Abstract
Previous proteomic analyses have shown that aminoacyl-tRNA synthetases in many organisms can be modified by acetylation of Lys. In this present study, leucyl-tRNA synthetase and arginyl-tRNA synthetase from Escherichia coli (EcLeuRS and EcArgRS) were overexpressed and purified and found to be acetylated on Lys residues by MS. Gln scanning mutagenesis revealed that Lys619, Lys624, and Lys809 in EcLeuRS and Lys126 and Lys408 in EcArgRS might play important roles in enzyme activity. Furthermore, we utilized a novel protein expression system to obtain enzymes harboring acetylated Lys at specific sites and investigated their catalytic activity. Acetylation of these Lys residues could affect their aminoacylation activity by influencing amino acid activation and/or the affinity for tRNA. In vitro assays showed that acetyl-phosphate nonenzymatically acetylates EcLeuRS and EcArgRS and suggested that the sirtuin class deacetylase CobB might regulate acetylation of these two enzymes. These findings imply a potential regulatory role for Lys acetylation in controlling the activity of aminoacyl-tRNA synthetases and thus protein synthesis.
Keywords: acetylation, amino acid, aminoacyl tRNA synthetase, catalysis, transfer RNA (tRNA), translation
Introduction
Aminoacyl-tRNA synthetase (aaRS)2 catalyzes esterification between its cognate amino acid and tRNA to produce aminoacyl-tRNA (aa-tRNA) in the initiation step of translation. A high level of accuracy is essential during aminoacylation to ensure quality control during protein synthesis. Disruption of translational fidelity can lead to mistranslation, with profound consequences for both prokaryotic and eukaryotic cells (1–3).
The 20 aaRSs can be divided into two classes, each with 10 members, based on sequence identity and characteristic structural motifs (4). Class I members have two signature peptides (HIGH and KMSK) located in the active site that form a characteristic dinucleotide binding fold (Rossmann fold, β-α-β-α-β). Both leucyl- and arginyl-tRNA synthetases (LeuRS and ArgRS) belong to class I aaRSs (5). Like 16 other aaRSs, LeuRS catalyzes aminoacylation of its cognate tRNA in a two-step reaction: (a) activation of the amino acid with ATP and formation of an aminoacyl adenylate and (b) transfer of the aminoacyl moiety from the aminoacyl adenylate to the cognate tRNA substrate (5, 6). However, ArgRS, together with glutamyl-tRNA synthetase and glutaminyl-tRNA synthetase, requires the presence of the cognate tRNA for amino acid activation (7, 8).
LeuRS consists of a Rossmann fold domain for aminoacylation, a helix bundle domain for binding the tRNA anticodon, a connective peptide 1 (CP1) domain for editing mischarged tRNA, a ZN1 module, a leucine-specific domain, and a C-terminal domain (CTD) for tRNA binding (9, 10). The aminoacylation and editing mechanisms of LeuRS from various species have been thoroughly investigated (11–13). ArgRS can be divided into five domains: an N-terminal additional domain (Add1) for tRNA D-loop recognition, a catalytic Rossmann fold domain, two domains (Ins-1 and Ins-2) inserted in the active site, and a C-terminal additional domain (Add2) that participates in the binding of the tRNA anticodon (14, 15). It is peculiar that the Add1 domain is conserved in ArgRS but not in other class I aaRSs. In most species, ArgRS lacks a canonical KMSK sequence, and a conserved lysine (Lys) upstream of the HIGH sequence motif in these enzymes stabilizes the transition state of the amino acid activation reaction (Arg-AMP formation) to compensate for the loss of the second Lys (K2) in the KMSK motif (16, 17).
Cells are constantly faced with the challenge of changing environmental conditions, and post-translational modification (PTM) is one method of dealing with this challenge. PTM can expand the genetic lexicon by endowing proteins with diversity beyond that can be achieved with the canonical 20 proteinogenic amino acids. Acetylation of the α-amino group of the N-terminal amino acid (irreversible) or the ϵ-amino group of internal Lys residues (reversible) is one type of PTM. In general, acetylation of Lys refers to reversible acetylation, and it can regulate fundamental cellular processes such as transcription, translation, pathways associated with central metabolism, and stress responses (18). Although the essential regulatory role of Lys acetylation in eukaryotes is widely accepted and relatively well understood, its function in bacteria and archaea remains more obscure (19, 20).
In Escherichia coli, although several putative protein acetylases are present in the genome, Gcn5-like YfiQ, which is highly similar to the acetyltransferase Pat in Salmonella enterica, is the only confirmed acetyltransferase to date (21–23), and CobB is the predominate deacetylase, which belongs the NAD+-dependent sirtuin family (22–24). Recently, the serine hydrolase YcgC was identified as a Zn2+- and NAD+-independent deacetylase that regulates a distinct set of substrates from CobB (25). One well studied target of protein acetylation is acetyl-CoA synthetase (Acs), which activates acetate to the high-energy intermediate acetyl coenzyme A (Ac-CoA; acetate + ATP + CoA → AMP + PPi + Ac-CoA). Reversible acetylation of a catalytic core Lys residue conserved in Acs enzymes from bacteria to human (Lys609 in S. enterica Acs) could regulate enzyme activity, because it blocks ATP-dependent adenylation of acetate, preventing the formation of acetyl-AMP and the subsequent production of Ac-CoA (18, 21). In S. enterica, Acs Lys609 is regulated by a protein acetylation/deacetylation system that includes Pat and CobB (21), and this system also coordinates carbon source utilization and metabolic flux by controlling the acetylation of metabolic enzymes (26). Interestingly, acetylation was recently found to be mediated nonenzymatically in mitochondria of both prokaryotes and eukaryotes (20, 23, 27–31). In E. coli, the majority of acetylation occurs independently of YfiQ, and the glycolysis intermediate acetyl-phosphate (AcP) is associated with a global shift in protein acetylation, whereas CobB regulates a subset of these chemical acetylation events (20, 23, 28).
Some aaRSs are modified by phosphorylation, which influences multidrug tolerance in E. coli and the reactive oxygen species defense mechanism in mammalian cells (32, 33). Despite growing knowledge, studies focusing on other forms of PTM of aaRSs are few in number. A large-scale proteomic survey demonstrated that some aaRSs from E. coli, S. enterica, Bacillus subtilis, Drosophila melanogaster, Mus musculus, Rattus norvegicus, and Homo sapiens are acetylated, and bioinformatics and network analysis of acetylation sites found aa-tRNA biosynthesis pathway enriched in some species (23, 26, 31, 34–42). Furthermore, some of the identified acetylated Lys residues are conserved, and it would be intriguing to decipher the exact role of acetylation of aaRSs.
Herein, we discovered that EcLeuRS and EcArgRS are modified by acetylation of Lys residues in vivo. By utilizing an engineered Methanosarcina barkeri pyrrolysyl-tRNA synthetase (MbPylRS)/MbtRNACUA pair (Nϵ-acetyllysyl-tRNA synthetase/tRNACUA pair, pAcKRS) system (43), we obtained enzymes harboring acetylation at specific sites and investigated the biochemical properties of EcLeuRS-KAcs and EcArgRS-KAcs. We also examined the molecular mechanism controlling regulation of the acetylation of EcLeuRS and EcArgRS and identified CobB and AcP as possible regulatory factors. Acetylation appears to be a mechanism for adjusting the activity of aaRSs and thereby controlling protein synthesis.
Results
MS revealed acetylation at 11 Lys residues in EcLeuRS
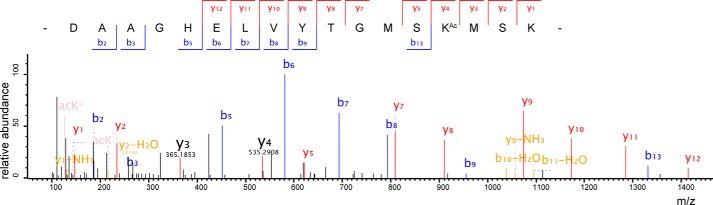
Previous studies demonstrated that LeuRS from E. coli, Saccharomyces cerevisiae, R. norvegicus, H. sapiens, and other species can be acetylated (38, 40, 41, 44). To identify the acetylation sites in EcLeuRS, we overexpressed ecleuS that encodes EcLeuRS with a N-terminal His6 tag in E. coli BL21 and purified the recombinant protein by Ni2+-NTA affinity chromatography. 11 Lys residues were detected to be acetylated in three independent MS analyses. They span the entire protein and include Lys619 and Lys624 in the KMSK signature sequence (Figs. 1 and 2, A and B).
Figure 1.
Identification of acetylation at Lys619 of EcLeuRS by MS. MS/MS spectrum of a tryptic peptide from EcLeuRS (DAAGHELVYTGMSKAcMSK) shows acetylation of Lys (KAc), confirmed as Lys619 by sequence alignment with the known sequence of EcLeuRS. Most major fragmentation ions matched predicted b or y ions.
