Abstract
Clostridium difficile has become one of the most common bacterial pathogens in hospital-acquired infections in the United States. Although C. difficile is strictly anaerobic, it survives in aerobic environments and transmits between hosts via spores. C. difficile spore germination is triggered in response to certain bile acids and glycine. Although glycine is the most effective co-germinant, other amino acids can substitute with varying efficiencies. Of these, l-alanine is an effective co-germinant and is also a germinant for most bacterial spores. Many endospore-forming bacteria embed alanine racemases into their spore coats, and these enzymes are thought to convert the l-alanine germinant into d-alanine, a spore germination inhibitor. Although the C. difficile Alr2 racemase is the sixth most highly expressed gene during C. difficile spore formation, a previous study reported that Alr2 has little to no role in germination of C. difficile spores in rich medium. Here, we hypothesized that Alr2 could affect C. difficile l-alanine-induced spore germination in a defined medium. We found that alr2 mutant spores more readily germinate in response to l-alanine as a co-germinant. Surprisingly, d-alanine also functioned as a co-germinant. Moreover, we found that Alr2 could interconvert l- and d-serine and that Alr2 bound to l- and d-serine with ∼2-fold weaker affinity to that of l- and d-alanine. Finally, we demonstrate that l- and d-serine are also co-germinants for C. difficile spores. These results suggest that C. difficile spores can respond to a diverse set of amino acid co-germinants and reveal that Alr2 can accommodate serine as a substrate.
Keywords: bacteria, bacterial genetics, bacterial pathogenesis, bacterial signal transduction, microbiology, physiology, clostridium difficile, germination, racemase, spore
Introduction
Clostridium difficile infection (CDI)2 has become one of the most common hospital acquired infections in the United States (1, 2). Antibiotic use is the major risk factor associated with CDI (2). Treatment of potential hosts with broad spectrum antibiotics leads to major alterations in the normally protective colonic microbiota and permits colonization of that host by C. difficile spores (2). Although antibiotics (e.g. vancomycin) are available to treat CDI, the continued perturbation to the colonic microbiota by these antibiotics leads to multiple rounds of recurring infections (3). It is for this reason that the Centers for Disease Control and Prevention have listed C. difficile as “an urgent threat” regarding the antibiotic-associated threats to the United States (1).
Although C. difficile is a strict anaerobe, it survives in aerobic environments and transmits between hosts in the spore form (4). Spores are metabolically dormant forms of bacteria and formed from vegetatively growing cells, often upon nutrient limitation. During spore formation, a vegetative cell asymmetrically divides into a mother cell compartment and a forespore compartment (5). Through coordinated gene expression, the two compartments mature the forespore into a multilayered spore that can resist harsh conditions (e.g. radiation, heat, antibiotics, etc.) (6). Spores are composed of a DNA-containing core where much of the water has been replaced with dipicolinic acid (often as a calcium chelate (CaDPA)), an inner membrane, a thin peptidoglycan layer, a thick specialized peptidoglycan-containing cortex layer, an outer membrane, a coat layer, and, in some organisms, an exosporium layer (5–7).
In a host, these spores must exit dormancy (germinate) to generate the actively growing, toxin-producing cells that elicit the primary symptoms of disease. C. difficile spore germination is stimulated by the combinatorial action of certain bile acids and certain amino acids (8–11). C. difficile spore germination is activated by cholic acid-class bile acids, whereas chenodeoxycholic acids competitively inhibit cholic acid-mediated germination (9, 10). Although bile acids are important for germination, they are not sufficient to activate germination on their own. Amino acids are also required for germination, and the most effective co-germinant is glycine (10). However, other amino acids can function as co-germinants with varying efficiencies (e.g. alanine) (11).
In most organisms, germinants interact with their cognate receptors at the inner membrane of the spore (7). These ger-type germinant receptors bind to germinants and lead to the release of CaDPA from the core in exchange for water (7). Subsequently, the spore cortex is degraded through the action of spore cortex lytic enzymes. C. difficile does not encode orthologues of the classical ger-type germinant receptor and instead uses a novel pathway for initiating spore germination where the CspC pseudoprotease is the bile acid germinant receptor (5, 12, 13). We hypothesized that CspC transmits a bile acid signal to the CspB protease, which then cleaves the spore cortex lytic enzyme, pro-SleC, to an active form (14–16). Activated SleC degrades the cortex leading to CaDPA release, because of osmotic changes in the core resulting from to cortex degradation (15, 17).
Recently, GerG and GerS were identified as important players in C. difficile spore germination. Encoded downstream of gerS is alr2, coding for an alanine racemase (18, 19). Alanine racemases enzymatically convert l-alanine to the d-alanine stereoisomer and are known to be involved in spore development and germination (20, 21). During spore development, the alanine racemase converts the l-alanine germinant to the d-alanine inhibitor to prevent germination of the developing spore within the mother cell (21–24). During Bacillus subtilis spore germination, the alanine racemase converts l-alanine to d-alanine to suppress spore germination, a process termed “autoinhibition” (24, 25); d-alanine does not inhibit germination by other B. subtilis germinants and is specific to the GerA germinant receptor (22). Autoinhibition of spore germination occurs, presumably, until the abundance of l-alanine accumulates to sufficient levels to overwhelm the racemase and stimulate spore germination (21). In C. difficile only the l-form of alanine can trigger spore germination when added with taurocholic acid (TA) (11). Unlike what is seen for B. subtilis spore germination, d-alanine does not inhibit germination by C. difficile spores (11).
Interestingly, in the same study that identified GerS as a novel regulator of spore germination, inactivation of C. difficile alr2 had minimal impact on C. difficile spore germination when analyzed in rich medium (18). However, we hypothesized that alr2 may play an important role during spore germination in defined medium in the presence of alanine and TA as germinants. Indeed, we found that C. difficile alr2 mutants play an important role during spore germination by converting d-alanine to the l-alanine co-germinant. Interestingly, we found that Alr2 also interconverts l- and d-serine and that these amino acids are germinants for C. difficile spores. Finally, we determine that the affinity of Alr2 for amino acids may be destabilized by the bound co-factor because an Alr2 mutant that does not bind to its co-factor has an affinity for l/d-alanine that is ∼80× greater than that of the wild-type protein. Our results shed new light on mechanisms of C. difficile spore germination and suggest that C. difficile spores are equipped to recognize more amino acids as co-germinants than previously thought.
