Abstract
Well-organized mitochondrial functions and dynamics are critical for early embryonic development and are operated via a large number of mitochondria-related genes (MtGs) encoded by both the nuclear and the mitochondrial genome. However, the mechanisms underlying mitochondrial modifications during the critical window between blastocyst implantation and postimplantation organogenesis are poorly understood. Herein, we performed high-resolution dynamic profiling of MtGs to acquire a more detailed understanding of mitochondrial modifications during early development. Our data suggest that the resumption of mitochondrial mass growth is not only facilitated by increased mitochondrial biogenesis and mitochondrial DNA (mtDNA) replication, but also by the appropriate balance between mitochondrial fission and fusion. In addition, increased levels of reactive oxygen species (ROS) resulting from enhanced mitochondrial functions may be the critical inducer for activating the glutathione (GSH)-based stress response system in early embryos. The appropriate balance between the mitochondrial stress response and apoptosis appears to be significant for cell differentiation and early organogenesis. Furthermore, we found that most MtGs undergo de novo promoter methylation, which may have functional consequences on mitochondrial functions and dynamics during early development. We also report that mtDNA methylation can be observed as early as soon after implantation. DNMT1, the predominant enzyme for maintaining DNA methylation, localized to the mitochondria and bound to mtDNA by the implantation stage. Our study provides a new insight into the involvement of mitochondria in early mammalian embryogenesis. We also propose that the epigenetic modifications during early development are significant for modulating mitochondrial functions and dynamics.
Keywords: DNA methylation, Early embryos, Glutathione, Mitochondria, Reactive oxygen species
Mitochondria not only provide the energy required to maintain cellular survival and growth but also participate in a number of pathways that maintain cellular homeostasis, including ion homeostasis, amino acid metabolism, glycolysis, fatty acid metabolism, signal transduction, and apoptosis [1]. Well-organized mitochondrial dynamics and functions are of prime importance for early embryonic development. During the period between the blastocyst and the postimplantation stages, which overlaps with implantation and organogenesis initiation, the mitochondrion undergoes dramatic modifications, including mitochondrial biogenesis and mitochondrial DNA (mtDNA) replication, a shift from anaerobic to aerobic metabolism, and changes in the mitochondrial ultrastructure [2]. These mitochondrial modifications are essential for successful implantation and survival. However, the comprehensive mechanism underlying mitochondrial dynamics during this critical developmental period remains largely elusive.
The orchestrated mitochondrial functions and dynamics are highly regulated by both the nuclear (nDNA) and mitochondrial genomes (nuclear-mitochondrial cross-talk). In addition to the 37 mtDNA-encoded genes, it is estimated that approximately 1,200 proteins encoded by nDNA can enter the mouse mitochondrial matrix or bind to its membrane [3]. These mitochondria-related genes (MtGs) have been targeted in high-resolution gene expression analysis to evaluate the involvement of mitochondrial dysfunctions in cardiac pathology [4] and cancer [5]. Recently, our own studies suggested that mitochondrial dysfunction may be the determining factor of impaired development of in vitro fertilized (IVF) embryos through disturbed oxidative phosphorylation (OXPHOS), reduced mitochondrial biogenesis, and dysregulated responses to oxidative stress [6].
In this study, we performed a dynamic high-resolution expression profiling of MtGs during early embryonic development, using samples from E3.5 blastocysts, epiblasts from E7.5 embryos (E7.5 epiblasts), and E10.5 embryos. The targeted profiling of a specific gene set leads to a more efficient enrichment of the involved mitochondrial functions, thereby providing a more detailed and comprehensive understanding of mitochondrial dynamics and functions during early development.
In addition to the expression profiling, we also highlighted the DNA methylation dynamics of MtGs for the period between the blastocyst and the postimplantation stages. One of the most important epigenetic events during this critical developmental period is the re-establishment of DNA methylation patterns, termed de novo DNA methylation. The progression of this. epigenetic event highly coincides with the mitochondrial modifications taking place by the implantation stage, such as the ‘embryonic shift’ from anaerobic to aerobic metabolism and the resumption of mitochondrial biogenesis. In addition, previous studies demonstrated that the DNA methylation status of nDNA-encoded MtGs underlies tissue- or cell type-dependent mitochondrial functions and dynamics [7, 8] as well as mitochondrial pathology [9,10,11]. These facts lead us to investigate whether de novo DNA methylation may play a regulatory role in the mitochondria during early development.
Materials and Methods
Animal preparation
F1 female mice (ICR, 5–6 weeks old) and F1 male mice (ICR, 8–9 weeks old) were fed ad libitum and housed in a room with a controlled light cycle (12L:12D). F1 females were naturally mated with F1 males. All studies were specifically approved by and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University.
Embryo preparation and collection
The criteria for sampling embryos were based on developmental progress and morphology. Embryos showing typical morphological features according to the well-established landmarks [12] were sampled for further analyses. All sampled embryos for RNA and DNA isolation were serially washed with phosphate-buffered saline (PBS; GIBCO, Life Technologies, NY, USA) and immediately stored in liquid nitrogen for future use. In addition, to avoid cross-contamination between embryonic and extraembryonic tissues, we controlled the efficiency of the dissection procedure by detecting the expression of markers specific to the extraembryonic ectoderm (ETS-related transcription factor, Elf5), ectoplacental cone (Achaete-scute like 2, ASCL2, also known as Mash2), and epiblasts (fibroblast growth factor 5, Fgf5) at E7.5 [13].