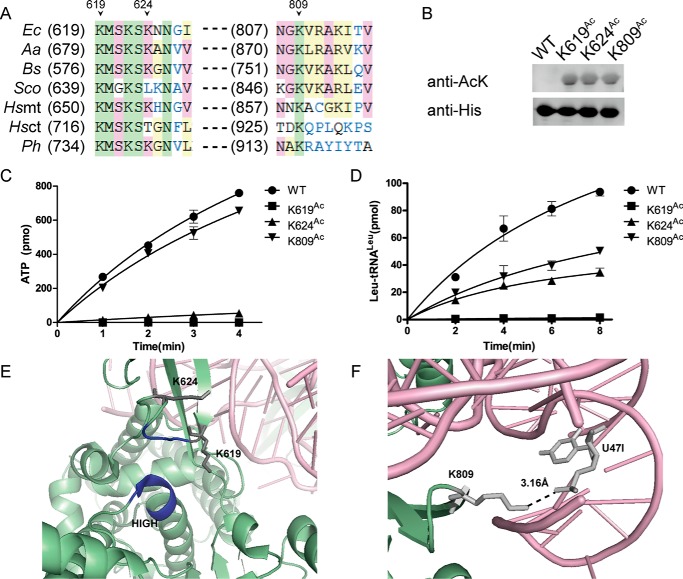
Figure 2.
Identification of Lys residues acetylated in EcLeuRS. A, the overall ternary structure of EcLeuRS and its cognate tRNALeu together with Leu in the editing conformation (PDB number 4ARC). B, schematic diagram of EcLeuRS. RF, Rossmann fold. C, aminoacylation assays screening potential crucial Lys residues. Left panel, mutation of Lys619, Lys624, and Lys809 to Gln damaged EcLeuRS canonical activities. Right panel, mutation of other Lys residues had a slightly negative effect on EcLeuRS canonical activities. The results are averages plus standard deviations from three independent experiments.
To understand the effect of acetylation on the aminoacylation activity of EcLeuRS, we separately mutated all 11 Lys residues to Gln, because this residue lacks a positive charge on the side chain and is a good mimic of acetylated Lys. We assayed the aminoacylation activity of the K-Q mutants and found that mutations at Lys619 and Lys624 in the amino acid activation active site and Lys809 in the CTD displayed decreased aminoacylation activity compared with WT EcLeuRS, whereas the aminoacylation activity of mutants at all other sites was unchanged (Fig. 2C). Lys402 is the only residue in the CP1 domain among these 11 residues, and the co-crystal structure of EcLeuRS with tRNALeu and Leu (PDB number 4ARC) indicated that Lys402 lies on the surface of the CP1 pocket and points away from the domain core, suggesting it is not likely to be essential for the editing function. Indeed, Ile-tRNALeu deacylation assays showed that the post-transfer editing activity of the EcLeuRS-K402Q mutant remained unchanged compared with native EcLeuRS (data not shown).
Characterization of KAc mutants reveals that Lys acetylation reduces EcLeuRS enzyme activity
To further explore the influence of acetylation on EcLeuRS, we used a previously described system to incorporate Nϵ-acetyl-l-Lys (AcK) at specific sites to generate EcLeuRS-KAcs in situ (Fig. 3) (43). We transformed E. coli BL21 cells with two plasmids: pAcKRS encoding the Nϵ-acetyllysyl-tRNA synthetase/tRNACUA pair that activates AcK and recognizes the UAG codon and another encoding EcLeuRS in which the Lys triplet codon was substituted with TAG.
Figure 3.
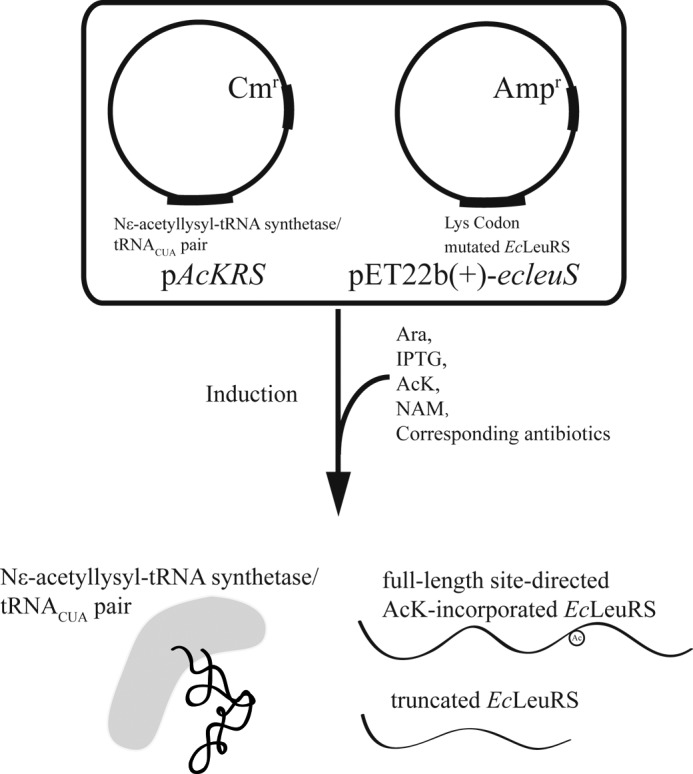
Flow diagram of the overexpression of site-directed AcK-incorporated proteins in E. coli BL21 (DE3). Taking EcLeuRS as an example, pAcKRS and pET22b(+)-ecleuS were co-transformed in E. coli BL21 (DE3), and engineered pAcKRS were induced by the addition of Ara. Subsequently, IPTG was added to induce the production of EcLeuRS in the presence of NAM, an inhibitor of CobB. With the assistance of pAcKRS, EcLeuRS was translated in full-length form with incorporation of AcK or in truncated form (terminating at the Lys codon mutation site). All other experimental details were as described previously (43).
Sequence alignment showed that Lys619 and Lys809 of EcLeuRS are conserved in LeuRSs from various species, whereas Lys624 is basically conserved in prokaryotic LeuRSs (Fig. 4A). Following overexpression as described above, EcLeuRS-K619Ac, EcLeuRS-K624Ac, and EcLeuRS-K809Ac were purified and confirmed to be 90% homogeneous by SDS-PAGE (data not shown). Western blotting confirmed the incorporation of AcK into EcLeuRS (Fig. 4B), and comparison of CD spectra of EcLeuRS-WT and EcLeuRSAcs confirmed that EcLeuRSAcs were properly folded (data not shown).
Figure 4.
Effect of acetylation of Lys residues on the Leu activation and aminoacylation activities of EcLeuRS. A, sequence alignment of LeuRSs from various species in regions homologous to Lys619, Lys624, and Lys809 in EcLeuRS. Ec, E. coli; Aa, Aquifex aeolicus; Bs, B. subtilis; Sco, Streptomyces coelicolor; Hs, H. sapiens; Ph, Pyrococcus horikoshii; mt, mitochondrial; ct, cytoplasmic. B, Western blotting confirming the incorporation of AcK in EcLeuRS-K619Ac, EcLeuRS-K624Ac, and EcLeuRS-K809Ac. C, Leu activation of EcLeuRS-K619Ac, EcLeuRS-K624Ac, and EcLeuRS-K809Ac. D, aminoacylation of EcLeuRS-K619Ac, EcLeuRS-K624Ac, and EcLeuRS-K809Ac resembling that of the K-Q mutants. E, closer view of the orientation of Lys619 and Lys624 relative to the conserved HIGH and KMSK motifs (HMGH and KMSK in EcLeuRS, depicted in dark blue; PDB code 4ARC). F, closer view of the interaction between EcLeuRS Lys809 and EctRNALeu U47I (PDB code 4ARC). The results are the averages and standard deviations from three independent experiments, and all Western blots were repeated.
Lys619 and Lys624 are located in or downstream of the conserved KMSK loop, which, together with the HIGH motif, is essential for the amino acid activation activity. EcLeuRS-K619Ac completely lost its Leu activation and leucylation activities (Fig. 4, C and D, and Tables 1 and 2). The amino acid activation and aminoacylation activities of EcLeuRS-K624Ac were also determined. Even though EcLeuRS-K624Ac severely lost its activation activity (Fig. 4C and Table 1), the catalytic efficiency (kcat/Km) in aminoacylation was not that severely damaged (Fig. 4D and Table 2). The total effect might be because tRNA charging is the rate-limiting step. Kd values between EcLeuRSs and tRNALeu calculated by fluorescence quenching showed that the binding affinity of EcLeuRS-K619Ac and-K624Ac with tRNALeu was not altered compared with that of EcLeuRS-WT (Table 3). These results suggest acetylation of these two Lys residues (especially Lys619) might lead to a conformational change in the synthetic active site pocket, decreasing the Leu activation and aminoacylation activities (Fig. 4E).
Table 1.
Observed rate constants (kobs) of EcLeuRSs in Leu activation
All parameters are average values from three independent determinations with standard deviations. nm, nonmeasurable (too low to be measured).
| Enzymes | kobs | Relative values |
|---|---|---|
| s−1 | % | |
| EcLeuRS-WT | 55.0 ± 3.6 | 100 |
| EcLeuRS-K619Ac | nm | |
| EcLeuRS-K624Ac | 4.4 ± 0.6 | 8 |
| EcLeuRS-K809Ac | 48.3 ± 5.1 | 88 |
Table 2.