Results
C. difficile alr2 is not required for spore germination in rich medium
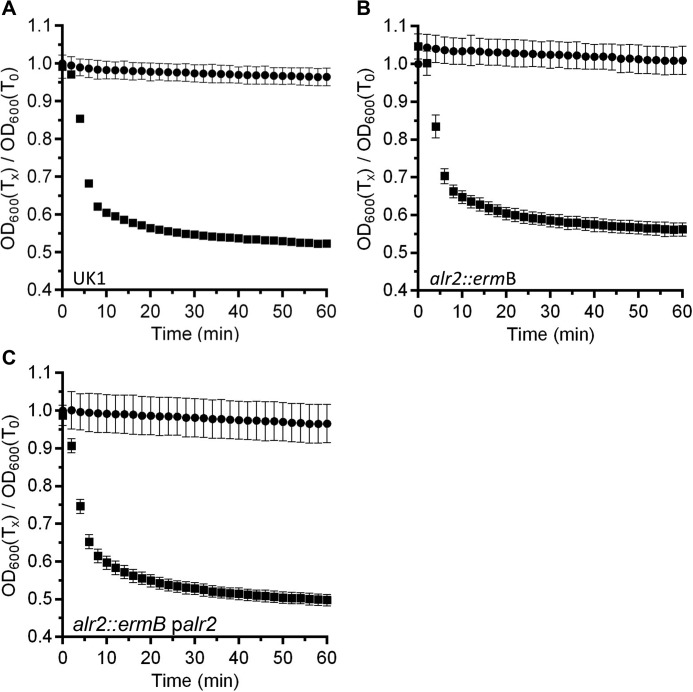
C. difficile alr2 (encoding an alanine racemase) is the sixth most expressed sporulation protein (26). Prior work has shown that although highly expressed, Alr2 seems to play little to no role during C. difficile spore germination in rich medium supplemented with TA germinant (18). However, given the diverse signals present in rich medium, we hypothesized that C. difficile alr2 may play a role similar to what is observed in other spore-forming bacteria by affecting germination in response to l-alanine. Toward this goal, we generated a TargeTron mutation in C. difficile UK1 alr2. When suspended in rich BHIS medium supplemented with TA (BHIS-TA), wild-type C. difficile UK1 spores rapidly germinate as shown as a decrease in the optical density of the suspension (Fig. 1A). Similarly, C. difficile RS07 (alr2::ermB) spores also rapidly germinated in BHIS-TA (Fig. 1B), and when the alr2 disruption was complemented by expressing alr2 from a plasmid, no difference was observed compared with the other tested strains (Fig. 1C). These results confirm prior observations that Alr2 plays little role during germination of C. difficile spores in rich medium.
Figure 1.
Alr2 does not contribute to germination by C. difficile spores in rich medium. Purified spores from wild-type C. difficile UK1 (A), C. difficile RS07 (alr2::ermB) (B), or C. difficile RS07 pRS89 (palr2) (C) were suspended in rich BHIS medium (●) or BHIS supplemented with 10 mm TA (■) at 25 °C. The A600 of the suspension was monitored over time. The data points represent the averages from three independent experiments, and the error bars represent the standard deviation of the mean.
C. difficile alr2 is dispensable for germination in response to l-alanine but essential for germination in response to d-alanine
To test the effects of an alr2 disruption on C. difficile spore germination, we analyzed spore germination in defined medium. C. difficile UK1 spores were suspended in buffer supplemented with TA and glycine, TA and l-alanine, or TA and d-alanine, and germination was monitored at 25 °C (supplemental Fig. S1). As expected, wild-type C. difficile UK1 germinated in response to glycine and in response to l-alanine (supplemental Fig. S1A). C. difficile UK1 spores did not initiate germination in response to TA and d-alanine. When C. difficile RS07 was tested for germination in response to TA and glycine or TA and l-alanine or TA and d-alanine, we observed a decrease in the ability of the strain to germinate in response to l-alanine but not in response to glycine (d-alanine, again, had no effect on C. difficile spore germination) (supplemental Fig. S1B). This defect in germination in response to l-alanine could be complemented to near wild-type levels (supplemental Fig. S1C).
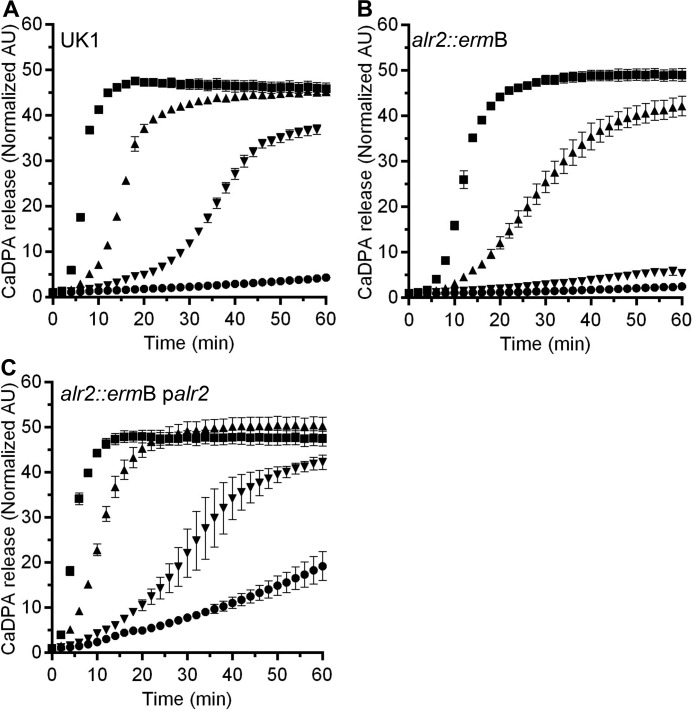
To date, most germination assays in our lab and others were performed at room temperature. Although germination often proceeds quickly at this temperature, we reasoned that some C. difficile spore enzymes may best function at 37 °C (a physiologically relevant temperature). Therefore, we again tested germination of wild-type C. difficile spores in response to TA and glycine or TA and l-alanine or TA and d-alanine. As expected, wild-type C. difficile UK1 spores germinated in response to TA and glycine or TA and alanine (Fig. 2A). Surprisingly, C. difficile UK1 spores also germinated using d-alanine as a co-germinant with TA (Fig. 2A). C. difficile RS07 spores germinated similar to wild-type UK1 spores in TA and glycine or TA and l-alanine (Fig. 2B). Interestingly, C. difficile RS07 spores were unable to respond to d-alanine as a co-germinant with TA at the concentration tested (Fig. 2B). This defect in germination could be restored by expressing alr2 from a plasmid (Fig. 2C). These results suggest that the Alr2 alanine racemase has a role in recognizing d-alanine as a co-germinant or that it converts d-alanine to l-alanine, which then functions as a co-germinant with TA.
