374N17 or RP23-158H14 from the C57BL/6J library as a template. The targeting vector was constructed by site-directed mutagenesis to replace Arg with Glu at the position of 753 (R753Q SNP) in exon 3 and was transfected into B6 embryonic stem cells. The targeting vector had a Neo selection cassette flanked by LoxP sites for antibiotic selection as well as diphtheria toxin A, which was used for negative selection to identify cells carrying the R753Q TLR2 SNP. After selection, the presence of the respective alleles was verified by Southern blot analysis and DNA sequencing of the PCR products generated using the primers 5′-TTTCTCTAGCCACCCTATGTCGCA-3′ (forward) and 5′-TTCCGGGCAAATGGATCATTG-3′ (reverse) (Fig. 2B and data not shown). Embryonic stem cells were injected into mouse blastocysts and transplanted into the uterus of Cre-expressing C57BL/6 mice to produce chimeric KI mice and to achieve removal of the Neo cassette. The chimeric heterozygous F1 mice were interbred to generate R753Q TLR2 homozygous mice. The mice were genotyped using DNA samples from tail snips and the primers 5′-AGTTCACACAAAAATGCAGCCTGTG-3′ (forward) and 5′-GGGTGGCTAGAGAAAGCATTTGCTA-3′ (reverse), and PCR products were run on a gel to identify wild-type (191 bp) or R753Q TLR2 species (316 bp) (Fig. 2C).
Mouse strains and macrophage isolation
WT B6 and TLR2 KO (B6.129-Tlr2tm1Kir/J) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All mice were housed in an environmentally controlled room in the UConn Health animal facility. Six-week-old female mice were used in the experiments. All animal experiments and procedures were approved by the Institutional Animal Care and Use Committee. Bone marrow cells were isolated from femora and tibiae of mice. To generate BMDMs, bone marrow cells were differentiated for 7 days in RPMI 1640 (Mediatech, Herndon, VA) supplemented with 10% FBS (HyClone), 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 15% L929 cell-conditioned, M-CSF-containing supernatant, as reported previously (30).
Bacterial cultures and infection
GFP-expressing M. smegmatis and M. bovis BCG were kindly provided by Drs. Volker Briken (University of Maryland College Park, College Park, MD) and Matyas Sandor (University of Wisconsin, Madison, WI), respectively. Bacteria were cultured in 7H9 broth containing 0.5% glycerol, 0.05% Tween 80, 10% oleic albumin dextrose complex enrichment, and 40 μg/ml kanamycin. Bacteria were grown to semiconfluent cultures (optical density at 600 nm ranging from 0.4–0.8), washed in PBS, allowed to settle for 10 min, and added to cells cultured in antibiotic-free DMEM at the respective m.o.i. for 2 h. Cells were washed in PBS and incubated in cDMEM supplemented with 100 μg/ml gentamycin (Thermo Fisher Scientific) for the period of the assay.
Plasmids and transfection
The pcDNA3-YFP-WT human (h) TLR2, pcDNA3-YFP-R753Q hTLR2, pELAM-luciferase (Luc), and p-thymidine kinase (TK)-Renilla-Luc plasmids were described previously (30). The R753Q SNP was introduced into the YFP-TLR2 expression vector by site-directed mutagenesis, and its presence was confirmed by sequencing, as published previously (30). For transient transfection, HEK293T cells were transfected with the respective plasmids, using the Lipofectamine 2000 transfection protocol as recommended by the manufacturer and as reported previously (30, 57). To generate stably transfected cell lines, HEK293 cells were transfected for 5 h with pcDNA3-YFP vectors encoding WT or R753Q TLR2s, recovered for 24 h, and selected for 2 weeks in cDMEM containing G418 (1 mg/ml). Cells were plated in 100-mm tissue culture dishes (for immunoprecipitation), 6-well plates (to generate cell extracts for immunoblotting), or 24-well plates (for gene expression, cytokine secretion, and reporter assays) as described previously (30, 57).
Assessment of bacterial uptake by flow cytometry
To examine bacterial uptake, BMDMs (1 × 106 cells/well in 24-well plates) were incubated for 2 h with GFP-expressing M. smegmatis (m.o.i. 10) in antibiotic-free RPMI 1640 medium, washed with PBS, and incubated in cDMEM containing 100 μg/ml gentamycin on ice or at 37 °C for the indicated times. Cells were detached by scraping, washed in ice-cold PBS containing 3% FBS, stained with Zombie® Aqua cell viability dye (BioLegend), fixed for 15 min in 3.7% formaldehyde, and analyzed by FACS on an LSRII flow cytometer (BD Biosciences) to measure GFP fluorescence. The data were analyzed using FlowJo software (Tree Star). BMDMs were gated using forward scatter versus side scatter, single cells were gated using forward scatter area versus forward scatter height, and then live cells were gated as Zombie® Aqua-negative events. Live BMDMs were then assessed for GFP-positive events as well as total GFP MFI.