Kinetic parameters of EcLeuRS and derived site-specific acetylated variants for EctRNALeu in aminoacylation
All parameters are average values from three independent determinations with standard deviations. nm, nonmeasurable (too low to be measured).
| Enzymes | Km | kcat | kcat/Km | Relative catalytic efficiency |
|---|---|---|---|---|
| μm | s−1 | s−1 μm−1 | % | |
| EcLeuRS-WT | 1.2 ± 0.3 | 5.7 ± 0.3 | 4.8 | 100 |
| EcLeuRS-K619Ac | nm | nm | ||
| EcLeuRS-K624Ac | 1.3 ± 0.2 | 2.8 ± 0.3 | 2.2 | 46 |
| EcLeuRS-K809Ac | 6.7 ± 0.7 | 5.0 ± 0.4 | 0.7 | 15 |
Table 3.
Kd values between tRNALeu and EcLeuRSs determined by fluorescence quenching
All parameters are average values from three independent determinations with standard deviations.
| Enzymes | Kd | Relative values |
|---|---|---|
| μm | -fold | |
| EcLeuRS-WT | 0.19 ± 0.01 | 1.0 |
| EcLeuRS-K619Ac | 0.21 ± 0.03 | 1.1 |
| EcLeuRS-K624Ac | 0.18 ± 0.01 | 0.9 |
| EcLeuRS-K809Ac | 0.27 ± 0.02 | 1.4 |
The flexibly linked CTD in LeuRS makes contacts with tertiary structural base pairs and the long variable arm of tRNALeu (10, 45). The ternary complex structure of EcLeuRS, tRNALeuUAA, and Leu in the editing conformation (PDB number 4ARC) revealed that Lys809 is located on the edge of one β-sheet in the CTD. The Leu activation activity of EcLeuRS-K809Ac was not changed compared with that of WT EcLeuRS; consistent with that, Lys809 is distant from the activation active site core region (Fig. 4C and Table 1). The crystal structure of EcLeuRS (PDB number 4ARC) showed that the side chain of Lys809 lies at a distance of 3.16 Å away from the phosphate group of the U47I ribose backbone of tRNALeu (Fig. 4F). EcLeuRS-K809Ac had a similar kcat toward EctRNALeu as did EcLeuRS-WT (5.0 s−1 for EcLeuRS-K809Ac, 5.7 s−1 for WT); nevertheless, the affinity for the cognate EctRNALeu (Km, 6.7 μm for EcLeuRS-K809Ac and 1.2 μm for WT) was decreased, and the catalytic efficiency (kcat/Km) of EcLeuRS-K809Ac (0.7 s−1 μm−1) was only 15% that of EcLeuRS-WT (4.8 s−1 μm−1; Table 2). In addition, the Kd of EcLeuRS-K809Ac with tRNALeu was 1.4-fold that of the native enzyme, implying a decrease in binding affinity between LeuRS and tRNALeu (Table 3). The interaction between enzyme and tRNALeu was partially disrupted by the acetylation of Lys809.
These results indicate that acetylation of Lys619, Lys624, and Lys809 could potentially inhibit either amino acid activation or tRNA-charging activities of EcLeuRS. Among these residues, Lys619 and Lys809 in EcLeuRS are the residues whose acetylation lead to a sharp reduction of catalytic efficiency. In addition, the aminoacylation activity of EcLeuRSAcs was comparable with the corresponding K-Q mutants (Figs. 2C and 4D), suggesting that Gln is a suitable mimic of acetylated Lys.
EcLeuRS is acetylated by AcP rather than YfiQ, and CobB can deacetylate EcLeuRSAc
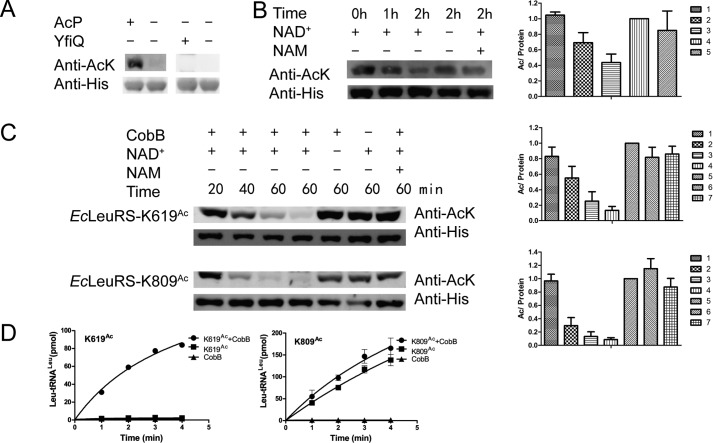
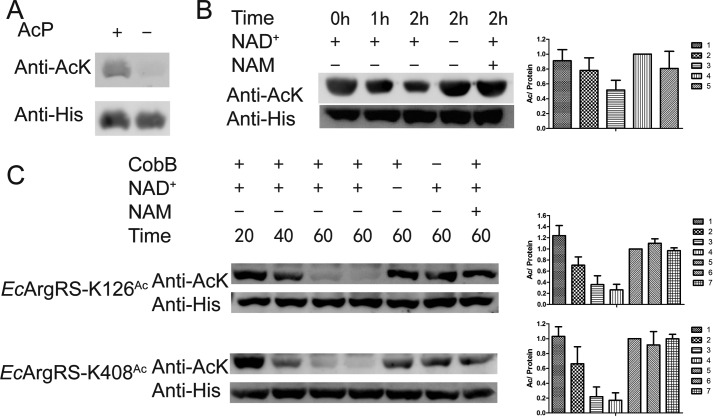
Determining the enzyme responsible for acetylating EcLeuRS is of particular interest. At present, the Gcn5-like acetyltransferase YfiQ is the only known enzyme that acetylates the ϵ-NH2 group of Lys in E. coli (23). However, purified YfiQ was unable to transfer the acetyl group of Ac-CoA to EcLeuRS in vitro (Fig. 5A). The metabolism of AcP, a high-energy intermediate between acetate and Ac-CoA, has been shown to alter global acetylation levels in vivo, and AcP acetylates proteins nonenzymatically at multiple Lys residues in vitro (23). Given that AcP is a critical regulator of acetylation in bacteria, we incubated purified EcLeuRS with AcP and detected an increase in EcLeuRS acetylation (no acetylation signal is detected on WT enzyme before AcP treatment), implying a potential role for AcP in the acetylation of LeuRS (Fig. 5A). We refer to this AcP-derived form of EcLeuRS as EcLeuRSAc.
Figure 5.
AcP and CobB regulate acetylation of EcLeuRS. A, AcP but not YfiQ acetylates EcLeuRS in vitro. B, CobB removal of the acetyl moiety of EcLeuRSAc. C, CobB deacetylation of EcLeuRS-K619Ac and EcLeuRS-K809Ac. In the presence of NAM or the absence of NAD+, CobB is inactivated. D, incubation with CobB recovers the aminoacylation activity of EcLeuRS-K619Ac (left panel) and EcLeuRS-K809Ac (right panel). All experiments were conducted at least twice. When quantifying of the relative amount of AcK signal/His signal, the sample without NAD+ and NAM was defined as 100%.
CobB, belonging to the sirtuin class, is the main deacetylase in E. coli (22–24). To determine whether CobB is involved in deacetylation of EcLeuRSAc, deacetylation assays were performed with EcLeuRSAc as the substrate for CobB. The results showed that CobB could deacetylate EcLeuRSAc, and the presence of the CobB inhibitor NAM, or the absence of its cofactor, NAD+, rendered CobB inactive (Fig. 5B). We also tested the activity of CobB on purified EcLeuRS-K619Ac and EcLeuRS-K809Ac, and Western blotting showed that CobB effectively decreased their acetylation in a time-dependent manner (Fig. 5C). Additionally, aminoacylation assays showed that after treatment with CobB, EcLeuRS-K619Ac and EcLeuRS-K809Ac recovered aminoacylation activity to some extent (Fig. 5D). The above results suggest CobB deacetylates EcLeuRS in vitro.