Figure 2.
Alr2 affects l/d-alanine-mediated germination by C. difficile spores. Purified spores from wild-type C. difficile UK1 (A), C. difficile RS07 (alr2::ermB) (B), or C. difficile RS07 pRS89 (palr2) (C) were suspended germination buffer supplemented with TA alone (●) or supplemented with glycine (■), l-alanine (▴), d-alanine (▾). CaDPA release from the germinating spores was monitored at 37 °C as an increase in Tb3+ fluorescence over time. The data points represent the average from three independent experiments, and the error bars represent the standard deviation of the mean.
C. difficile RS07 spores more readily germinate in response to l-alanine as a co-germinant with TA
Alanine racemases are known to be involved in spore germination, but all studied to date are thought to covert l-alanine to d-alanine, which then acts as an inhibitor of l-alanine-mediated germination, and it is unclear whether d-alanine conversion to l-alanine is important in these organisms. During germination, C. difficile alr2 may convert d-alanine to l-alanine, which would stimulate spore germination in combination with TA. However, because l-alanine is a germinant (and d-alanine does not inhibit C. difficile spore germination), if Alr2 converts l-alanine to d-alanine during germination, C. difficile RS07 spores may be more sensitive to the l-alanine co-germinant. To quantify this, we determined the EC50 values for l-alanine in C. difficile UK1 spores, C. difficile RS07 spores, and C. difficile RS07 palr2 spores. Although spore germination is a multienzyme process, these kinetic measurements allow for the quantitative interaction of the spores with activators and inhibitors of germination (27–34). When C. difficile UK1 spores were suspended in buffer supplemented with TA and increasing concentrations of l-alanine, we observed an increase in the germination rate. Using the rates and concentrations tested, we found that wild-type C. difficile spores had an EC50 of ∼5.5 mm for l-alanine (Table 1). C. difficile RS07 spores were more sensitive to l-alanine as a co-germinant with an EC50 value of 2.7 mm, suggesting that Alr2 is able to convert l-alanine to d-alanine (Table 1). When the complemented strain was analyzed, the EC50 value was higher than wild-type, 10.7 mm, suggesting that overexpression of Alr2 drives l-alanine to d-alanine more than what is observed in wild type (Table 1).
Table 1.
EC50 values (mm) for amino acids and the C. difficile spore
The averages and standard deviations for three biological replicates are shown. CND, could not determine; EC50, concentration that achieves the half-maximum germination rate.
|
C. difficile strain |
|||
|---|---|---|---|
| UK1 (wild type) | RS07 (alr2::ermB) | RS07 pRS89 (alr2::ermB palr2) | |
| l-Alanine | 5.5 ± 1.1 | 2.7 ± 1.8 | 10.7 ± 2.5 |
| d-Alanine | 11.5 ± 2.3 | 24.2 ± 6.6 | 6.6 ± 1.0 |
| l-Serine | 8.2 ± 0.8 | 2.5 ± 1.7 | 4.9 ± 0.5 |
| d-Serine | 7.8 ± 1.2 | 19.1 ± 5.0 | CND |
Next, we determined the EC50 values for d-alanine during spore germination. C. difficile UK1 demonstrates an EC50 of 11.5 mm for d-alanine. If Alr2 is converting d-alanine to l-alanine during germination, the EC50 value should increase in C. difficile RS07. Indeed, C. difficile RS07 spores were less sensitive to d-alanine as a germinant and yielded an EC50 value of 24.2 mm, weaker than wild-type. When alr2 was expressed from a plasmid, the EC50 decreased to 6.6 mm because of its conversion to l-alanine. The EC50 of C. difficile RS07 palr2 with d-alanine was similar to the EC50 of wild-type spores and l-alanine. Importantly, because Alr2 affected germination in response to l-alanine and d-alanine, the EC50 values for C. difficile UK1 are an approximation for these amino acids (some of the l-alanine is converted to d-alanine and vice versa during germination). Taken together, these results suggest that d-alanine can be converted to l-alanine, which functions as a co-germinant. Moreover, these results demonstrate that d-alanine itself is a co-germinant for C. difficile spores.
Alr2 interconverts l-serine and d-serine
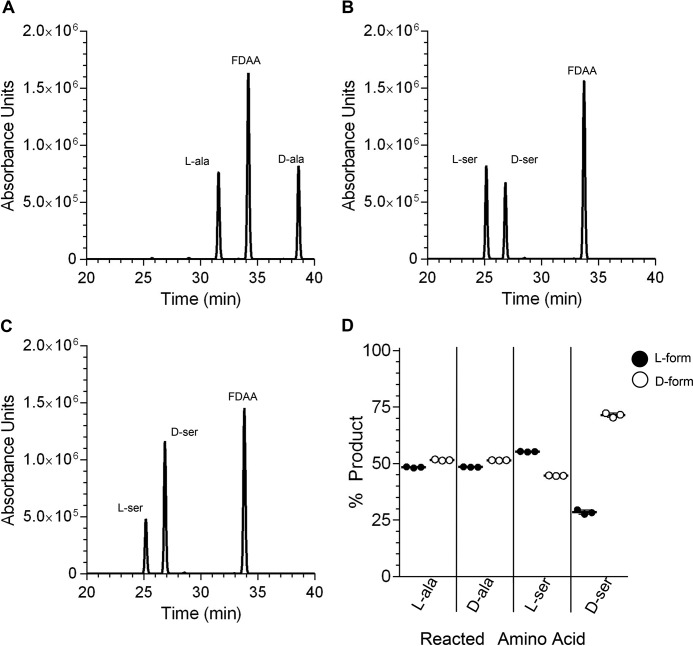
A prior study determined a crystal structure of a C. difficile alanine racemase (35). Although not known at the time, this racemase is Alr2. When recombinantly expressed and purified, Alr2 (supplemental Fig. S2) was suspended in reaction buffer and reacted with l- or d-alanine before labeling and separating by HPLC. As shown in Fig. 3A, when incubated with l-alanine, Alr2 converted l-alanine to an approximately equal amount of d-alanine (Fig. 3D); we use >98% pure d- or l-amino acid for these studies, and the opposite enantiomer is extremely low in these studies (supplemental Fig. S3). During the course of these experiments, we included l-serine as a would-be negative control for conversion. To our surprise, Alr2 converted l-serine to an approximately equal amount of d-serine (Fig. 3B). When tested with d-serine, Alr2 did not convert as much, and nearly 75% of the d-serine (Fig. 3C, quantified in Fig. 3D) remained in the d-form. Because Alr2 converted l- and d-serine, we tested every other soluble amino acid (data not shown). No other amino acid was converted by Alr2, indicating that Alr2 has specificity to the l- and d-forms of alanine and serine.