Assessment of cell surface TLR2 expression by flow cytometry
To examine surface TLR2 expression, BMDMs were stained for 30 min on ice with PE-labeled anti-mouse CD282 (TLR2) or PE-labeled rat IgG2a κ isotype control Abs. Cells were washed with PBS, stained with Zombie® Aqua cell viability dye, fixed for 15 min in 3.7% formaldehyde, and analyzed by FACS on a LSRII flow cytometer (BD Biosciences). The data were analyzed using FlowJo software (Tree Star). Briefly, BMDMs were gated using forward scatter versus side scatter, then single cells were gated using forward scatter area versus forward scatter height, and then live cells were gated as Zombie® Aqua-negative events. Live BMDMs were then assessed for TLR2 expression by measuring PE expression as seen by MFI.
Isolation of RNA and real-time PCR
Total RNA was prepared using the TRIzol protocol (Thermo Fisher Scientific), residual genomic DNA was digested with DNase, and RNA was repurified as recommended by the manufacturer. cDNA was prepared from 1 μg of total RNA using the reverse transcription system (Promega) and examined by real-time PCR with the following primers: human IL-8, 5′-CACCGGAAGGAACCATCTCACT-3′ (forward) and 5′-TGCACCTTCACACAGAGCTGC-3′ (reverse); human hypoxanthine-guanine phosphoribosyltransferase, 5′-ACCAGTCAACAGGGGACATAAAAG-3′ (forward) and 5′-GTCTGCATTGTTTTGCCAGTGTC-3′ (reverse); mycobacterial acid-fast bacilli-16S, 5′-GCGTGCTTAACACATGCAAGTC-3′ (forward) and 5′-TCCTCCTGATATCTGCGCATTC-3′ (reverse); mouse TLR2, 5′-CCGAAACCTCAGACAAAGCG-3′ (forward) and 5′-TCACACACCCCAGAAGC ATC-3′ (reverse); mouse GAPDH, 5′-GCTGACCTGCTGGATTACATT-3′ (forward) and 5′-GTTGAGAGATCATCTCCACCA-3′ (reverse); mouse TNF-α, 5′-CCCAGGCAGTCAGATCATCTTC-3′ (forward) and 5′-GCTTGAGGGTTTGCTACAACATG-3′ (reverse); mouse IL-1β, 5′-CTGGTGTGTGACGTTCCCAT-3′ (forward) and 5′-GATTCTTTCCTTTGAGGCCCA-3′ (reverse); mouse IL-6, 5′-TCAGGAAATTTGCCTATTGAAAATTT-3′ (forward) and 5′-GCTTTGTCTTTCTTGTTAT CTTTTAAGTTGT-3′ (reverse); and mouse IL-10, 5′-ATTTGAATTCCCTGGGTGAGAAG-3′ (forward) and 5′-CACAGGGGAGAAATCG ATGACA-3′ (reverse). Reaction mixtures were processed on a MyIQ real-time PCR machine (Bio-Rad). The data were analyzed by the 2−ΔΔCT method (58) or expressed as a ratio of gene of interest normalized to the respective housekeeping gene (hypoxanthine-guanine phosphoribosyltransferase or GAPDH) as reported previously (57).