Acetylation of EcArgRS influences its catalytic rate
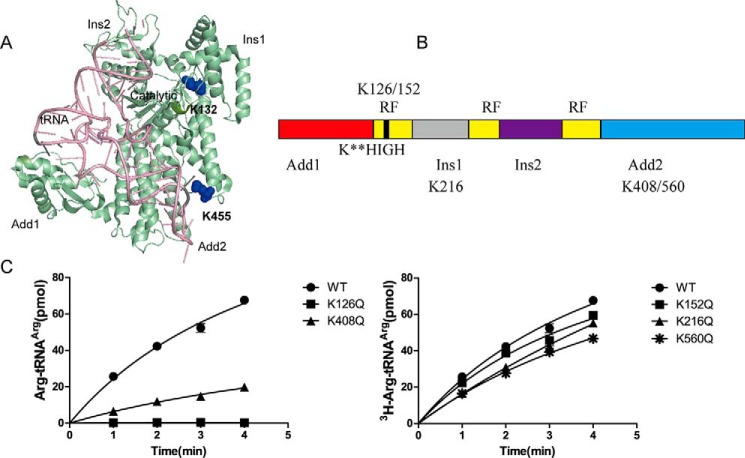
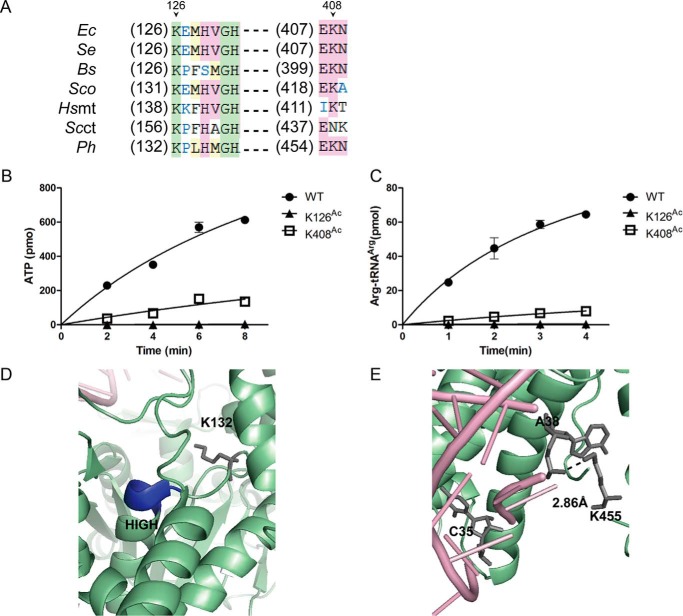
To investigate whether EcArgRS is regulated by acetylation, similar experiments to those described above were performed on EcArgRS. MS (repeated three times) detected acetylation at five Lys residues in EcArgRS including Lys126, which is upstream of the HIGH motif in the activation site (Figs. 6 and 7) (16, 17). As described above for EcLeuRS, we performed Gln-scanning mutagenesis on these five Lys residues of EcArgRS and found that K-Q mutants of Lys126 and Lys408 displayed a decrease in aminoacylation activity (Fig. 7C). Sequence alignment showed that Lys126 and Lys408 are highly conserved among prokaryotic and eukaryotic ArgRSs (Fig. 8A). As described above for EcLeuRS, we utilized the pAcKRS system to generate EcArgRS-KAcs, and CD spectra confirmed that their secondary structures were not altered by the point mutations (data not shown).
Figure 6.
Identification of acetylation at Lys126 of EcArgRS by MS. MS/MS spectrum of a tryptic peptide from EcArgRS (QTIVVDYSAPNVAKAcEMHVGHLR) showing acetylation of Lys (KAc), confirmed as Lys126 by sequence alignment of the peptide with the known sequence of EcArgRS. Most major fragmentation ions matched predicted b or y ions.
Figure 7.
Lys residues acetylated in EcArgRS. A, crystal structure of PhArgRS complexed with PhtRNAArg and ANP (PDB code 2ZUE). B, schematic diagram of EcArgRS. RF, Rossmann fold. C, aminoacylation assay screening of potentially crucial Lys residues. Left panel, mutation of Lys126 and Lys408 damages the enzymatic activities of EcArgRS. Right panel, mutation of other Lys residues has a slight negative effect on the activities EcArgRS. The results are the averages and standard deviations from three independent experiments.
Figure 8.
Effect of acetylation of Lys on the Leu activation and aminoacylation activities of EcArgRS. A, sequence alignment of ArgRSs from various species in regions homologous to Lys126 and Lys408 of EcArgRS. The abbreviations are the same as those in Fig. 4, except for two species (Se, S. enterica; Sc, S. cerevisiae). B, Arg activation of EcArgRS-KAcs. C, aminoacylation of EcArgRS-KAcs resembling that of the K-Q mutants. D, orientation of PhArgRS Lys132 (homologous to Lys126 in EcArgRS) relative to the conserved HIGH motif (HMGH in PhArgRS, depicted in dark blue). E, closer view of the interaction between PhArgRS Lys455 (corresponding to Lys408 in EcArgRS) and PhtRNAArg A38. The results are the averages and standard deviations from three independent experiments.
EcArgRS-K126Ac lost Arg activation and arginylation activities and its affinity with tRNAArg did not change compared with WT enzyme (Fig. 8, B and C; see Table 6). Lys126 lies in the upstream of the HIGH (HVGH in EcArgRS) sequence within the catalytic pocket of EcArgRS, where it compensates for the lack of a canonical KMSK (especially the second Lys, K2) in ArgRS in most species and thus makes an important contribution to the aminoacylation reaction (Fig. 8D) (16, 17). Acetylation of Lys126 in EcArgRS led to the complete loss of arginylation, consistent with previous experiments on TtArgRS-K116G that also showed a complete loss of activation and aminoacylation activities (17).
Table 6.
Kd values between tRNAArg and EcArgRSs determined by fluorescence quenching
All parameters are average values from three independent determinations with standard deviations.
| Enzymes | Kd | Relative value |
|---|---|---|
| μm | -fold | |
| EcArgRS-WT | 0.24 ± 0.03 | 1.0 |
| EcArgRS-K126Ac | 0.26 ± 0.02 | 1.1 |
| EcArgRS-K408Ac | 0.40 ± 0.03 | 1.7 |
We next focused on the Lys408 residue in the Add2 domain that is implicated in the binding of the tRNAArg anticodon region (15, 46). EcArgRS-K408Ac displayed a low Arg activation activity (Fig. 8B and Table 4) and aminoacylation activity (Fig. 8C and Table 5), and kinetic parameters revealed a weaker affinity for EctRNAArg (Km = 9.0 μm) compared with WT enzyme (Km = 2.7 μm). The kcat (6.9 s−1) was also decreased to 24% that of WT (28.4 s−1), and the overall effect was a decrease in the catalytic efficiency (kcat/Km, 0.8 s−1 μm−1) to only 8% that of the native enzyme (10.5 s−1 μm−1) (Table 5). The Kd value of EcArgRS-K408Ac with tRNAArg (0.40 μm) was also increased by ∼1.7-fold compared with EcArgRS-WT (0.24 μm; Table 6). Lys408 is located in the hairpin following helix α13 of EcArgRS, corresponding to Lys455 in PhArgRS. In the crystal structure of PhArgRS (PDB number 2ZUE), Lys455 lies within an α-helix adjacent to the phosphate group of A38 in the anticodon loop of tRNAArg (Fig. 8E). In yeast and E. coli, C35 is one of the major identity elements of tRNAArg (47, 48). Acetylation of Lys408 may distort the neighboring region that interacts with the tRNAArg anticodon and consequently decrease the affinity of EcArgRS for tRNAArg. Given that EcArgRS requires tRNAArg as the activator during Arg activation, the decrease in the EcArgRS-K408Ac activation activity could be partly due to a loss in affinity with tRNAArg. Overall, the in vitro data show that the acetylation of EcArgRS (Lys126 and Lys408) could severely decrease its aminoacylation activity.
Table 4.
Observed rate constants (kobs) of EcArgRSs in the presence of EctRNAArg in Arg activation
All parameters are average values from three independent determinations with standard deviations. nm, nonmeasurable (too low to be measured).
| Enzymes | kobs | Relative values |
|---|---|---|
| s−1 | % | |
| EcArgRS-WT | 43.5 ± 2.7 | 100 |
| EcArgRS-K126Ac | nm | |
| EcArgRS-K408Ac | 10.7 ± 0.6 | 25 |
Table 5.
Kinetic parameters of EcArgRS and derived site-specific acetylated variants for EctRNAArg in aminoacylation
All parameters are average values from three independent determinations with standard deviations. nm, nonmeasurable (too low to be measured).
| Enzymes | Km | kcat | kcat/Km | Relative catalytic efficiency |
|---|---|---|---|---|
| μm | s−1 | s−1 μm−1 | % | |
| EcArgRS-WT | 2.7 ± 0.1 | 28.4 ± 3.4 | 10.5 | 100 |
| EcArgRS-K126Ac | nm | nm | ||
| EcArgRS-K408Ac | 9.0 ± 0.6 | 6.9 ± 0.5 | 0.8 | 8 |
EcArgRS acetylation appears to be regulated by AcP and CobB
We also attempted to identify enzymes or other molecules involved in acetylation of EcArgRS. In vitro assays showed that AcP acetylated purified EcArgRS (Fig. 9A), but YfiQ did not (data not shown) (no acetylation signal is detected on WT enzyme before AcP treatment). Furthermore, in vitro CobB deacetylated EcArgRSAc, the product of EcArgRS following treatment with AcP (Fig. 9B). Purified EcArgRS-K126Ac and EcArgRS-K408Ac variants were also deacetylated by CobB, and as described above, addition of NAM or the absence of NAD+ caused CobB to lose its deacetylation activity (Fig. 9C). AcP and CobB therefore appear to control acetylation and deacetylation of EcArgRS, consistent with the results discovered above for EcLeuRS.