Figure 3.
Alr2 interconverts l/d-alanine and serine. A–C, recombinantly expressed and purified Alr2 was incubated with l-alanine (A), l-serine (B), or d-serine (C) for 1 h. The reaction was labeled with FDAA and separated by HPLC. The HPLC retention times of each amino acid were determined using labeled amino acid standards. D, peak areas from each reaction were quantified and presented as percentages of product generated from a reaction involving either l-alanine or d-alanine, etc., and each data point is shown. Each bar represents the average peak area from three technical replicates. The error bars represent the standard deviation from the mean and in most cases are smaller than the bars.
l- and d-serine are co-germinants for C. difficile spores
Because Alr2 interconverted l- and d-serine, we reasoned that C. difficile spores may respond to l-serine as a germinant. Thus, we determined the EC50 values for l- and d-serine during C. difficile spore germination, as described above (Table 2). Wild-type C. difficile UK1 displayed an EC50 for l-serine of 8.2 mm. This EC50 value decreased to 2.5 mm in the absence of alr2. Complementation of C. difficile RS07 led to a decrease in EC50 from the mutant levels to 4.9 mm but did not restore the EC50 to wild-type levels. Interestingly, C. difficile spores showed an EC50 for d-serine similar to that of l-serine. Also, similar to C. difficile RS07 and d-alanine, d-serine was a poor germinant in the absence of alr2 (EC50 = 19.1 mm). Unfortunately, we were unable to complement the phenotype in response to d-serine, for unknown reasons. These results suggest that both l- and d-serine are co-germinants for C. difficile spores.
Table 2.
ITC analysis of Alr2 and Alr2K39A binding to l/d-alanine and serine
The averages and standard errors of the mean for at least three technical replicates are shown. The differences in binding between l-alanine and d-alanine or l-serine and d-serine are not statistically significant. The difference in the binding of Alr2 to l-alanine and l-serine or d-alanine and d-serine are significant (p < 0.05 and p < 0.01, respectively). CND, could not determine.
| Alr2WT |
Alr2K39A |
|||
|---|---|---|---|---|
| KD | ΔH | KD | ΔH | |
| mm | μm | |||
| l-Alanine | 9.2 ± 0.7 | −7 ± 0.7 | 64 ± 6 | −1.1 ± 0.1 |
| d-Alanine | 9.2 ± 0.4 | −7.8 ± 0.7 | 46 ± 9 | −1.7 ± 0.1 |
| l-Serine | 14 ± 1 | −16 ± 1 | CND | CND |
| d-Serine | 17 ± 1 | −21 ± 0.2 | CND | CND |
Determining the affinity of Alr2 and Alr2K39A to L/D alanine and serine
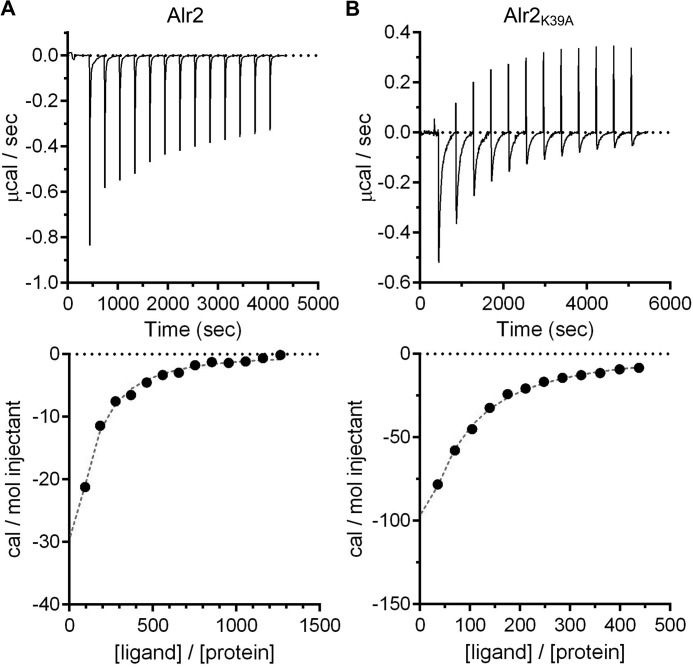
Previously, kinetic analysis of l- to d-alanine conversion (and vice versa) was used to determine the affinities of Alr2 to the alanine isoforms (35). Because we found that Alr2 can convert l- and d-serine, we tested the affinity of Alr2 to alanine and serine by isothermal titration calorimetry (ITC) (Fig. 4). As shown in Fig. 4A, when l-alanine was injected into the ITC cell, we observed exothermic binding between Alr2 and l-alanine, which could be saturated. We found that Alr2 had similar affinities for l-alanine (9.2 mm) and d-alanine (9.2 mm). The affinities for l-serine (14 mm) and d-serine (17 mm) were weaker (Table 2). Because the ITC method injects ligand into the ITC cell, Alr2 converted the injected ligand to its isomer (supplemental Fig. S4).
Figure 4.
Binding of Alr2WT and Alr2K39A to l-alanine. Recombinantly expressed and purified Alr2WT (A) or Alr2K39A (B) was tested for their ability to bind to l-alanine by ITC. The thermograms are shown in the top panels. A binding isotherm was generated from the total heat following each ligand injection and is plotted in the bottom panels. The data are representative of at least three technical experiments. The summarized data can be found in Table 2.
Because of the conversion of the l- to the d-form (and vice versa) in the ITC cell, the affinity of Alr2 for each of the tested amino acids could be an average of the affinities for tested and converted amino acids. For this reason, we engineered a mutation that disrupts the binding of the co-factor that is essential for catalysis (pyridoxal 5′-phosphate). The Alr2K39A mutant was confirmed to be enzymatically dead by HPLC analysis of Alr2K39A reactions with l- and d-alanine (data not shown). When the binding of Alr2K39A to l- or d-alanine was tested by ITC, we observed significantly less signal (Fig. 4B) compared with that of wild-type Alr2 (Fig. 4A). However, we observed a signal that could be saturated (Fig. 4B). From this data, we determined that Alr2K39A had an affinity of 65 μm for l-alanine and 50 μm for d-alanine (140 times that of wild-type for l-alanine and 184 times that of wild-type for d-alanine). Unfortunately, we could not determine the affinity of AlrK39A and l-serine or d-serine, and the signal-to-noise ratio was much too great to yield usable data.