Co-immunoprecipitation and Western blot analyses
Cell lysates were prepared as described previously (30, 57). In brief, cells were lysed in lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 5 mm NaF, 2 mm sodium orthovanadate, 1 mm PMSF, and complete protease inhibitors). Cell lysates were cleared by centrifugation at 13,000 rpm for 10 min, and insoluble debris was discarded. For co-immunoprecipitation, cell lysates were precleared for 4 h at 4 °C with protein G-agarose beads (25 μl of 50% slurry per sample, Roche Applied Science) upon rotation. Precleared cell extracts were incubated overnight at 4 °C with the respective Abs in lysis buffer containing 20 mm HEPES (pH 7.4), 0.5% Triton X-100, 150 mm NaCl, 12.5 mm β-glycerophosphate, 50 mm NaF, 1 mm DTT, 1 mm sodium orthovanadate, 2 mm EDTA, 1 mm PMSF, and protease inhibitor mixture (Roche Applied Science). Thereafter, protein G-agarose beads were added (45 μl/sample), and incubation continued for 4 h. Beads were washed five times with lysis buffer and suspended in Laemmli sample buffer (50 mm Tris-Cl (pH 6.8), 10% glycerol, 2% SDS, 0.1% bromphenol blue, and 5% 2-mercaptoethanol). Proteins were separated by SDS-PAGE on 4–20% mini gels (Thermo Fisher Scientific), transferred to polyvinylidene fluoride membranes (Bio-Rad), blocked, and probed with the respective Abs, as described previously (30). The protein bands were quantified using the National Institutes of Health ImageJ software package and expressed as values of phosphorylated proteins normalized to total species or to β-actin or tubulin.
Assessment of intracellular bacterial cfu
BMDMs were plated in 24-well plates (1 × 106 cells/well), grown overnight, incubated for 2 h with M. smegmatis (m.o.i. 10) in antibiotic-free RPMI 1640 medium, washed with PBS, and incubated in cDMEM containing 100 μg/ml gentamycin on ice or at 37 °C for the indicated times. Cells were lysed for bacterial counts in PBS containing 0.05% Tween 80 (59). The number of cfu per 1 × 106 cells was determined by plating 10-fold serial dilutions of cell lysates on Middlebrook 7H10 plates complemented with 0.5% glycerol and 10% oleic albumin dextrose complex enrichment (59, 60). For selective medium, 40 μg/ml kanamycin was added. Plates were incubated at 37 °C, and cfu were counted after 2 days.
Analyses of cytokine secretion
Supernatants from cell stimulated with TLR agonists or infected with M. smegmatis were cleared by centrifugation, and cytokine levels were determined using ELISA kits (BioLegend) according to the recommendations of the manufacturer. The ELISA kits had the following limits of detection: IL-1β, 16 pg/ml; IL-6, 4 pg/ml; IL-10, 16 pg/ml; and TNF-α, 4 pg/ml.
NF-κB reporter assays
NF-κB reporter assays were performed as described previously (30, 57). In brief, 293/TLR2 cells expressing WT or R753Q TLR2 species were grown in 24-well plates to reach 70% confluence and transfected with pELAM-Luc (300 ng/well) and pTK-RL (10 ng/well) plasmids (the total plasmid amount was adjusted to 400 ng/well with pcDNA3.1) using Lipofectamine 2000. After recovery for 48 h, cells were treated for 16 h as indicated in the figure legends and lysed in a passive lysis buffer (Promega). Firefly versus Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega) on an LB 9507 luminometer (Berthold Technologies).
Statistical analysis
Experimental results were processed by the GraphPad Prism software package (GraphPad Software, San Diego, CA) using one-way analysis of variance with Tukey's post hoc test, and the data were expressed as mean ± S.D.
Author contributions
G. P. conducted the experiments, analyzed the results, and wrote the manuscript. R. P. performed several of the NF-κB reporter experiments shown in Fig. 1. A. E. M. conceived the idea for the project, conceptualized the research, analyzed the data, and edited and finalized the manuscript written by G. P.
Acknowledgments
We thank Drs. Volker Briken (University of Maryland College Park, College Park, MD) and Matyas Sandor (University of Wisconsin, Madison, WI) for kindly providing us with GFP-expressing M. smegmatis and M. bovis BCG, respectively. We also thank Sarah Ahlbrand (University of Maryland College Park) for help and advice regarding M. smegmatis protocols.
This work was supported in whole by National Institutes of Health Grant R56 AI097210 (to A. E. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Mtb
- Mycobacterium tuberculosis
- TLR
- Toll-like receptor
- BCG
- bacillus Calmette-Guérin
- TIR
- Toll-IL-1R
- IRAK
- IL-1R-associated kinase
- KI
- knock-in
- m.o.i.
- multiplicity of infection
- BMDM
- bone marrow-derived macrophage
- MFI
- mean fluorescence intensity
- Ab
- antibody
- Pam3CSK4
- S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH
- cDMEM
- complete DMEM
- Luc
- luciferase
- TK
- thymidine kinase
- PE
- phycoerythrin.