Figure 9.
AcP and CobB regulate acetylation of EcArgRS. A, AcP acetylates EcArgRS in vitro. B, CobB removes the acetyl moiety of EcArgRSAc. C, CobB deacetylates EcArgRS-K126Ac and other acetylated variants. CobB deacetylation activity is lost in the presence of NAM or the absence of NAD+. All experiments were performed at least twice. When quantifying of the relative amount of AcK signal/His signal, the group without NAD+ and NAM was defined as 100%.
Discussion
AaRSs are found to be acetylated
Some aaRSs are post-translationally modified. In E. coli, tRNAGlu-bound glutamyl-tRNA synthetase can be phosphorylated at Ser239 in the KMSK motif by the eukaryote-like serine-threonine kinase HipA. This PTM results in a loss of aminoacylation activity, which increases uncharged tRNAGlu loading at the A site of the ribosome, triggering (p)ppGpp formation and facilitating multidrug tolerance (32).
Acetylation regulates proteins involved in transcription, amino acid, nucleotide and protein biosynthesis, protein folding, and detoxification responses in various species. We focused on aaRSs and found that many are acetylated on Lys residues located both on the surface and within the catalytic core or the tRNA binding domain in others' papers. In the present work, we confirmed the acetylation of two class Ia aaRSs in E. coli by three independent MS experiments. The residues found to be acetylated on these two aaRSs are not exactly the same as what were found in the studies of Weinert et al. (23) and Kuhn et al. (28). This might result from the use of different E. coli strains under various growth stages. Differences of nutrients in the media used might also influence the acetylation. In addition, we purified overexpressed proteins, rather than endogenous aaRSs. However, acetylation of some crucial residues that had been identified by the above authors (like Lys619 and Lys624 in EcLeuRS and Lys126 and Lys408 in EcArgRS) were consistently identified in our studies. We screened several Lys residues at which acetylation may negatively regulate the aminoacylation activity of enzyme by using a Gln scanning mutagenesis approach. Furthermore, we used the pAcKRS system to express and purify EcLeuRS and EcArgRS acetylated at specific sites (43) and characterized the effect of acetylation on the catalytic properties.
The catalytic activity of aaRSs is regulated by acetylation
Acetylation of Lys619 and Lys809 greatly impacted the aminoacylation activity of EcLeuRS, and both residues are highly conserved among LeuRS in various species (Fig. 4A). Lys622 is the second Lys in the KMSK motif (K2), which is the key residue that directly interacts with Leu-AMP and believed to stabilize the negatively charged transition state of the first reaction step in class I aaRSs. Mutation of this residue led to a severe loss of enzymatic activity (49–51). Acetylation of the adjacent residues, Lys619 and Lys624 caused significant inhibition of the first step of aminoacylation. In Bacillus stearothermophilus tyrosyl-tRNA synthetase (TyrRS), another class I aaRS, 230KFGK233 corresponds to the signature sequence KMSK. Previous data showed that the BsTyrRS-K230N mutant lost its activation activity, indicating that the first Lys in the KMSK is crucial to the activation of tyrosine (49). Lys619 is the first Lys in the signature sequence of EcLeuRS, and Lys624 is adjacent to the signature sequence. Acetylation of the two Lys residues should inhibit the active site of amino acid activation and thus influence the activation and aminoacylation activities of EcLeuRS. Hsu et al. (52) utilized Ala scanning mutagenesis to identify specific sites in the CTD that may be important for RNA-protein interactions and found that mutation of Lys809 had a negligible effect on aminoacylation catalytic efficiency. By contrast, our results suggest acetylation of Lys809 has a marked negative effect on the affinity for EctRNALeu.
Previously, relatively less attention has been paid to EcArgRS. Lys126, the residue upstream the signature sequence HIGH (HVGH), makes up for the absence of K2 in the KMSK motif in EcArgRS (17), and acetylation of key residues in the activation pocket leads to the complete loss of activation activity and consequent aminoacylation activity. Lys408 (PhArgRS Lys455) is in the vicinity of the tRNAArg anticodon region that harbors the identity element C35 (PDB number 2ZUE), and acetylation of Lys408 inhibited amino acid activation and transfer of the arginyl group from Arg-AMP to the 3′end of tRNAArg. These results suggest for the first time that acetylation regulates the amino acid activation and aminoacylation activities of EcArgRS and EcLeuRS.
Acetylation of EcArgRS and EcLeuRS was first reported in previous MS studies, including acetylation of Lys residues surrounding or within the conserved HIGH and KMSK motifs (23, 40, 42). We noticed that the conserved Lys residue upstream of the HIGH motif was also acetylated in various ArgRSs (Lys156 in the yeast cytosol, Lys139 in R. norvegicus mitochondria, and Lys205 in the H. sapiens, R. norvegicus, and M. musculus cytosol; Fig. 10A) (35, 38, 39, 54). These results indicate that Lys acetylation is a conserved mechanism for regulating the catalytic activity of aaRSs in particular conditions.
Figure 10.
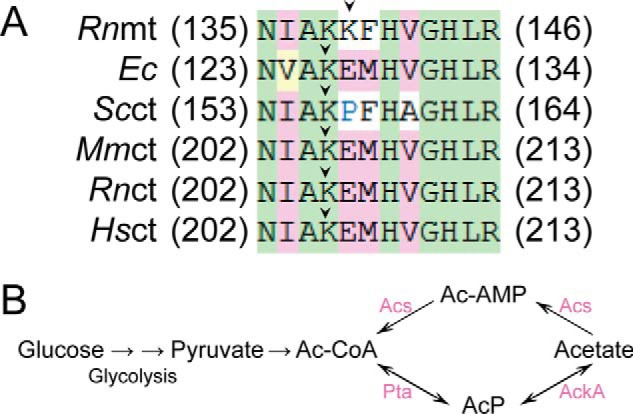
Sequence alignment of the HIGH region and the acetate metabolism pathway. A, sequence alignment of the HIGH region with ArgRSs from various species. Lys residues preceding the HIGH motif (indicated by an arrow, Lys126 in EcArgRS) were found to be acetylated by MS. Abbreviations are the same as Figs. 4 and 8, except for two species (Rn, R. norvegicus; Mm, M. musculus). B, acetate metabolism pathway in E. coli (23). Pta, AckA, and Acs are crucial enzymes involved in the interconversion of Ac-CoA and acetate.
Recently, TyrRS has also been shown to be highly acetylated in response to oxidative stress. This aaRS is primarily acetylated on Lys244 near the nuclear localization signal, and the acetylation inhibits the aminoacylation activity of TyrRS. Acetylation, which is regulated by PCAF and sirtuin 1, also promotes TyrRS translocation from cytoplasm to the nucleus and protects against DNA damage caused by oxidative stress in mammalian cells and zebrafish. This study provided us with other perspectives about the biological role of aaRS acetylation (55).
AcP and CobB appear to regulate acetylation of EcArgRS and EcLeuRS
Our results showed that AcP and CobB may regulate acetylation of EcLeuRS and EcArgRS in vitro. We also tried to explore the acetylation state of these two endogenous aaRSs, but haven't yet found a physiological state that leads to an obvious increase in the acetylation of aaRSs. In a previous study (23), the global acetylation state of E. coli was found to be elevated in growth-arrested (GE) cells compared with those in the exponential phase (EP). This accumulation required AcP, and most acetylation was independent of YfiQ. Additionally, CobB can suppress chemical acetylation by AcP in growing and GE cells (23), and proteins functioning in translation, transcription and central metabolism are acetylated, with a considerable number of the acetylated sites regulated by AcP (28).
The supplementary data in the study of Weinert et al. (23) showed that acetylation of Lys126 in EcArgRS can differ in E. coli cells cultured in M9 minimal medium on different genetic background and under different metabolic states. Acetylation of EcArgRS Lys126 in stationary phase BL21 cells was ∼10-fold higher compared with EP cells, indicating elevation of acetylation during this stage. Furthermore, acetylation of this site in EP ΔackA BW25113 cells was increased ∼10-fold compared with EP control cells, suggesting that elevation of AcP levels (ΔackA) by genetic manipulation could stimulate acetylation at specific sites (Fig. 10B). Acetylation of EcArgRS Lys126 in EP Δpta BW25113 cells was 62.5% that of the level measured in EP WT cells, indicating that a decrease in AcP in Δpta cells reduces the acetylation of Lys126 (Fig. 10B). In addition, deletion of the yfiQ gene had no direct effect on the acetylation of Lys126 in EP ΔyfiQ MG1655 cells compared with EP control MG1655 cells, consistent with our in vitro assay results that similarly indicated that YfiQ is not involved in the acetylation of the equivalent residue in EcArgRS. Further studies should focus on investigating the ability of CobB to deacetylate EcLeuRSAc and EcArgRSAc in vivo, as well as the physiological significance of acetylation. In addition, questions about whether there are some undiscovered acetyltransferases that can take AcP as a substrate and catalyze acetylation should be addressed.