Discussion
Spore germination is most often triggered by l-alanine (5). In the model spore-forming organism, B. subtilis, l-alanine interacts with the GerAA-AB-AC germinant receptor (7). This interaction leads to the release of CaDPA from the spore core and subsequent cortex degradation. The GerA germinant receptor is conserved across most spore-forming bacteria (36). In Bacillus anthracis, and other organisms, l-alanine is a germinant whose action is dependent on the presence of the GerA germinant receptor. Importantly, C. difficile does not encode orthologues of the gerA germinant receptor but germinates through a novel spore germination pathway (5, 12, 13, 15, 17).
The bile acid germinant receptor, CspC, activates germination through an unknown mechanism (12). Subsequently, the CspB protease activates pro-SleC by cleaving the N terminus from the protein, generating active cortex hydrolase (14). SleC then degrades the cortex resulting in osmotic changes that are perceived at the inner spore membrane (15, 17). This change in osmotic pressure at the inner spore membrane is relieved by the SpoVAC mechanosensing protein and results in CaDPA release from the core (17). Although bile acids are necessary to trigger germination, they are not sufficient. An amino acid signal is required to function as a co-germinant (10).
The most effective amino acid co-germinant is glycine (10). However, other amino acids can function as co-germinants (5). Previously, Howerton et al. (11) found that l-alanine could function as a co-germinant with TA for C. difficile spores. In their study, they found that d-alanine did not inhibit l-alanine-mediated spore germination, unlike what is observed in other spore-forming bacteria. Importantly, the authors found that d-alanine did not function as an activator of spore germination at 30 °C (11). Herein, we found that d-alanine can stimulate spore germination in combination with TA at 37 °C. Moreover, in the same study, l-serine was nearly inactive as a co-germinant (l-serine generated ∼5% germination when compared with glycine); d-serine was not tested (11). However, we found that both l-serine and d-serine can function as co-germinants for C. difficile spores at 37 °C. Our data suggest that temperature is an important consideration when determining activators and inhibitors of C. difficile spore germination.
Surprisingly, C. difficile Alr2 converted l-serine to d-serine (and vice versa). The affinities of Alr2 to l-serine and d-serine were similar, but the difference was not statistically significant. When tested for the ability to convert l-serine to d-serine, we observed that Alr2 generated nearly 50% d-serine, suggesting that Alr2 efficiently converts l-serine to d-serine. When Alr2 was given with d-serine as a substrate, Alr2 only generated ∼25% l-serine from 100% d-serine. Because the binding affinities of Alr2 and l- or d-serine are similar, this could indicate that the rate of conversion of d-serine to l-serine is much slower than the reverse reaction. Because of the conversion of l/d-alanine and serine during ITC, we tested the binding of Alr2K39A to these amino acids. Although the KD for Alr2K39A and l- and d-serine could not be determined because of a low signal-to-noise ratio, we found the KD for l- and d-alanine to be much lower than the KD for the wild-type protein (Table 2). This suggests that the pyridoxyl 5′-phosphate (PLP) co-factor may destabilize the interaction of Alr2 with its substrate, potentially as part of the catalytic mechanism.
Alanine racemases play important roles during the growth of Gram-negative and Gram-positive bacteria (20). These racemases are used to convert l-alanine to the d-alanine that is used to synthesize stem peptides during cell wall synthesis (20). As such, these proteins can be targeted by antibiotic treatment to prevent cell wall synthesis. d-Cycloserine is a commonly used antibiotic that targets alanine racemase, indicating that the racemization of alanine is important for cellular physiology (35). Alr2 can be inactivated by d-cycloserine, but C. difficile is naturally resistant to the antibiotic action of d-cycloserine, and d-cycloserine is included in medium that is selective for C. difficile growth (35, 37). This suggests that the Alr2 that is incorporated into the C. difficile spores is unlikely to be used during cell wall synthesis. C. difficile encodes other amino acid racemases. For example, CDR20291_2507 is annotated as a putative alanine racemase. However, there are several other putative amino acid racemases, including a putative VanTG orthologue (serine/alanine racemase). It is likely that one of these other racemases is insensitive, or has reduced sensitivity, to d-cycloserine. We tested the effects of pre-exposure of d-cycloserine to C. difficile spores as a means to inactivate Alr2 to prevent the conversion of amino acids in the C. difficile spore. Unfortunately, d-cycloserine functioned as a weak co-germinant (data not shown). Because of the background spore germination generated by d-cycloserine, we could not inactivate Alr2 chemically.
Autoinhibition of spore germination is important for the development of a spore and ensuring that the spore germinates under appropriate conditions (21, 24). Alanine racemase is important for autoinhibition by preventing premature germination of the developing spore within the mother cell or under less-than-suitable environmental conditions. We did not observe a difference between spores produced by the wild-type UK1 strain and spores produced by the alr2 mutant strain. Because C. difficile spore germination is activated in response to bile acids and an amino acid co-germinant, it is not surprising that the alr2 mutation did not affect spore formation. The alr2 mutation resulted in an increased sensitivity to l-alanine and a decreased sensitivity to d-alanine (Table 1). Because d-alanine does not inhibit C. difficile spore germination, autoinhibition is minimal during C. difficile spore germination and only applies to the conversion of a very good germinant to a weak germinant.
A prior study analyzed the proteome of C. difficile strain 630 spores (38). In this study, Alr2 was found to be extracted by incubating spores in 2-mercaptoethanol (a condition that solubilizes coat proteins) (38). Although the location of Alr2 has not been determined, this would suggest that it resides outside of the spore core where it would be available to convert l/d-alanine and serine. In this same study, no serine racemase was found (38). However, the authors found that C. difficile 630 CD3237 (a putative proline racemase) was present in the spore extracts. Although proline has not been described as a germinant for C. difficile spores, the inclusion of a proline racemase into the spore could signal the importance of proline racemization for C. difficile physiology, potentially for use in Stickland metabolism (39).