References
- 1. Kaufmann S. H. (2006) Tuberculosis: back on the immunologists' agenda. Immunity 24, 351–357 [DOI] [PubMed] [Google Scholar]
- 2. Stamm C. E., Collins A. C., and Shiloh M. U. (2015) Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol. Rev. 264, 204–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell D. G., Cardona P.-J., Kim M.-J., Allain S., and Altare F. (2009) Foamy macrophages and the progression of the human TB granuloma. Nat. Immunol. 10, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flynn J. L., Chan J., and Lin P. L. (2011) Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 4, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briken V., Ahlbrand S. E., and Shah S. (2013) Mycobacterium tuberculosis and the host cell inflammasome: a complex relationship. Front. Cell. Infect. Microbiol. 3, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berrington W. R., and Hawn T. R. (2007) Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol. Rev. 219, 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Divangahi M., Mostowy S., Coulombe F., Kozak R., Guillot L., Veyrier F., Kobayashi K. S., Flavell R. A., Gros P., and Behr M. A. (2008) NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J. Immunol. 181, 7157–7165 [DOI] [PubMed] [Google Scholar]
- 8. Ferwerda G., Girardin S. E., Kullberg B. J., Le Bourhis L., de Jong D. J., Langenberg D. M., van Crevel R., Adema G. J., Ottenhoff T. H., Van der Meer J. W., and Netea M. G. (2005) NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 1, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGettrick A. F., and O'Neill L. A. (2010) Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr. Opin. Immunol. 22, 20–27 [DOI] [PubMed] [Google Scholar]
- 10. Akira S., Takeda K., and Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 [DOI] [PubMed] [Google Scholar]
- 11. Underhill D. M., Ozinsky A., Smith K. D., and Aderem A. (1999) Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. U.S.A. 96, 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reiling N., Hölscher C., Fehrenbach A., Kröger S., Kirschning C. J., Goyert S., and Ehlers S. (2002) Cutting edge: Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J. Immunol. 169, 3480–3484 [DOI] [PubMed] [Google Scholar]
- 13. Means T. K., Wang S., Lien E., Yoshimura A., Golenbock D. T., and Fenton M. J. (1999) Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. 163, 3920–3927 [PubMed] [Google Scholar]
- 14. Drage M. G., Pecora N. D., Hise A. G., Febbraio M., Silverstein R. L., Golenbock D. T., Boom W. H., and Harding C. V. (2009) TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell. Immunol. 258, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heldwein K. A., Liang M. D., Andresen T. K., Thomas K. E., Marty A. M., Cuesta N., Vogel S. N., and Fenton M. J. (2003) TLR2 and TLR4 serve distinct roles in the host immune response against Mycobacterium bovis BCG. J. Leukocyte Biol. 74, 277–286 [DOI] [PubMed] [Google Scholar]
- 16. Sampaio E. P., Elloumi H. Z., Zelazny A., Ding L., Paulson M. L., Sher A., Bafica A. L., Shea Y. R., and Holland S. M. (2008) Mycobacterium abscessus and M. avium trigger Toll-like receptor 2 and distinct cytokine response in human cells. Am. J. Respir. Cell Mol. Biol. 39, 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujiwara N., Naka T., Ogawa M., Yamamoto R., Ogura H., and Taniguchi H. (2012) Characteristics of Mycobacterium smegmatis J15cs strain lipids. Tuberculosis 92, 187–192 [DOI] [PubMed] [Google Scholar]
- 18. O'Neill L. A., Golenbock D., and Bowie A. G. (2013) The history of Toll-like receptors: redefining innate immunity. Nat. Rev. Immunol. 13, 453–460 [DOI] [PubMed] [Google Scholar]
- 19. Li X. (2008) IRAK4 in TLR/IL-1R signaling: possible clinical applications. Eur. J. Immunol. 38, 614–618 [DOI] [PubMed] [Google Scholar]
- 20. Lin S. C., Lo Y. C., and Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawai T., and Akira S. (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 [DOI] [PubMed] [Google Scholar]
- 22. Thuong N. T., Dunstan S. J., Chau T. T., Thorsson V., Simmons C. P., Quyen N. T., Thwaites G. E., Thi Ngoc Lan N., Hibberd M., Teo Y. Y., Seielstad M., Aderem A., Farrar J. J., and Hawn T. R. (2008) Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 4, e1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogus A. C., Yoldas B., Ozdemir T., Uguz A., Olcen S., Keser I., Coskun M., Cilli A., and Yegin O. (2004) The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur. Respir. J. 23, 219–223 [DOI] [PubMed] [Google Scholar]