In vitro fertilization and culture
To generate in vitro fertilized embryos, female ICR mice were superovulated by an intraperitoneal (i.p.) injection of 5 IU pregnant mare serum gonadotropin (PMSG; Ningbo Hormone Product, Ningbo, China), followed by an i.p. injection of 5 IU human chorionic gonadotropin (hCG; Ningbo Hormone Product) 48 h later. At 14 h after hCG treatment, cumulus-enclosed oocyte complexes were recovered from oviducts and cumulus cells were removed by digestion with hyaluronidase (Sigma-Aldrich, St. Louis, MO, USA) for 3–5 min. The oocytes were rinsed in human tubal fluid (HTF) medium (Sage, Bedminster, NJ, USA), and placed into 60 μl drops of HTF medium covered with paraffin oil, before being equilibrated overnight at 37°C and 5% CO2. Sperm was collected from the cauda epididymis and capacitated for 1 h in HTF medium at 37°C and 5% CO2. Oocytes were inseminated 15 h post-hCG with 106 spermatozoa. After 4 h in the incubator, oocytes and zygotes were washed several times in potassium simplex optimization medium containing amino acids (KSOM + AA; Millipore, Billerica, MA, USA) and then transferred to 60 µl drops of KSOM medium. The zygotes, determined by the presence of two pronuclei, were cultured to the blastocyst stage at 37°C in a 5% CO2 atmosphere. Well-developed late-cavitating blastocysts of similar morphology were sampled at 106–112 h post-hCG after culturing in KSOM medium [14]. Pseudopregnant female mice were mated with vasectomized males 3.5 days prior to embryo transfer. Twelve IVF blastocysts were transferred to a single recipient, with six embryos placed in each uterine horn.
High-throughput RNA sequencing (RNA-seq) and biological analysis
Total RNAs were extracted from embryos at different stages with the TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Polyadenylated RNAs were isolated using the Oligotex mRNA Midi Kit (Qiagen, Valencia, CA, USA). The RNA-seq libraries were constructed using the SOLiD Whole Transcriptome Analysis Kit (Applied Biosystems, Carlsbad, CA, USA) following the standard protocol and sequenced on the SOLiD platform (Applied Biosystems) to generate high-quality single-end reads. The raw reads were aligned to genome sequences, trimming off a nucleotide from each of the 5′ and 3′ ends and allowing up to two mismatches. Reads mapped to multiple locations were discarded and only uniquely mapped reads were used for the subsequent analysis. Gene expression levels were measured in reads per kilobase of exon model per million mapped reads (RPKM). A single pooling strategy for relatively large sample sizes was used, as described previously [15, 16].
DAVID v6.7 (http://david.abcc.ncifcrf.gov) was used to annotate biological themes (gene ontology, GO). REVIGO analysis (http://revigo.irb.hr) was also performed to visualize the interactive relationship among enriched processes. The KEGG database (http://www.genome.jp/kegg) was used to determine the associated pathways. Data on MtGs were fed into STRING (http://string.embl.de) to build an interaction network. The phenotype annotations of MtGs were analyzed based on the Mouse Genome Informatics (MGI; http://www.informatics.jax.org/phenotypes.shtml) database.
Apoptosis analysis
Collected blastocysts were washed three times with 0.1% PVA/PBS and then transferred to PBS supplemented with 4% (v/v) paraformaldehyde and 0.5% Triton X-100 for simultaneous fixation and permeabilization at 37°C for 45 min. Apoptotic nuclei were detected using the In Situ Cell Death Detection Kit (Roche, Mannheim, Germany). The number of apoptotic nuclei was determined under a BX51epifluorescence microscope (Olympus, Tokyo, Japan).
RNA isolation and quantitative real-time RT-PCR (qRT-PCR)
Total RNA extraction from embryos was performed using TRIzol according to the manufacturer’s instructions. Reverse transcription (RT) was carried out using a commercially available first-strand cDNA synthesis kit (iScript cDNA Synthesis Kit; Bio-Rad Laboratories, USA). Real-time PCR was performed in the Bio-Rad CFX96 Real-Time PCR System using SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Used primers are listed in Supplementary Table 1 (online only).
MtDNA copy number determination by qPCR
The nuclear-encoded β-actin gene and the mitochondria-encoded ND5 gene were used to determine the mtDNA copy numbers with the primers listed in Supplementary Table 1. Standard curves were constructed using recombinant plasmids that were serially diluted 103–108 times. The mtDNA and nDNA copy numbers were calculated from the threshold cycle number (CT) and corrected using the standard curve. The overall number of mtDNA copies per cell was calculated using the following formula: mtDNA copies per cell = ND5 copy number / (β-actin copy number/2).
Measurement of reduced glutathione (GSH) and oxidized glutathione (GSSG)
The contents of intracellular reduced glutathione (GSH) and oxidized glutathione (GSSG) in each blastocyst were determined using commercially available kits (Beyotime, Jiangsu, China) according to the manufacturer’s instructions. Absorbance was measured at 412 nm using the Infinite M200 PRO NanoQuant microplate reader (Tecan, Männedorf, Switzerland). Approximately 30 E3.5 blastocysts and 20 E7.5 epiblasts were used for measurements.