This study builds upon the hypothesis that the unidentified amino acid germinant receptor has broad specificity. Prior work has shown that the amino acid germinant receptor may require both a free carboxylate and a free amino group (11). In this study, some amino acids did not function as co-germinants (e.g. l-histidine or l-serine). Our findings suggest either that there are multiple receptors that each recognize different amino acids as co-germinants or that the amino acid germinant receptor can accommodate a diverse range of amino acids (both l- and d-forms). Because studies on C. difficile spore germination have been conducted at lower than physiological temperatures, it would be worthwhile to revisit the requirements of the amino acids to function as co-germinants at 37 °C.
Materials and methods
Strains and growth conditions
C. difficile UK1 and derivatives were routinely grown at 37 °C in an anaerobic environment (10% H2, 5% CO2, 85% nitrogen) in BHIS medium. Escherichia coli was routinely grown in LB medium at 37 °C. B. subtilis was grown in LB medium. Antibiotics were added when necessary (20 μg/ml chloramphenicol, 10 μg/ml thiamphenicol, 20 μg/ml lincomycin, 50 μg/ml kanamycin, and 100 μg/ml ampicillin).
Molecular biology
A mutation in C. difficile alr2 mutant was generated by retargeting the pJS107 TargeTron plasmid, as described previously (12). Primers to generate C. difficile RS07 and its complementing plasmid (pRS89) are listed in Table 3. Briefly, potential insertion sites for the group II intron were determined using the Targetronics algorithm, and a potential insertion site was identified in the sense orientation at nucleotide 138 of the alr2 coding sequence based upon the sequenced C. difficile R20291 strain (C. difficile UK1 is not a sequenced strain but is closely related to the R20291 strain). A gBlock (Integrated DNA technologies) was synthesized and cloned into the TOPO-ZeroBlunt cloning vector and transformed into E. coli DH5α, generating pRS85. The retargeting fragment was subcloned into pJS107 at the HindIII and BsrGI restriction sites, transformed into E. coli DH5α, yielding pRS86. The alr2 complementing plasmid (pRS89) was generated by amplifying the P1 promoter and the alr2 coding sequences using Phusion DNA polymerase (New England Biolabs). The resulting fragments were cloned into the B. subtilis-C. difficile shuttle vector, pJS116, at the XbaI and XhoI restriction sites using Gibson assembly (40). The plasmid used to express recombinant Alr2 protein was created by amplifying the alr2 gene from C. difficile UK1 using primers 5′pET_alr and 3′pET_alr (Table 3) and cloned into expression vector pET222b at the XhoI and NdeI restriction sites. The resulting plasmid was named pJS164. Similarly, the plasmid for the expression of AlrK39A was created by using overlapping PCR using primers 5′alr L39A and 3′alr L39A (Table 3) to create a change AA into GC such that the resulting amino acid would change from lysine to alanine. The amplified fragment with the mutation was stitched together using 5′ alr and 3′ alr primers and cloned into pET22b expression vector at XhoI and NdeI site, and the resulting plasmid was named pRS100. The DNA sequences for all plasmids were verified by sequencing (Eurofins).
Table 3.
Oligonucleotides used in this study
| Primer name | Primer sequence |
|---|---|
| R20291_alr 138s (gBlock) | TTCCCCTCTAGAAAAAAGCTTATAATTATCCTTATATGGCCATGGTGTGCGCCCAGATAGGGTGTTAAGTCAAGTAGTTTAAGGTACTACTCTGTAAGATAACACAGAAAACAGCCAACCTAACCGAAAAGCGAAAGCTGATACGGGAACAGAGCACGGTTGGAAAGCGATGAGTTACCTAAAGACAATCGGGTACGACTGAGTCGCAATGTTAATCAGATATAAGGTATAAGTTGTGTTTACTGAACGCAAGTTTCTAATTTCGATTCCATATCGATAGAGGAAAGTGTCTGAAACCTCTAGTACAAAGAAAGGTAAGTTAGCACCATGGACTTATCTGTTATCACCACATTTGTACAATCTG |
| 5′ alr | CATGCAAAAAATAACAGTG |
| 3′ alr | TTATTTTAGCAAATAACTGTTT |
| 5′ pJS116_XbaI_P1-prom | TACGAATTCGAGCTCGGTACCCGGGGATCCTCTAGACCAGTTGTAGATTCAGAGAATA |
| 3′ alr_P1-Prom | TGCCCATGTAGGCACTGTTATTTTTTGCATCACACCTCCTACTTCAGTTT |
| 5′ P1-Prom_alr | TACAATGAATAAACTGAAGTAGGAGGTGTGATGCAAAAAATAACAGTGC |
| 3′ pJS116_XhoI_alr | CCAGTGCCAAGCTTGCATGTCTGCAGGCCTCGAGTTATTTTAGCAAATAACTGTTTATTT |
| 5′ pET_alr | GTTTAACTTTAAGAAGGAGATATACATATGCAAAAAATAACAGTGCCTACATG |
| 3′ pET_alr | ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTTTTAGCAAATAACTGTTTATTTGTAC |
| 5′ alr L39A | AAGATTTGTGGAGTAATAGCAGCTGATGCATATGG |
| 3′ alr L39A | GTCCATATGCATCAGCTGCTATTACTCCACAAAT |
Conjugation and mutant selection
Both pRS86 and pRS89 were inserted into C. difficile UK1 via conjugation with the Tn916-containing B. subtilis BS49 as a donor. B. subtilis was transformed with these plasmids following standard protocols, and transformants were confirmed using PCR. B. subtilis BS49 containing pRS86 or pRS89 was conjugated with C. difficile UK1, as described previously (12). After screening for tetracycline-sensitive and thiamphenicol-resistant colonies (transposon-negative, plasmid-positive), the colonies were confirmed to have the desired plasmid using PCR. To isolate a TargeTron insertion into alr2, the above isolates were spread onto BHIS plates containing 20 μg/ml lincomycin and incubated for 24–36 h to obtain colonies. Lincomycin-resistant colonies were isolated and tested for insertion of TargeTron by PCR using the primers 5′alr and 3′alr (Table 3). An isolate with the desired group II intron insertion was identified, and this strain was renamed C. difficile RS07.
Preparation of spores
All spores were purified from BHIS agar medium as described previously (8, 10, 12). Briefly, both wild-type C. difficile UK1 and C. difficile RS07 were grown on BHIS agar medium, and C. difficile RS07 pRS89 was grown on BHIS agar medium supplemented with 5 μg/ml thiamphenicol. Strains were grown for 4–5 days before isolating the growth and suspending in 1 ml of sterile water/plate. The suspension was stored overnight at 4 °C to promote release of the spores from the mother cells. The suspensions were then washed five times with sterile water to remove cell debris. The resulting spores were purified using a bed of 60% sucrose by centrifuging at 4,000 × g for 30 min. After centrifugation, the sucrose was removed, and the spores at the bottom of the tube were isolated and washed five times with sterile water to remove the sucrose. The spores were >99% pure and heat-activated at 65 °C for 30 min before use.