- 24. Ben-Ali M., Barbouche M. R., Bousnina S., Chabbou A., and Dellagi K. (2004) Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin. Diagn. Lab Immunol. 11, 625–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berdeli A., Celik H. A., Ozyürek R., Dogrusoz B., and Aydin H. H. (2005) TLR-2 gene Arg753Gln polymorphism is strongly associated with acute rheumatic fever in children. J. Mol. Med. 83, 535–541 [DOI] [PubMed] [Google Scholar]
- 26. Bhide M. R., Mucha R., Mikula I. Jr., Kisova L., Skrabana R., Novak M., and Mikula I. Sr. (2009) Novel mutations in TLR genes cause hyporesponsiveness to Mycobacterium avium subsp. paratuberculosis infection. BMC Genet. 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang T. J., Yeum C. E., Kim B. C., You E. Y., and Chae G. T. (2004) Differential production of interleukin-10 and interleukin-12 in mononuclear cells from leprosy patients with a Toll-like receptor 2 mutation. Immunology 112, 674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yim J. J., Kim H. J., Kwon O. J., and Koh W. J. (2008) Association between microsatellite polymorphisms in intron II of the human Toll-like receptor 2 gene and nontuberculous mycobacterial lung disease in a Korean population. Hum. Immunol. 69, 572–576 [DOI] [PubMed] [Google Scholar]
- 29. Pöyhönen L., Nuolivirta K., Vuononvirta J., Kröger L., Huhtala H., Mertsola J., He Q., and Korppi M. (2015) Toll-like receptor 2 subfamily gene polymorphisms are associated with bacillus Calmette-Guerin osteitis following newborn vaccination. Acta Paediatr. 104, 485–490 [DOI] [PubMed] [Google Scholar]
- 30. Xiong Y., Song C., Snyder G. A., Sundberg E. J., and Medvedev A. E. (2012) R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J. Biol. Chem. 287, 38327–38337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleinnijenhuis J., Oosting M., Joosten L. A., Netea M. G., and Van Crevel R. (2011) Innate immune recognition of Mycobacterium tuberculosis. Clin. Dev. Immunol. 2011, 405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ladel C. H., Blum C., Dreher A., Reifenberg K., Kopf M., and Kaufmann S. H. (1997) Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect. Immun. 65, 4843–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dorhoi A., and Kaufmann S. H. (2016) Pathology and immune reactivity: understanding multidimensionality in pulmonary tuberculosis. Semin. Immunopathol. 38, 153–166 [DOI] [PubMed] [Google Scholar]
- 34. Blander J. M., and Medzhitov R. (2004) Regulation of phagosome maturation by signals from Toll-like receptors. Science 304, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 35. Kapetanovic R., Nahori M. A., Balloy V., Fitting C., Philpott D. J., Cavaillon J. M., and Adib-Conquy M. (2007) Contribution of phagocytosis and intracellular sensing for cytokine production by Staphylococcus aureus-activated macrophages. Infect. Immun. 75, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen Y., Kawamura I., Nomura T., Tsuchiya K., Hara H., Dewamitta S. R., Sakai S., Qu H., Daim S., Yamamoto T., and Mitsuyama M. (2010) Toll-like receptor 2- and MyD88-dependent phosphatidylinositol 3-kinase and Rac1 activation facilitates the phagocytosis of Listeria monocytogenes by murine macrophages. Infect. Immun. 78, 2857–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Letiembre M., Echchannaoui H., Bachmann P., Ferracin F., Nieto C., Espinosa M., and Landmann R. (2005) Toll-like receptor 2 deficiency delays pneumococcal phagocytosis and impairs oxidative killing by granulocytes. Infect. Immun. 73, 8397–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luther K., Torosantucci A., Brakhage A. A., Heesemann J., and Ebel F. (2007) Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 β-glucan receptor and Toll-like receptor 2. Cell. Microbiol. 9, 368–381 [DOI] [PubMed] [Google Scholar]
- 39. Doyle S. E., O'Connell R. M., Miranda G. A., Vaidya S. A., Chow E. K., Liu P. T., Suzuki S., Suzuki N., Modlin R. L., Yeh W.-C., Lane T. F., and Cheng G. (2004) Toll-like receptors induce a phagocytic gene program through p38. J. Exp. Med. 199, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yam W.-C., and Siu K.-H. G. (2013) Rapid identification of mycobacteria and rapid detection of drug resistance in Mycobacterium tuberculosis in cultured isolates and in respiratory specimens in. Methods Mol. Biol. 943, 171–199 [DOI] [PubMed] [Google Scholar]