Acylation of aaRSs might regulate the metabolic state of cells
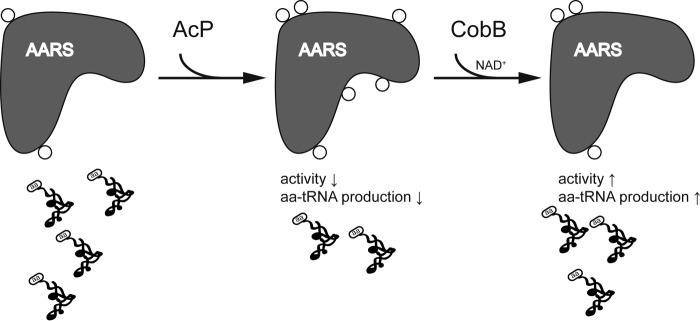
In E. coli and yeast, acetylation is much less abundant than phosphorylation, and succinylation of aaRSs (Lys619 and Lys624 on EcLeuRS) has recently been reported (56). Nonenzymatic protein acylation has been linked to negative regulation of protein function because carbon stress and deacylases are evolved to reverse this form of PTM in both prokaryotes and eukaryotes (29). Our results, together with those of previous studies, suggest that acetylation and other forms of acylation may inhibit the activity of aaRSs in response to environmental stresses. Under normal conditions, aaRSs may endure basal-level acetylation. When there is a stimulus that requires cells to reduce their growth rates, cells could utilize an economical way to slow down protein synthesis. Environmental stresses may cause AcP-mediated acetylation of aaRSs, which inhibits their aminoacylation activities, and the deacetylase CobB may reverse this PTM to recover aa-tRNA biosynthesis when conditions improve (Fig. 11). It is very interesting to understand exactly how acetylation specificity is achieved. To our understanding, first, even though AcP can acetylate peptides nonenzymatically at high concentrations, with some salt and Mg2+, selectivity and specificity can increase (28). Second, it is possible that there might be an/some undiscovered acetyltransferase(s) that can utilize AcP as a substrate and catalyze acetylation (23); lastly, deacetylase could preferentially remove acetylation on some specific sites. Moreover, cross-talk between different types of acylation may be important and should be investigated. Whether acetylation could also affect other functions of aaRSs beyond translation, as demonstrated by TryRS, is a fascinating to question to be answered (55).
Figure 11.
Proposed acetylation mechanism for aaRSs. In this model, AcP nonenzymatically acetylates aaRSs, which negatively regulates their aminoacylation activities, and this PTM is removed by CobB to recover aaRS function and maintain cellular homeostasis (29).
Concluding remarks
Herein, we confirmed acetylation of EcLeuRS and EcArgRS in vivo and identified the Lys residues involved. To investigate the significance of this form of PTM, we engineered K-Q mutants to identify residues that may affect the aminoacylation activity and employed a novel site-directed AcK incorporation system to prepare EcLeuRS and EcArgRS acetylated at specific sites. Characterization of the amino acid activation and tRNA-charging activities of these EcLeuRS-KAc and EcArgRS-KAc variants confirmed that acetylation of several Lys residues negatively regulates their catalytic activities. Subsequent in vitro assays suggest that AcP might be the source of the nonenzymatic acetylation of EcLeuRS and EcArgRS, and CobB is likely to be responsible for deacetylation. This work extends our understanding of acetylation of aaRSs. Whether this type of PTM is prevalent and physiologically important remains to be elucidated.
Experimental procedures
Materials
l-Leu, l-Arg, l-Ile, AcP (potassium lithium salt lithium potassium acetyl phosphate), Ac-CoA, AcK (Nϵ-acetyl-l-Lys), NaBu, NAD+, nicotinamide (NAM), MgCl2, NaCl, KCl, KF, ATP, Tris-HCl, HEPES, Na4PPi, inorganic pyrophosphate, DTT, activated charcoal, and His6-tagged monoclonal antibody were purchased from Sigma-Aldrich. Polyclonal antibody recognizing AcK was bought from Cell Signaling Technology (Danvers, MA). Amicon ultra-15 centrifugal filters and nitrocellulose membranes (0.22 μm) were obtained from Merck Millipore. A Bradford protein assay kit was bought from Bio-Rad. Isopropyl-1-thio-β-d-galactopyranoside (IPTG) and peptone were purchased from Amresco (Solon, OH). l-[3H]Leu, l-[3H]Ile, l-[3H]Arg, [32P]Na4PPi, and [α-32P]ATP were obtained from Perkin-Elmer. Nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography agarose was purchased from Qiagen. Superdex 75 resin and PVDF membranes were purchased from GE Healthcare. dNTP mixtures, arabinose (Ara), Tween 20, Triton X-100, BSA, Na4PPi, KH2PO4, and K2HPO4 were purchased from Sangon (Shanghai, China). Oligonucleotide primers were synthesized by Invitrogen. A DNA fragment rapid purification kit and a plasmid extraction kit were obtained from Yuanpinghao Biotech (Tianjin, China). Protein standard markers, T4 ligase, restriction endonucleases and Zeba spin desalting columns were obtained from Thermo Scientific (Waltham, MA). The KOD-plus mutagenesis kit and KOD-plus Neo enzyme were purchased from TOYOBO (Osaka, Japan), and DNA sequencing was performed by Biosune (Shanghai, China). The pAcKRS system was gift from Prof. Jiang-yun Wang. Competent E. coli Top10 and BL21 (DE3) cells were prepared in our laboratory.
Gene cloning, mutagenesis, protein expression, and purification
Plasmid pET30a(+)-ecleuS encoding EcLeuRS with a N-terminal His6 tag was constructed previously in our lab (57). Mutation of ecleuS was performed by PCR as reported (57). EcLeuRS and its K-Q mutants were purified by Ni-NTA affinity chromatography as reported (58). To obtain site-directed EcLeuRS-KAc, we constructed pET22b(+)-ecleuS encoding EcLeuRS with a C-terminal His6 tag, in which target Lys codons were separately mutated to TAG. We co-transformed E. coli BL21 (DE3) cells with pAcKRS and pET22b(+)-ecleuS with a C-terminal His6 tag. As described previously (43), EcLeuRS-KAcs were overexpressed in E. coli BL21 (DE3) following the addition of Ara and IPTG, in the presence of ampicillin, chloramphenicol, 10 mm NAM, and AcK (Fig. 3). EcLeuRS-KAc was purified by Ni-NTA affinity chromatography, followed by gel-filtration chromatography with a Superdex 75. The purity of the preparations was assessed by SDS-PAGE, and protein concentration was determined using the Bradford protein assay kit. EcLeuRS enzymes were stored in buffer containing 20 mm potassium phosphate (pH 6.8) and 1 mm DTT (58).
The gene encoding EcArgRS was amplified from plasmid pUC18-ecargS and inserted into pET28a(+) (59). WT EcArgRS and K-Q mutants were purified by Ni-NTA affinity chromatography as described above for EcLeuRS. Because EcArgRS with a C-terminal His6 tag displayed no activity (data not shown), EcArgRS-KAc enzymes were also expressed using the pET28a(+) vector. EcArgRS-KAc enzymes were purified by two-step chromatography as described above for EcLeuRS and stored in buffer containing 20 mm potassium phosphate (pH 7.5) and 1 mm DTT (59). Cloning of genes encoding YfiQ, CobB, and YcgC and purification of the recombinant enzymes was performed as reported previously (25, 26).
CD spectroscopy
The secondary structures of EcLeuRS, EcArgRS, and their variants were determined by CD spectroscopy as described previously (9).
Acquisition of tRNAs
E. coli tRNALeuCAG (EctRNALeu) and E. coli tRNAArgICG (EctRNAArg) were isolated from the corresponding overexpression strains as previously described (60, 61). The charging level of EctRNALeu and EctRNAArg was measured with 1 μm corresponding EcLeuRS and EcArgRS for more than 20 min. Both tRNAs harbored ∼1400 pmol/A260 units of accepting activity.
Mass spectrometry
E. coli BL21 (DE3) cells harboring the corresponding plasmids containing ecleuS or ecargS were inoculated from overnight culture in a ratio of 1:100 to 2× YT medium. When the A600 reached 0.6–0.8, ecleuS and ecargS genes were induced by IPTG in the presence of 10 mm NAM in 22 °C for 6 h. The cells pellets were lysed by sonication in the presence of PMSF. After centrifugation, the supernatants were applied to the Ni-NTA column. Then His6-taggged EcLeuRS and EcArgRS were purified by the affinity chromatography. After SDS-PAGE, the target bands were separated, excised, and sent to Shanghai Applied Protein Technology (Shanghai, China). Protein bands were in-gel digested, subjected to nanoLC to separate the resultant peptides, and identified by mass spectrometry (Thermo Finnigan, Silicon Valley, CA). Data processing and analysis of raw files were conducted using Proteomics Tools 3.1.6 and Mascot 2.2.