Methylated DNA immunoprecipitation-sequencing (MeDIP-seq)
Global genomic DNA, including both nDNA and mtDNA, was isolated from embryonic samples using the DNeasy Kit (Qiagen) according to the manufacturer’s instructions. The quality of each DNA sample with respect to integrity, purity, and concentration was assessed using a DN-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Genomic DNA was then fragmented using a Covarias sonication system (Covarias, Woburn, MA, USA). After sonication, the fragments were denatured to produce single-stranded DNA (ssDNA). Following denaturation, the ssDNA was incubated with antibodies recognizing 5-methylcytosine (5mC). Magnetic beads conjugated to anti-mouse-IgG were then used to bind the anti-5mC antibodies and unbound DNA was removed along with the supernatant. Finally, DNA was released by digesting the antibodies with proteinase K and collected. Sequencing was carried out in 50 bp reads on the Illumina Hiseq 2000 (Illumina, San Diego, CA, USA) by the Beijing Genomics Institute (BGI, Shenzhen, Guangdong, China) following the standard protocol.
MeDIP-qPCR
Global DNA was isolated using the Solar MicroElute Genomic DNA Kit (Solar Technologies, Gaithersburg, MD, USA). Sonicated DNA (fragment length: 100–600 bp) was treated with the EpiQuik MeDIP Ultra Kit (Epigentek, Farmingdale, NY, USA) to enrich methylated DNA fragments. Briefly, samples were precipitated with anti-5-mC, while samples precipitated with non-immunized IgG served as a negative control. The enriched methylated DNA was quantified by qPCR using specific primers (Supplementary Table 1) and the enrichment efficiency of MeDIP-qPCR was measured as fold enrichment (FE) with the following formula: FE (%) = 2(IgG CT –Sample CT) × 100%, as presented in the product manual.
Results
General dynamic profiling of MtGs during early development
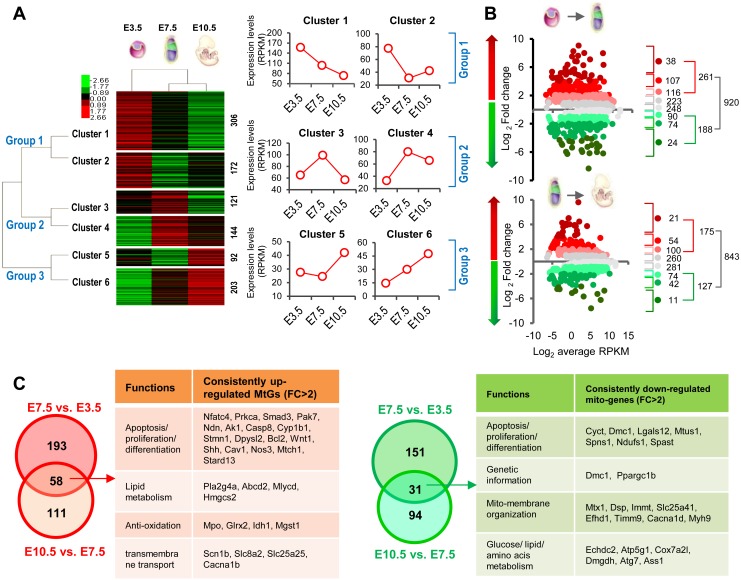
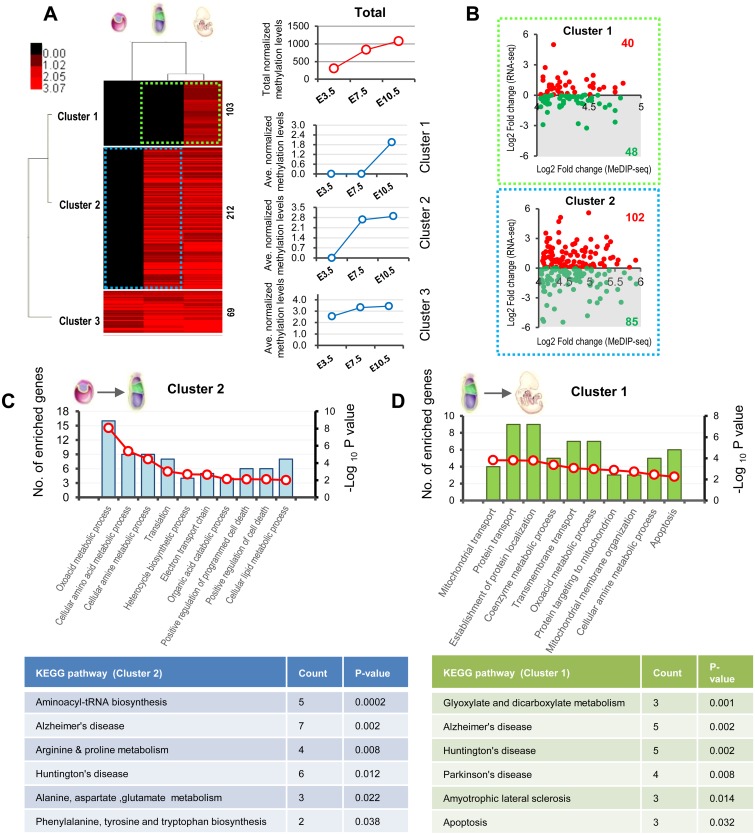
We gathered information for over 1000 MtGs, as reported by Mootha’s study [3], from the transcriptomic data obtained from E3.5 blastocysts, E7.5 epiblasts, and E10.5 embryos (Supplementary Table 2: online only). The hierarchical clustering heatmap showed that all MtGs can be sorted into three main groups based on expression dynamics, namely whether they were most abundant at E3.5, E7.5, or E10.5. Each group can be further divided into two clusters (Fig. 1A). Compared with E3.5 blastocysts, E7.5 epiblasts and E10.5 embryos were clustered more tightly, with similar MtG expression patterns. Functional analysis showed that each cluster was significantly associated with many aspects of mitochondrial functions or related processes, including OXPHOS, oxoacid and amino acid metabolism, and apoptosis (Supplementary Fig. 1A: online only), implying that mitochondrial dynamics during early development are orchestrated via complicated expression patterns involving MtGs (Supplementary Fig. 1B).