- 41. Yim J. J., Ding L., Schäffer A. A., Park G. Y., Shim Y. S., and Holland S. M. (2004) A microsatellite polymorphism in intron 2 of human Toll-like receptor 2 gene: functional implications and racial differences. FEMS Immunol. Med. Microbiol. 40, 163–169 [DOI] [PubMed] [Google Scholar]
- 42. Schröder N. W., Diterich I., Zinke A., Eckert J., Draing C., von Baehr V., Hassler D., Priem S., Hahn K., Michelsen K. S., Hartung T., Burmester G. R., Göbel U. B., Hermann C., and Schumann R. R. (2005) Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J. Immunol. 175, 2534–2540 [DOI] [PubMed] [Google Scholar]
- 43. Soylu A., Ate H., Cingöz S., Türkmen M., Demir B. K., Tunca M., Sakızlı M., Cirit M., Ersoy R., Ulgenalp A., and Kavukçu S. (2011) TLR polymorphisms in FMF: association of TLR-2 (Arg753Gln) and TLR-4 (Asp299Gly, Thre399Ile) polymorphisms and myeloid cell TLR-2 and TLR-4 expression with the development of secondary amyloidosis in FMF. Inflammation 34, 379–387 [DOI] [PubMed] [Google Scholar]
- 44. Gianni T., and Campadelli-Fiume G. (2014) The epithelial αvβ3-integrin boosts the MYD88-dependent TLR2 signaling in response to viral and bacterial components. PLoS Pathog. 10, e1004477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rahman A., Sobia P., Gupta N., Kaer L. V., and Das G. (2014) Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol. PLoS ONE 9, e86886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y., Wang Y., Yamakuchi M., Isowaki S., Nagata E., Kanmura Y., Kitajima I., and Maruyama I. (2001) Upregulation of Toll-like receptor 2 gene expression in macrophage response to peptidoglycan and high concentration of lipopolysaccharide is involved in NF-κB activation. Infect. Immun. 69, 2788–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T., Lafuse W. P., and Zwilling B. S. (2000) Regulation of Toll-like receptor 2 expression by macrophages following Mycobacterium avium infection. J. Immunol. 165, 6308–6313 [DOI] [PubMed] [Google Scholar]
- 48. Rehli M. (2002) Of mice and men: species variations of Toll-like receptor expression. Trends Immunol. 23, 375–378 [DOI] [PubMed] [Google Scholar]
- 49. Haehnel V., Schwarzfischer L., Fenton M. J., and Rehli M. (2002) Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J. Immunol. 168, 5629–5637 [DOI] [PubMed] [Google Scholar]
- 50. Musikacharoen T., Matsuguchi T., Kikuchi T., and Yoshikai Y. (2001) NF-κB and STAT5 play important roles in the regulation of mouse Toll-like receptor 2 gene expression. J. Immunol. 166, 4516–4524 [DOI] [PubMed] [Google Scholar]
- 51. Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., and Aderem A. (2000) The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., and Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 53. Bulut Y., Faure E., Thomas L., Equils O., and Arditi M. (2001) Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167, 987–994 [DOI] [PubMed] [Google Scholar]
- 54. Chattopadhyay S., and Sen G. C. (2014) Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 25, 533–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., and Knaus U. G. (2000) Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 56. Herrtwich L., Nanda I., Evangelou K., Nikolova T., Horn V., Sagar Erny D., Stefanowski J., Rogell L., Klein C., Gharun K., Follo M., Seidl M., Kremer B., Münke N., et al. (2016) DNA Damage signaling instructs polyploid macrophage fate in granulomas. Cell 167, 1264–1280 e1218 [DOI] [PubMed] [Google Scholar]
- 57. Figueroa L., Xiong Y., Song C., Piao W., Vogel S. N., and Medvedev A. E. (2012) The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J. Immunol. 188, 4506–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 59. Anes E., Peyron P., Staali L., Jordao L., Gutierrez M. G., Kress H., Hagedorn M., Maridonneau-Parini I., Skinner M. A., Wildeman A. G., Kalamidas S. A., Kuehnel M., and Griffiths G. (2006) Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell Microbiol. 8, 939–960 [DOI] [PubMed] [Google Scholar]
- 60. Singh R. P., Jhamb S. S., and Singh P. P. (2009) Effect of morphine on Mycobacterium smegmatis infection in mice and macrophages. Indian J. Microbiol. 49, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]