Amino acid activation, aminoacylation, misaminoacylation, and deacetylation assays
Assays of EcLeuRS were performed as previously described (62). The amino acid activation of EcLeuRS and its mutants was assayed by monitoring ATP-PPi exchange reactions at 37 °C in reaction mixture containing 100 mm HEPES (pH 7.8), 10 mm MgCl2, 10 mm KF, 4 mm ATP, 2 mm [32P] NaPPi, 5 mm Leu, 0.1 mg/ml BSA, and 10 nm enzyme. The aminoacylation activity of EcLeuRS was monitored at 37 °C in reaction mixtures containing 100 mm Tris-HCl (pH 7.8), 30 mm KCl, 12 mm MgCl2, 2 mm DTT, 4 mm ATP, 10 μm tRNALeu, 40 μm [3H] Leu, 0.1 mg/ml BSA, and 1 nm enzymes. In aminoacylation reaction, kinetic constants for EcLeuRS and its mutants were determined in the presence of tRNALeu at concentrations between 0.5 and 30 μm. Misacylation assays were performed in a similar system to that of aminoacylation, except 40 μm [3H] Ile (30 Ci/mmol) and 1 μm WT-EcLeuRS were used instead of 40 μm [3H] Leu and 1 nm enzyme. Deacylation assays with [3H] Ile-EctRNALeu were carried out in buffer containing 100 mm Tris-HCl (pH 7.5), 30 mm KCl, 12 mm MgCl2, 2 mm DTT, and 1 μm [3H] Ile-EctRNALeu. 20 nm enzymes were used to initiate reactions at 37 °C.
ATP-PPi exchange assays of EcArgRSs were performed at 37 °C in reaction mixtures containing 130 mm Tris-HCl (pH 7.2), 6 mm MgCl2, 2 mm ATP, 2 mm [32P] NaPPi, 2 mm Arg, 0.1 mg/ml BSA, and tRNAArg at saturating concentrations. 5 nm EcArgRSs was used to initiate ATP-PPi exchange reactions. Aminoacylation assays of EcArgRS and its variants were performed in reaction mixtures containing 50 mm Tris-HCl (pH 7.8), 80 mm KCl, 8 mm MgCl2, 0.5 mm DTT, 4 mm ATP, 10 μm tRNAArg, 100 μm [3H] Arg, 0.1 mg/ml BSA, and 1 nm enzymes at 37 °C (59). In aminoacylation reaction, kinetic constants for EcArgRS and its mutants were determined in the presence of tRNAArg between 0.5 and 80 μm (63).
Determination of the tRNALeu dissociation constant (Kd) by fluorescence quenching assays
0.1 μm proteins in 400 μl of equilibrium titration buffer containing 100 mm Tris-HCl (pH 8.2), 12 mm MgCl2, and 0.5 mm DTT were excited at 280 nm in a quartz cuvette, and the appropriate emission wavelength was monitored at room temperature as described previously (64). Maximum emission of EcLeuRS and EcArgRS was observed at 340 nm, and this was used to measure the fluorescence intensity of enzymes titrated with their cognate tRNAs. The final concentration of tRNAs ranged from 0.06 to 4.21 μm, and the total volume of titrations containing tRNA was less than 20 μl (1/20 of the original volume). We calculated the Kd values by plotting the fluorescence intensity change (a.u.) against final tRNA concentration (μm) according to the “one site-specific binding” option in GraphPad Prism software. BSA was used as a control.
In vitro acetylation/deacetylation assays
YfiQ-mediated acetylation assays were performed at 37 °C in reaction mixtures containing 50 mm Tris-HCl (pH 8.0), 1 mm DTT, 10% glycerol, 0.2 mm Ac-CoA, and 5 mm NaBu (26). Acetylation of EcLeuRS and EcArgRS by AcP was carried out with freshly prepared 10 mm AcP for 1 h in reaction buffer containing 40 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 40 mm KCl, 1 mm DTT, and enzyme at 37 °C.
In vitro deacetylation reactions were carried out as described previously with purified EcLeuRS or EcArgRS and purified CobB in reaction mixtures containing 40 mm HEPES (pH 7.0), 6 mm MgCl2, 1 mm NAD+, 1 mm DTT, and 10% glycerol at 37 °C (26). To investigate the effect of deacetylation by CobB on the aminoacylation activities of EcLeuRS-KAc, enzymes were incubated with or without CobB in deacetylation buffer for 1 h at 37 °C and subsequently diluted to 5 nm to initiate the aminoacylation reaction. For deacetylation of EcLeuRSAcs and EcArgRSAcs (corresponding enzymes preincubated with AcP), proteins were pretreated (desalted) using spin desalting columns.
Western blotting
For the detection of AcK, PVDF membranes were blocked in buffer containing 50 mm Tris-HCl (pH 7.5), 1% peptone, and 10% (v/v) Tween 20 and subsequently incubated with diluted primary antibody in 50 mm Tris-HCl and 0.1% peptone (53). All other Western blotting assays were conducted using conventional methods.
Author contributions
Q. Y. and E.-D. W. designed the study, analyzed the data, and wrote the paper. Q.-Q. J. assisted with the obtaining of clones and the preparation of samples for mass spectrometry. W. Y. participated in determination of the secondary structure of EcLeuRS and EcArgRS by CD analysis. F. Y. helped in the preparation of proteins. QY performed all the other experiments. All of the authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Professor Jiang-Yun Wang (Institute of Biophysics, Chinese Academy of Sciences) for providing pAcKRS plasmid. We also thank Prof. Guo-Ping Zhao, Dr. Wei Zhao, Dr. Xu-Feng Cen, and Dr. Xiao-Biao Han (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) and Gisela Geoghegan (School of Medicine, University of Utah) for valuable advice.
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB19000000 and National Natural Science Foundation of China Grants 31570792, 31130064, and 91440204. The authors declare that they have no conflicts of interest with the contents of this article.
- aaRS
- aminoacyl-tRNA synthetase
- LeuRS
- leucyl-tRNA synthetase
- ArgRS
- arginyl-tRNA synthetase
- Acs
- acetyl-CoA synthetase
- WT
- wild type
- AcK
- Nϵ-acetylated lysine
- AcP
- acetyl-phosphotate
- PDB
- Protein Data Bank
- PTM
- post-translational modification
- K-Q mutants
- proteins that with a substitution of Q for K
- KAc mutants
- proteins that with a substitution of AcK for K
- CP1
- connective peptide 1
- CTD
- C-terminal domain
- Ac-CoA
- acetyl coenzyme A
- TyrRS
- tyrosyl-tRNA synthetase
- NAM
- nicotinamide
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- Ara
- arabinose.