Fig. 1.
General dynamic profiling of mitochondria-related genes (MtGs) during early development. (A) Hierarchical clustering analysis based on the dynamic expression levels of MtGs of embryos from pre- to postimplantation. Normalized expression levels (RPKM) are represented by different colors: red indicates high abundance and green indicates low abundance. The number of MtGs in each cluster is presented on the right side. General trends in expression changes are indicated by the average RPKM value of MtGs in each cluster. (B) Fold change (FC) distribution analysis of MtGs between E3.5 blastocysts to E7.5 epiblasts as well as between E7.5 epiblasts and E10.5 embryos. Only MtGs with statistically significant changes in expression were included (P < 0.05). (C) Venn diagrams of upregulated (left panel, red) and downregulated (right panel, green) MtGs for two developmental transitions. Tables show the major functions of MtGs that are consistently upregulated (left) or downregulated (right) during early development.
To further explore the involvement of mitochondrial functions and dynamics during early embryonic development, the expression patterns of MtGs at E3.5, E7.5, and E10.5 were compared consecutively, i.e., E7.5 versus E3.5 and E10.5 versus E7.5. These two transitions encompass a series of important developmental events, such as implantation, cellular differentiation, and organogenesis. In the first transition, i.e., from E3.5 blastocysts to E7.5 epiblasts, 920 MtGs displayed significant changes in expression (P < 0.05), accounting for approximately 80% of identified MtGs. During the transition from E7.5 epiblasts to E10.5 embryos, the expression of 843 MtGs changed significantly (P < 0.05), accounting for approximately 73% of identified MtGs. Both percentages were higher than the corresponding values of the global transcriptome, which were approximately 70% and 64%, respectively (Fig. 1B), implying that expression of MtGs is highly dynamic during early development. These observations also suggest that mitochondrial features may undergo a more dramatic transition from E3.5 blastocysts to E7.5 epiblasts, in accordance with the results of the hierarchical clustering.
A Venn diagram analysis was performed to screen MtGs consistently up- or downregulated (FC > 2) during the transition E3.5 to E10.5. According to the results of the analysis, 58 MtGs showed consistent upregulation while 31 MtGs were consistently downregulated (Fig. 1C). Considering that the expression of these MtGs consistently changed during early development, they may contribute significantly to the highly dynamic mitochondrial functions taking place during this stage.
Detailed functional profiling of MtGs during early development
To further understand the comprehensive mechanism underlying mitochondrial functions and dynamics during early development, high-resolution functional profiling was performed for the MtGs whose expression significantly changed (P < 0.05, FC > 2) during the transition from E3.5 to E7.5 (Supplementary Table 3: online only) as well as from 7.5 to E10.5 (Supplementary Table 4: online only).
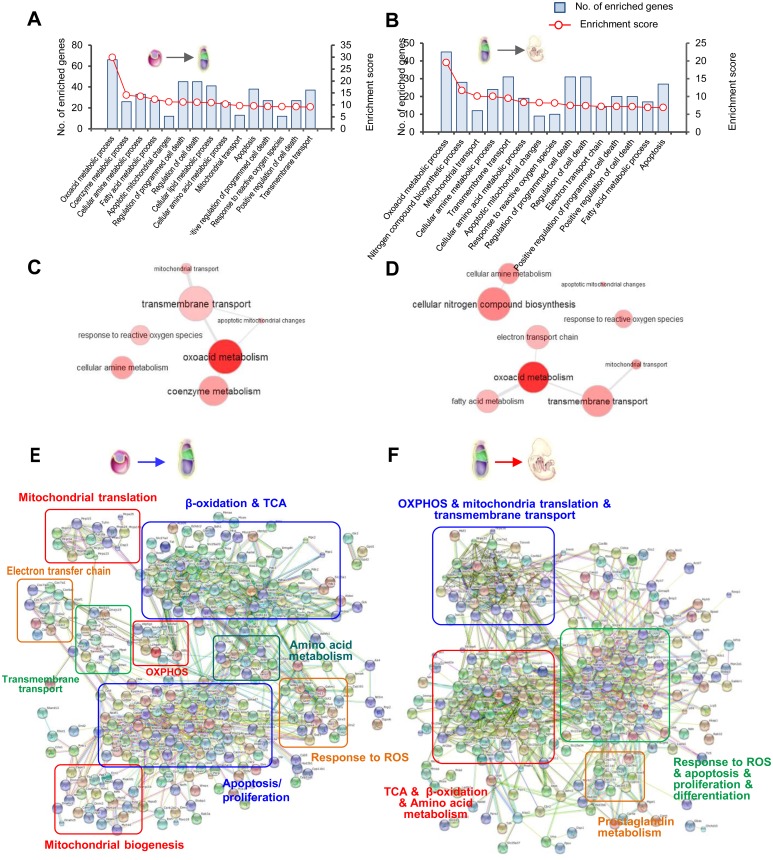
GO classification of biological processes indicated that ‘Oxoacid metabolic process’, ‘Coenzyme metabolic process’, ‘Cellular amine metabolic process’, and ‘Fatty acid metabolic process’ were the four most significantly enriched terms in the transition from E3.5 blastocysts to E7.5 epiblasts, suggesting that metabolic changes are the most evident feature of mitochondrial modifications during this period. In addition, ‘Apoptotic mitochondrial changes’ and ‘Response to ROS’ may also be significantly involved in this transition. Similar processes were enriched in the transition from E7.5 epiblasts to E10.5 embryos (Fig. 2A and B). Next, we performed REVIGO analysis to visualize the interactive relationship among the enriched processes. Results from both transitions suggest that ‘Transmembrane transport’ may play a key role in regulating mitochondrial metabolism and mitochondria-mediated apoptosis (Fig. 2C and D).