Spore germination
Spore germination was analyzed at 25 or 37 °C using both a DPA release assay and an optical density assay both. All assays were carried out in 100 μl of total volume using buffer containing a final concentration of 50 mm HEPES, 100 mm NaCl (pH 7.5), 10 mm TA, and varying concentrations of amino acids. The DPA release assay was performed as described previously using a final spore density of 0.5 optical density with 30 mm of amino acids and 250 μm of TbCl3 in a 96-well plate reader at low PMT settings (excitation at 270, emission at 420 nm) (17). The optical density-based germination assay was also carried out using a plate reader at 595 nm with final 0.5 optical density spores and 30 mm of amino acids.
For the calculation of EC50, the DPA release assay was used at 37 °C with increasing concentrations of amino acids (0–200 mm and a final optical density at 600 nm of 0.3 spores). The rates of germination were determined using the slopes of the linear portions of the germination plots. The data are reported as the averages from three independent experiments with one standard deviation from the mean.
Purification of Alr2 and AlrK39A
pJS164 and pRS100 were transformed into E. coli BL21(DE3) slyD::kan. To express and purify Alr26His, the cells were grown at 37 °C to an A600 = 0.8 in 2× TY medium supplemented with ampicillin before inducing for 4 h at 37 °C in the presence of 500 μm isopropyl β-d-thiogalactopyranoside. The cells were pelleted at 6,000 × g for 15 min in a Sorvall centrifuge (Beckman Coulter). The pellets were suspended in lysis buffer (LIB) containing 50 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm PLP, 1 mm tris-(2-carboxyethyl) phosphine hydrochloride, 15 mm imidazole and protease inhibitor (1 mm PMSF) before freezing at −80 °C.
The cell pellets were thawed and suspended in LIB (25 ml/liter of cell pellet). Lysozyme and DNase were added, and the cell suspension was incubated on ice for 30 min. The cells were lysed using sonication. The sonicated extract was clarified by centrifugation at 15,000 × g for 30 min at 4 °C. Alr26his was purified from the extract by nickel-affinity chromatography. Beads were washed once in wash buffer LIB containing 30 mm imidazole. The recombinant protein was eluted from the beads using the wash buffer with 500 mm imidazole. The eluted protein was dialyzed in LIB for 3 h and further purified by gel filtration chromatography (Akta Pure, GE Healthcare) on a Sephedex G200 size-exclusion column. Protein was injected and separated at 1 ml/min using LIB. Protein was detected at 280 nm and the eluted protein was collected (supplemental Fig. S3).
Enzymatic assays and HPLC separation of stereoisomers
l- to d-alanine and l- to d-serine conversion (and vice versa) was initiated by adding 500 ng of purified Alr2 to 50 mm amino acid in 50 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm PLP, and 1 mm tris-(2-carboxyethyl) phosphine hydrochloride (total reaction volume was 100 μl (155 nm)). The reactions were incubated at 37 °C for 1 h.
To assay for the presence of l- and d-forms of the tested amino acids, 200 μl of 1% FDAA (Marfey's reagent) in acetone and 40 μl of 1 m sodium bicarbonate was added to the reaction conditions above and incubated for 1 h at 40 °C (41). After the 1-h incubation, 20 μl of 2 m HCl was added to stop the reaction. The reacted material was separated by reverse-phase HPLC (Prominence system; Shimadzu) using a Thermo Scientific Synchronis C18 column. 3 μl of the reacted material was injected and separated at a 0.65 ml/min flow rate of buffer A (50 mm triethylammonium phosphate, pH 3.5, in 25% methanol) and a gradient of 0% buffer B (50 mm triethylammonium phosphate, pH 3.5, in 75% methanol) to 100% buffer B in 40 min before returning to 0% buffer B for another 30 min (70 min total time). Elution of the FDAA and FDAA-labeled compounds was detected by absorbance at 340 nm, and the peak area was quantified.
Isothermal titration calorimetry
All ITC experiments were done using a MicroCal iTC200 System (Malvern) at a constant temperature of 25 °C. The ITC base buffer was 50 mm Tris-HCl (pH 7.5), 250 mm NaCl, and 1 mm PLP. In the ITC chamber, wild-type Alr2 was used at a concentration of 32–45 μm in the ITC base buffer, whereas Alr2K39A was at 100 μm. In the syringe, d- and l-alanine ligands were dissolved in ITC base buffer at 200 mm for wild-type protein or 20 mm for the Alr2K39A experiments, and d- and l-serine were made to 200 mm. All solutions were filtered or centrifuged at 18,000 × g for 5 min prior to use. Thermograms were processed and fit using a single-ligand model (with n fixed to unity), using Origin software, to obtain K (association constant) and ΔH (enthalpy). The dissociation constant (KD) was calculated from the relationship KD = 1/K.
Author contributions
R. S. and J. A. S. designed the study and wrote the manuscript. R. S. and S. W. L. performed the experiments. S. W. L. provided comments regarding the paper. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Sorg laboratory and Dr. Leif Smith's laboratory at Texas A&M University for helpful comments during the preparation of this manuscript. We also thank Dr. Tadhg Begley's laboratory for the generous use of the ITC machine.
This work was supported by awards 5R01AI116895 and 1U01AI124290 from the NIAID, National Institutes of Health (to J. A. S.) The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S4.
- CDI
- C. difficile infection
- TA
- taurocholic acid
- CaDPA
- calcium dipicolinic acid
- BHIS-TA
- brain-heart infusion broth supplemented with taurocholic acid
- ITC
- isothermal titration calorimetry
- PLP
- pyridoxal 5′-phosphate
- FDAA
- 1-fluoro-2–4-dinitrophenyl-5-l-alanine amide.