References
- 1. Nangle L. A., De Crecy Lagard V., Doring V., and Schimmel P. (2002) Genetic code ambiguity: cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 277, 45729–45733 [DOI] [PubMed] [Google Scholar]
- 2. Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., Davisson M. T., Sundberg J. P., Schimmel P., and Ackerman S. L. (2006) Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 [DOI] [PubMed] [Google Scholar]
- 3. Nangle L. A., Motta C. M., and Schimmel P. (2006) Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 13, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 4. Eriani G., Delarue M., Poch O., Gangloff J., and Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 5. Ibba M., and Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 6. Ling J., Reynolds N., and Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 7. Mehler A. H., and Mitra S. K. (1967) The activation of arginyl transfer ribonucleic acid synthetase by transfer ribonucleic acid. J. Biol. Chem. 242, 5495–5499 [PubMed] [Google Scholar]
- 8. Shimada A., Nureki O., Dohmae N., Takio K., and Yokoyama S. (2001) Gene cloning, expression, crystallization and preliminary X-ray analysis of Thermus thermophiles arginyl-tRNA synthetase. Acta Crystallogr. D Biol. Crystallogr. 57, 272–275 [DOI] [PubMed] [Google Scholar]
- 9. Ye Q., Wang M., Fang Z. P., Ruan Z. R., Ji Q. Q., Zhou X. L., and Wang E. D. (2015) Degenerate connective polypeptide 1 (CP1) domain from human mitochondrial leucyl-tRNA synthetase. J. Biol. Chem. 290, 24391–24402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palencia A., Crépin T., Vu M. T., Lincecum T. L. Jr., Martinis S. A., and Cusack S. (2012) Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu B., Zhao M. W., Eriani G., and Wang E. D. (2007) A present-day aminoacyl-tRNA synthetase with ancestral editing properties. RNA 13, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan M., Zhu B., Zhou X. L., He R., Chen X., Eriani G., and Wang E. D. (2010) tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 285, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan M., Zhu B., Liu R. J., Chen X., Zhou X. L., Wang E. D. (2013) Inter-domain communication modulates the tRNA-dependent pre-transfer editing of leucyl-tRNA synthetase. Biochem. J. 449, 123–131 [DOI] [PubMed] [Google Scholar]
- 14. Cavarelli J., Delagoutte B., Eriani G., Gangloff J., and Moras D. (1998) l-Arginine recognition by yeast arginyl-tRNA synthetase. EMBO J. 17, 5438–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delagoutte B., Moras D., and Cavarelli J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 19, 5599–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J., Yao Y. N., Liu M. F., and Wang E. D. (2003) Arginyl-tRNA synthetase with signature sequence KMSK from Bacillus stearothermophilus. Biochem. J. 376, 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekine S., Shimada A., Nureki O., Cavarelli J., Moras D., Vassylyev D. G., and Yokoyama S. (2001) Crucial role of the high-loop lysine for the catalytic activity of arginyl-tRNA synthetase. J. Biol. Chem. 276, 3723–3726 [DOI] [PubMed] [Google Scholar]
- 18. Kim G. W., and Yang X. J. (2011) Comprehensive lysine acetylomes emerging from bacteria to humans. Trends. Biochem. Sci. 36, 211–220 [DOI] [PubMed] [Google Scholar]
- 19. Soppa J. (2010) Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010, 820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye Q., and Wang E. D. (2016) Lysine acetylation of proteins in prokaryotes. Chin. Bull. Life Sci. 28, 427–435 [Google Scholar]
- 21. Starai V. J., and Escalante-Semerena J. C. (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 22. Hu L. I., Lima B. P., and Wolfe A. J. (2010) Bacterial protein acetylation: the dawning of a new age. Mol. Microbiol. 77, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weinert B. T., Iesmantavicius V., Wagner S. A., Schölz C., Gummesson B., Beli P., Nyström T., and Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272 [DOI] [PubMed] [Google Scholar]
- 24. AbouElfetouh A., Kuhn M. L., Hu L. I., Scholle M. D., Sorensen D. J., Sahu A. K., Becher D., Antelmann H., Mrksich M., Anderson W. F., Gibson B. W., Schilling B., and Wolfe A. J. (2015) The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiology Open 4, 66–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tu S., Guo S. J., Chen C. S., Liu C. X., Jiang H. W., Ge F., Deng J. Y., Zhou Y. M., Czajkowsky D. M., Li Y., Qi B. R., Ahn Y. H., Cole P. A., Zhu H., and Tao S. C. (2015) YcgC represents a new protein deacetylase family in prokaryotes. eLife pii, e05322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., et al. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner G. R., and Payne R. M. (2013) Widespread and enzyme-independent Nϵ-acetylation and Nϵ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuhn M. L., Zemaitaitis B., Hu L. I., Sahu A., Sorensen D., Minasov G., Lima B. P., Scholle M., Mrksich M., Anderson W. F., Gibson B. W., Schilling B., and Wolfe A. J. (2014) Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9, e94816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner G. R., and Hirschey M. D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mizuno Y., Nagano-Shoji M., Kubo S., Kawamura Y., Yoshida A., Kawasaki H., Nishiyama M., Yoshida M., and Kosono S. (2016) Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. Microbiology Open 5, 152–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., and Yoshida M. (2015) Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10, e0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Germain E., Castro-Roa D., Zenkin N., and Gerdes K. (2013) Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254 [DOI] [PubMed] [Google Scholar]
- 33. Lee J. Y., Kim D. G., Kim B. G., Yang W. S., Hong J., Kang T., Oh Y. S., Kim K. R., Han B. W., Hwang B. J., Kang B. S., Kang M. S., Kim M. H., Kwon N. H., and Kim S. (2014) Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 127, 4234–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., and Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 36. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. R., Olsen J. V., Jensen L. J., and Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal. 4, ra48. [DOI] [PubMed] [Google Scholar]
- 38. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sol E. M., Wagner S. A., Weinert B. T., Kumar A., Kim H. S., Deng C. X., and Choudhary C. (2012) Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One 7, e50545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang K., Zheng S., Yang J. S., Chen Y., and Cheng Z. (2013) Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 12, 844–851 [DOI] [PubMed] [Google Scholar]
- 41. Weinert B. T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schilling B., Christensen D., Davis R., Sahu A. K., Hu L. I., Walker-Peddakotla A., Sorensen D. J., Zemaitaitis B., Gibson B. W., and Wolfe A. J. (2015) Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98, 847–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neumann H., Peak-Chew S. Y., and Chin J. W. (2008) Genetically encoding Nϵ-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 [DOI] [PubMed] [Google Scholar]
- 44. Hebert A. S., Dittenhafer-Reed K. E., Yu W., Bailey D. J., Selen E. S., Boersma M. D., Carson J. J., Tonelli M., Balloon A. J., Higbee A. J., Westphall M. S., Pagliarini D. J., Prolla T. A., Assadi-Porter F., Roy S., et al. (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell 49, 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng Y. G., Wei H., Ling C., Martin F., Eriani G., and Wang E. D. (2004) Two distinct domains of the beta subunit of Aquifex aeolicus leucyl-tRNA synthetase are involved in tRNA binding as revealed by a three-hybrid selection. Nucleic Acids Res. 32, 3294–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Geslain R., Bey G., Cavarelli J., and Eriani G. (2003) Limited set of amino acid residues in a class Ia aminoacyl-tRNA synthetase is crucial for tRNA binding. Biochemistry 42, 15092–15101 [DOI] [PubMed] [Google Scholar]
- 47. Schulman L. H., and Pelka H. (1989) The anticodon contains a major element of the identity of arginine transfer RNAs. Science 246, 1595–1597 [DOI] [PubMed] [Google Scholar]
- 48. Sissler M., Giegé R., and Florentz C. (1996) Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J. 15, 5069–5076 [PMC free article] [PubMed] [Google Scholar]
- 49. Fersht A. R., Knill-Jones J. W., Bedouelle H., and Winter G. (1988) Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: a mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry 27, 1581–1587 [DOI] [PubMed] [Google Scholar]
- 50. Mechulam Y., Dardel F., Le Corre D., Blanquet S., and Fayat G. (1991) Lysine 335, part of the KMSKS signature sequence, plays a crucial role in the amino acid activation catalysed by the methionyl-tRNA synthetase from Escherichia coli. J. Mol. Biol. 217, 465–475 [DOI] [PubMed] [Google Scholar]
- 51. Perona J. J., Rould M. A., and Steitz T. A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry 32, 8758–8771 [DOI] [PubMed] [Google Scholar]
- 52. Hsu J. L., Rho S. B., Vannella K. M., and Martinis S. A. (2006) Functional divergence of a unique C-terminal domain of leucyl-tRNA synthetase to accommodate its splicing and aminoacylation roles. J. Biol. Chem. 281, 23075–23082 [DOI] [PubMed] [Google Scholar]
- 53. Zhang T., Wang S., Lin Y., Xu W., Ye D., Xiong Y., Zhao S., and Guan K. L. (2012) Acetylation negatively regulates glycogen phosphorylase by recruiting protein phosphatase 1. Cell Metab. 15, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., and Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell Proteomics 11, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao X., Li C., Xiao S., Tang Y., Huang J., Zhao S., Li X., Li J., Zhang R., and Yu W. (2017) Acetylation promotes TyrRS nuclear translocation to prevent oxidative damage. Proc. Natl. Acad. Sci. U.S.A. 114, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Colak G., Xie Z., Zhu A. Y., Dai L., Lu Z., Zhang Y., Wan X., Chen Y., Cha Y. H., Lin H., Zhao Y., and Tan M. (2013) Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell Proteomics 12, 3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yan W., Ye Q., Tan M., Chen X., Eriani G., and Wang E. D. (2015) Modulation of aminoacylation and editing properties of leucyl-tRNA synthetase by a conserved structural module. J. Biol. Chem. 290, 12256–12267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen J., Li Y., Wang E., and Wang Y. (1999) High-level expression and single-step purification of leucyl-tRNA synthetase from Escherichia coli. Protein Expr. Purif. 15, 115–120 [DOI] [PubMed] [Google Scholar]
- 59. Liu M., Huang Y., Wu J., Wang E., and Wang Y. (1999) Effect of cysteine residues on the activity of arginyl-tRNA synthetase from Escherichia coli. Biochemistry 38, 11006–11011 [DOI] [PubMed] [Google Scholar]
- 60. Li Y., Wang E. D., and Wang Y. L. (1998) Overproduction and purification of Escherichia coli tRNALeu. Sci. China C life Sci. 41, 225–231 [DOI] [PubMed] [Google Scholar]
- 61. Wu J. F., Wang E. D., Wang Y. L., Gilbert E., and Jean G. (1999) Gene Cloning, overproduction and purification of Escherichia coli tRNA(Arg)(2). Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 31, 226–232 [PubMed] [Google Scholar]
- 62. Chen J. F., Guo N. N., Li T., Wang E. D., and Wang Y. L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39, 6726–6731 [DOI] [PubMed] [Google Scholar]
- 63. Lin S. X., Shi J. P., Cheng X. D., and Wang Y. L. (1988) Arginyl-tRNA synthetase from Escherichia coli, purification by affinity chromatography, properties, and steady-state kinetics. Biochemistry 27, 6343–6348 [DOI] [PubMed] [Google Scholar]
- 64. Hu Q. H., Huang Q., and Wang E. D. (2013) Crucial role of the C-terminal domain of Mycobacterium tuberculosis leucyl-tRNA synthetase in aminoacylation and editing. Nucleic Acids Res. 41, 1859–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]