Fig. 2.
General functional profiling of MtGs whose expression significantly changed (P < 0.05, FC > 2) during early development. (A–B) Classification of GO terms based on the functional annotation of biological processes (BPs) enriched in the transition from E3.5 blastocysts to E7.5 epiblasts, as well as from E7.5 epiblasts to E10.5 embryos. The left ordinate represents the number of enriched MtGs corresponding to each term and the right ordinate represents the enrichment score (defined as −Log10 P-value). (C–D) Graph visualization of enriched BPs in A and B based on REVIGO analysis. Bubble color indicates P-value. Functionally associated BPs are linked. (E–F) Interaction networks of MtGs during early development created by a web-based search of the STRING database. Boxed regions represent tightly interconnected functional clusters.
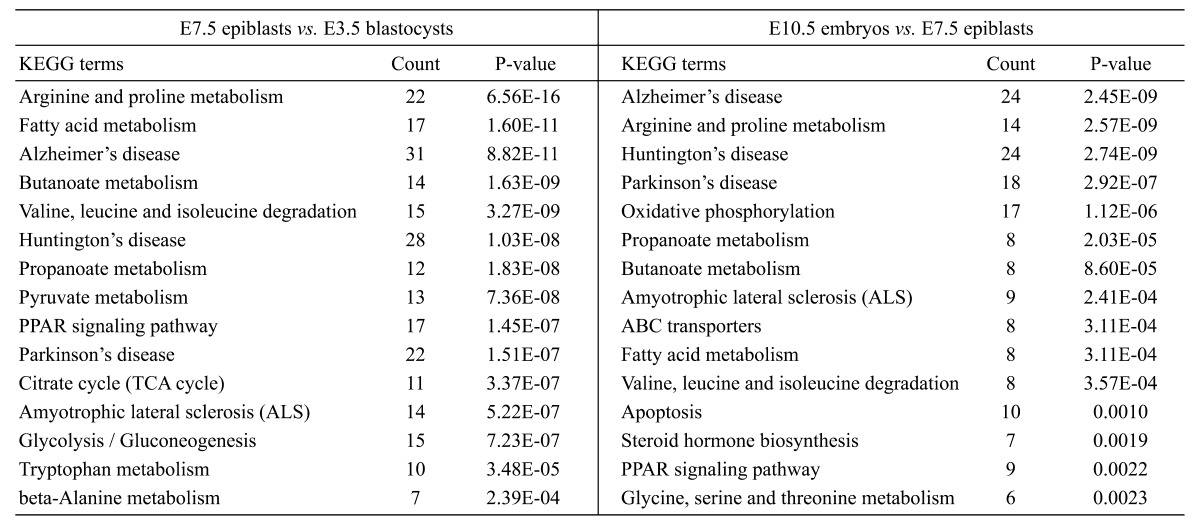
KEGG pathway analysis provided us with more detailed insights into mitochondrial modifications. For the transition from E3.5 blastocysts to E7.5 epiblasts, the altered mitochondrial energy metabolism included ‘TCA cycle’, ‘OXPHOS’, and ‘Fatty acid β-oxidation’, while the changed amino acid metabolism involved arginine, proline, valine, leucine, glutamate, and glycine. It should be noted that some pathways of neurodegenerative diseases, including ‘Alzheimer’s disease (AD)’, ‘Parkinson’s disease (PD)’ and ‘Huntington’s disease (HD)’ were also enriched, mainly because of MtGs associated with ‘OXPHOS’ and ‘apoptosis’. Likewise, changes in energy metabolism and amino acid metabolism were also significant during the transition from E7.5 to E10.5 embryos (Table 1).
Table 1. Enriched KEGG pathways (Top 15) for MtGs significantly changed during early development.
Using dynamically changed MtGs (P < 0.05, FC > 2) as seed nodes, interaction networks were constructed. In addition to functional clusters similar to those identified by the GO and KEGG analyses, other processes or functions, such as ‘mitochondrial translation’ and ‘mitochondrial biogenesis’, were also significantly clustered. Another interesting observation was that some well-known genes responsible for the regulation of cell pluripotency and differentiation, such as members of the MAPK (Mapk8 and Mapk12), WNT (Wnt1 and Wnt3), and SMAD (Smad3 and Smad7) families, were clustered tightly with other MtGs, suggesting an essential role of mitochondrial functions in the regulation of pluripotency and differentiation in early embryos (Fig. 2E and F).