References
- 1. Centers for Disease Control and Prevention (2013) Antibiotic/antimicrobial resistance: biggest threats, http://www.cdc.gov/drugresistance/biggest_threats.html
- 2. Theriot C. M., and Young V. B. (2015) Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 69, 445–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shields K., Araujo-Castillo R. V., Theethira T. G., Alonso C. D., and Kelly C. P. (2015) Recurrent Clostridium difficile infection: from colonization to cure. Anaerobe 34, 59–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deakin L. J., Clare S., Fagan R. P., Dawson L. F., Pickard D. J., West M. R., Wren B. W., Fairweather N. F., Dougan G., and Lawley T. D. (2012) The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 80, 2704–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhattacharjee D., McAllister K. N., and Sorg J. A. (2016) Germinants and their receptors in Clostridia. J. Bacteriol. 198, 2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Setlow P. (2014) Spore resistance properties. Microbiol. Spectr. 2, 10.1128/microbiolspec.TBS-0003-2012 [DOI] [PubMed] [Google Scholar]
- 7. Setlow P. (2014) Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 196, 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Francis M. B., Allen C. A., and Sorg J. A. (2013) Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One 8, e73653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sorg J. A., and Sonenshein A. L. (2009) Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191, 1115–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sorg J. A., and Sonenshein A. L. (2008) Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howerton A., Ramirez N., and Abel-Santos E. (2011) Mapping interactions between germinants and Clostridium difficile spores. J. Bacteriol. 193, 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francis M. B., Allen C. A., Shrestha R., and Sorg J. A. (2013) Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 9, e1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sebaihia M., Wren B. W., Mullany P., Fairweather N. F., Minton N., Stabler R., Thomson N. R., Roberts A. P., Cerdeño-Tárraga A. M., Wang H., Holden M. T., Wright A., Churcher C., Quail M. A., Baker S., et al. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786 [DOI] [PubMed] [Google Scholar]
- 14. Adams C. M., Eckenroth B. E., Putnam E. E., Doublié S., and Shen A. (2013) Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 9, e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francis M. B., Allen C. A., and Sorg J. A. (2015) Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J. Bacteriol. 197, 2276–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gutelius D., Hokeness K., Logan S. M., and Reid C. W. (2014) Functional analysis of SleC from Clostridium difficile: an essential lytic transglycosylase involved in spore germination. Microbiology 160, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Francis M. B., and Sorg J. A. (2016) Dipicolinic acid release by germinating Clostridium difficile spores occurs through a mechanosensing mechanism. mSphere 1, e00306–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fimlaid K. A., Jensen O., Donnelly M. L., Francis M. B., Sorg J. A., and Shen A. (2015) Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog. 11, e1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donnelly M. L., Li W., Li Y. Q., Hinkel L., Setlow P., and Shen A. (2017) A Clostridium difficile-specific, gel-forming protein required for optimal spore germination. mBio 8, e02085–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Radkov A. D., and Moe L. A. (2014) Bacterial synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 98, 5363–5374 [DOI] [PubMed] [Google Scholar]
- 21. Chesnokova O. N., McPherson S. A., Steichen C. T., and Turnbough C. L. Jr. (2009) The spore-specific alanine racemase of Bacillus anthracis and its role in suppressing germination during spore development. J. Bacteriol. 191, 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yasuda Y., and Tochikubo K. (1984) Relation between d-glucose and l- and d-alanine in the initiation of germination of Bacillus subtilis spore. Microbiol. Immunol. 28, 197–207 [DOI] [PubMed] [Google Scholar]
- 23. Titball R. W., and Manchee R. J. (1987) Factors affecting the germination of spores of Bacillus anthracis. J. Appl. Bacteriol. 62, 269–273 [DOI] [PubMed] [Google Scholar]
- 24. McKevitt M. T., Bryant K. M., Shakir S. M., Larabee J. L., Blanke S. R., Lovchik J., Lyons C. R., and Ballard J. D. (2007) Effects of endogenous d-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infect. Immun. 75, 5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anmuth M., Harding J., Kravitz E., and Stedman R. L. (1956) Autoinhibition of bacterial endospore germination. Science 124, 403–405 [DOI] [PubMed] [Google Scholar]
- 26. Fimlaid K. A., Bond J. P., Schutz K. C., Putnam E. E., Leung J. M., Lawley T. D., and Shen A. (2013) Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 9, e1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhattacharjee D., Francis M. B., Ding X., McAllister K. N., Shrestha R., and Sorg J. A. (2015) Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J. Bacteriol. 198, 777–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramirez N., and Abel-Santos E. (2010) Requirements for germination of Clostridium sordellii spores in vitro. J. Bacteriol. 192, 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sorg J. A., and Sonenshein A. L. (2010) Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramirez N., Liggins M., and Abel-Santos E. (2010) Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J. Bacteriol. 192, 4215–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Girinathan B. P., Monot M., Boyle D., McAllister K. N., Sorg J. A., Dupuy B., and Govind R. (2017) Effect of tcdR mutation on sporulation in the epidemic Clostridium difficile strain R20291. mSphere 2, e00383–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dodatko T., Akoachere M., Jimenez N., Alvarez Z., and Abel-Santos E. (2010) Dissecting interactions between nucleosides and germination receptors in Bacillus cereus 569 spores. Microbiology 156, 1244–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akoachere M., Squires R. C., Nour A. M., Angelov L., Brojatsch J., and Abel-Santos E. (2007) Indentification of an in vivo inhibitor of Bacillus anthracis spore germination. J. Biol. Chem. 282, 12112–12118 [DOI] [PubMed] [Google Scholar]
- 34. Stoltz K. L., Erickson R., Staley C., Weingarden A. R., Romens E., Steer C. J., Khoruts A., Sadowsky M. J., and Dosa P. I. (2017) Synthesis and biological evaluation of bile acid analogues inhibitory to Clostridium difficile spore germination. J. Med. Chem. 60, 3451–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Asojo O. A., Nelson S. K., Mootien S., Lee Y., Rezende W. C., Hyman D. A., Matsumoto M. M., Reiling S., Kelleher A., Ledizet M., Koski R. A., and Anthony K. G. (2014) Structural and biochemical analyses of alanine racemase from the multidrug-resistant Clostridium difficile strain 630. Acta Crystallogr. D. Biol. Crystallogr. 70, 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paredes-Sabja D., Setlow P., and Sarker M. R. (2011) Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19, 85–94 [DOI] [PubMed] [Google Scholar]
- 37. George W. L., Sutter V. L., Citron D., and Finegold S. M. (1979) Selective and differential medium for isolation of Clostridium difficile. J. Clin. Microbiol. 9, 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawley T. D., Croucher N. J., Yu L., Clare S., Sebaihia M., Goulding D., Pickard D. J., Parkhill J., Choudhary J., and Dougan G. (2009) Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191, 5377–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouillaut L., Self W. T., and Sonenshein A. L. (2013) Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd, Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 41. Bhushan R., and Brückner H. (2004) Marfey's reagent for chiral amino acid analysis: a review. Amino Acids 27, 231–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.