Mitochondrial modifications required for successful implantation
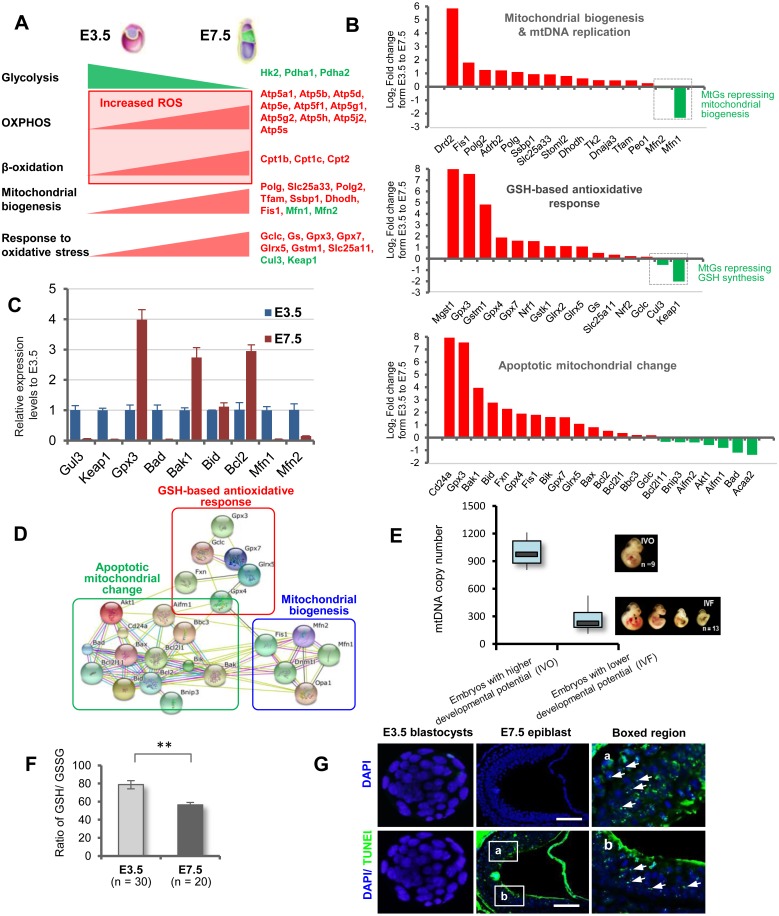
The above-mentioned functional profiling revealed that many mitochondrial modifications had undergone evident temporal changes by the implantation stage. The observed changes in the expression of some key genes of energy metabolism were in accordance with the ‘embryonic shift’ from anaerobic to aerobic metabolism. Accordingly, MtGs associated with mitochondrial biogenesis showed changes that may accommodate the enhanced aerobic metabolism. It is noteworthy that the expression of MtGs underlying GSH biosynthesis and functions changed correspondingly, suggesting a physiological response to the increased ROS levels resulting from the enhanced mitochondrial functions (Fig. 3A).
Fig. 3.
Mitochondrial modifications during implantation stage. (A) Model illustrating the ‘embryonic shift’ from anaerobic to aerobic metabolism, as well as the resumption of mitochondrial biogenesis and the enhanced response to oxidative stress. Red text indicates upregulated MtGs, while green text indicates downregulated MtGs. (B) Fold changes of representative MtGs involved in essential mitochondrial modifications by the implantation stage. (C) Relative expression levels of some representative MtGs in E3.5 blastocysts and E7.5 epiblasts determined by qRT-PCR. (D) Interaction networks of MtGs responsible for essential mitochondrial modifications taking place during the implantation stage. (E) Boxplot quantitative comparison of mtDNA copy numbers between E10.5 embryos with higher and lower survival rates. (F) Ratios of reduced glutathione (GSH) to oxidized glutathione (GSSG) in E3.5 blastocysts and E7.5 epiblasts; ** P < 0.01. (G) Representative images of apoptosis detected by TUNEL in E3.5 blastocysts and E7.5 epiblasts. Rightmost panels: higher magnification of boxed regions in E7.5 epiblasts. The nuclei of blastomeres were labeled with DAPI. Representative TUNEL-positive apoptotic nuclei are indicated with arrows. Scale bars, 100 μm.
To gain further insight into the comprehensive mechanism underlying the increased mitochondrial biogenesis and GSH production in early embryonic development, we analyzed the dynamics of MtGs involved in these processes based on the gene list provided by Quick GO (https://www.ebi.ac.uk/QuickGO) and Wikipathways (http://www.wikipathways.org/index.php/WikiPathways). Results showed that the expression of most of the MtGs associated with these processes correspondingly changed from E3.5 to E7.5 (Fig. 3B). The changes of some selected MtGs were validated using qRT-PCR (Fig. 3C). Next, to understand the interactive relationship of these processes, we constructed a small network. This revealed a high level of association among these processes (Fig. 3D). Indeed, the involvement of mitochondrial biogenesis and GSH-based stress response in mitochondria-mediated apoptosis can also be deduced by the fact that many MtGs are shared among these terms.
To assess the association between the resumption of mitochondrial biogenesis and mtDNA replication on the embryonic developmental potential, we compared the mtDNA copy numbers between in vivo conceived (IVO) and IVF embryos, which are characterized by higher and lower developmental rates, respectively. Our previous studies showed that IVF embryos have lower survival rate and weight, as well as greater variability in size and increased growth defects, compared to their IVO counterparts [6, 13, 17]. Quantitative analysis showed that mtDNA copy numbers in IVF embryos laid in a significantly lower range compared to those in IVO embryos (Fig. 3E), suggesting that depressed mitochondrial resumption is strongly associated with impaired embryonic developmental potential.
The presence of enhanced mitochondrial mass and metabolism during the implantation stage led us to speculate that postimplantation embryos face increased oxidative stress compared to preimplantation embryos. To this end, we determined the GSH/GSSG ratio, which is a well-accepted indicator for evaluating oxidative stress status [18], in E3.5 blastocysts and E7.5 epiblasts. We found the latter to display a significantly higher GSH/GSSG ratio, indicating increased oxidative stress soon after implantation (Fig. 3F).
Subsequently, because of the significant changes in the expression patterns of apoptotic MtGs that had taken place by the implantation stage, we decided to compare the occurrence of apoptosis in E3.5 blastocysts and E7.5 epiblasts. TUNEL analysis showed that E7.5 epiblasts displayed a high rate of apoptosis compared to E3.5 blastocysts (Fig. 3G). Possible false-negative apoptotic nuclei in E3.5 blastocysts can be excluded by the parallel observation of TUNEL-positive apoptotic nuclei in IVF blastocysts, which have been reported to have a relative higher apoptotic rate [6].
The involvement of MtGs in embryonic survival and organogenesis
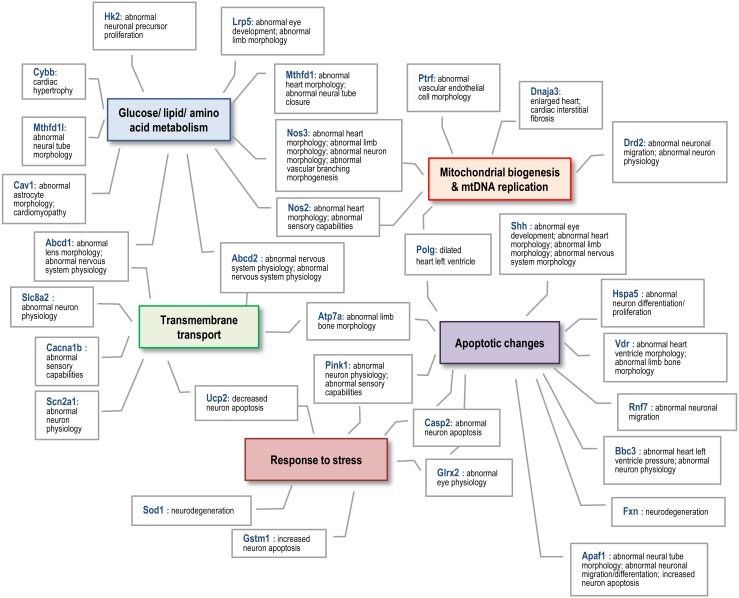
To test the potential role of MtGs in embryonic survival and organogenesis, we performed an MGI analysis using dynamically changed MtGs (P < 0.05, FC > 2; Supplementary Tables 5 and 6: online only). Many MtGs were annotated with phenotypes associated with aberrant embryonic/fetal development, such as ‘Complete embryonic lethality’, ‘Embryonic growth arrest’, ‘Failure of somite differentiation’, and ‘Failure of embryo implantation’. In addition, many MtGs were associated with abnormal morphology or aberrant development of organs, especially of the neurosensory and cardiovascular systems, as well as with lethality during organogenesis. These observations suggested that orchestrated mitochondrial functions and dynamics are essential for embryonic survival and normal organogenesis both during and after implantation. In addition, we found that different mitochondrial modifications might functionally correlate with each other to regulate early organogenesis (Fig. 4).
Fig. 4.
Schematic diagram of the functional associations among MtGs that are annotated with aberrant organogenesis during the postimplantation stage. Colored boxes indicate mitochondrial modifications that may be essential for embryonic survival and normal organogenesis during the implantation and postimplantation periods.
Dynamics of promoter DNA methylation of nDNA-encoded MtGs during early development
The dramatic changes of mitochondrial functions and dynamics during early development, including the resumption of mitochondrial biogenesis and the shift from anaerobic to aerobic metabolism, coincide with de novo DNA methylation, which is one of the most important epigenetic events during early development. This led us to hypothesize that the de novo DNA methylation of MtGs may have an essential regulatory role in the mitochondrial modifications taking place during early development. To test this hypothesis, nDNA-encoded MtGs whose promoters were methylated at least once at three time-points (Supplementary Table 7: online only) were hierarchically clustered based on their promoter DNA methylation levels, as described previously [19]. The resulting heatmap showed a generally progressive increase in promoter DNA methylation of MtGs during early development, especially of those belonging to clusters 1 and 2, which correspond to genes that are unmethylated in E3.5 blastocysts and gain promoter DNA methylation before E7.5 or E10.5, respectively. On the other hand, promoter DNA methylation increased slightly in cluster 3, which includes genes whose promoters appear methylated throughout early development (Fig. 5A).
Fig. 5.
Dynamics of promoter DNA methylation of nDNA-encoded MtGs and functional consequences during early development (A) Hierarchical clustering analysis based on dynamic methylation levels of MtGs during the transition from pre- to postimplantation embryos. Normalized methylation levels are represented by different colors: red indicates relatively hypermethylated promoters and black indicates relatively hypomethylated promoters. The numbers of MtGs in each cluster are displayed on the right side of the plot. Graphs next to the plot display promoter methylation levels during early development. The red line (upper graph) indicates total normalized methylation values while blue lines (three lower graphs) correspond to average normalized methylation levels in each cluster. (B) Scatter plots of expression changes versus mean differences of methylation for the MtGs of cluster 1 (upper plot, boxed by green dotted line) and cluster 2 (lower plot, boxed by blue dotted line) whose promoters underwent de novo methylation before E7.5 and E10.5, respectively. Green dots indicate MtGs that are downregulated during the process of promoter de novo methylation, while red dots indicate upregulated MtGs (C–D) Functional profiling of MtGs corresponding to the green dots of the shaded regions of the scatter plots in (B) and may be negatively regulated by promoter de novo methylation taking place between E3.5 and E7.5 or between E7.5 and E10.5.
To test the correlation between promoter de novo methylation and the expression levels of MtGs during early development, we integrated the promoter DNA methylation profiles to the expression profiles of MtGs in clusters 1 and 2 (Fig. 5B; boxed by green and blue dotted lines, respectively). In cluster 1, 48 MtGs (54.5%) were downregulated during the transition from E7.5 to E10.5, with increased levels of promoter DNA methylation. In cluster 2, 85 MtGs (45.5%) showed decreased expression at E7.5 compared to E3.5, and the reduction in expression coincided with promoter de novo methylation taking place before E7.5. This result suggests that promoter de novo methylation may have a considerable regulatory effect on the expression of approximately half of MtGs, during early development.
To further analyze the functional consequences of de novo methylation on mitochondrial modifications during early development, GO and KEGG analyses were performed for MtGs that were presumably regulated by increased promoter DNA methylation (indicated by green dots in Fig. 5A). The results showed that these MtGs were significantly associated with most mitochondrial functions taking place during both transitions. Amongst others, these included ‘Oxoacid metabolic process’, ‘Cellular amino acid metabolic process’, ‘Electron transport chain’, ‘Mitochondrial transport’, and ‘OXPHOS’ (Fig. 5C and D). It should be noted that although the number of enriched MtGs was very low (2–7), the levels of significance were very high, which implied that MtG promoter de novo methylation may profoundly affect mitochondrial functions and dynamics during early development.
MtDNA methylation in postimplantation embryos
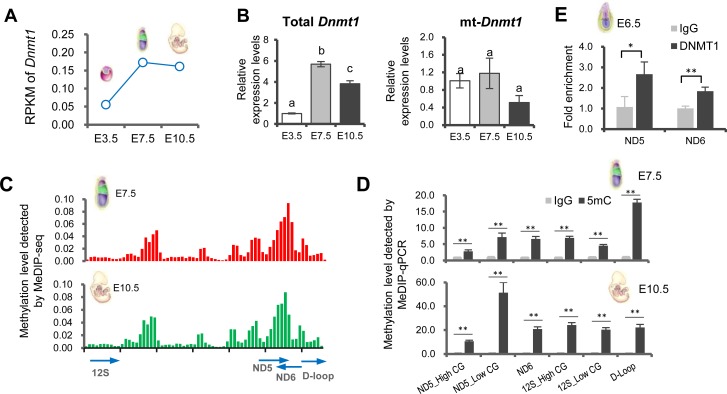
Dnmt1 expression was found to be significantly higher in postimplantation than preimplantation embryos (Fig. 6A). This observation was confirmed by qRT-PCR analysis. As Dnmt1 can also translocate to the mitochondria for maintaining mtDNA methylation [20], we further investigated the expression dynamics of a transcript variant of Dnmt1 specifically targeted to mitochondria (mt-Dnmt1), which is not fully synchronized with the levels of total Dnmt1 (Fig. 6B). Based on the relatively stable expression of mt-Dnmt1 up to the implantation stage, we hypothesized that embryonic mtDNA may undergo methylation after implantation. This idea was supported by the enrichment of MeDIP reads on mtDNA in E7.5 epiblasts and E10.5 embryos (Fig. 6C, Supplementary Table 8: online only). We validated this observation by performing MeDIP-qPCR with a highly sensitive and specific anti-5mC antibody. It should be mentioned that the primers used for detection were blasted against the mouse nuclear genome to prevent a possible contamination from nuclear sequences of mitochondrial origin.
Fig. 6.
MtDNA methylation in postimplantation embryos. (A) Expression dynamics of Dnmt1 during early development as assessed by RNA-seq. (B) Relative expression levels of total Dnmt1 and mt-Dnmt1 during early development as determined by qRT-PCR. (C) MtDNA methylation levels are presented as enriched MeDIP-seq reads (y-axis) on mtDNA positions (x-axis) in E7.5 epiblasts (upper plot) and E10.5 embryos (lower plot). (D) Relative methylation levels at selected regions of mtDNA as detected by MeDIP-qPCR in E7.5 epiblasts (upper plot) and E10.5 embryos (lower plot). (E) Relative enrichment of DNMT1 at selected regions (Nd5 and Nd6) of mtDNA as determined by CHIP-qPCR in E6.5 embryos. Values indicated by different letters are significantly different (P < 0.05). * P < 0.05; ** P < 0.01.
Focusing on the MeDIP-qPCR results from both hyper- (ND5, ND6) and hypomethylated (12S rRNA) regions, as well as a control region (D-loop), we observed that each of these regions was significantly methylated, with an increasing trend during the transition from E7.5 to E10.5 (Fig. 6D). Moreover, to confirm that mt-Dnmt1 can translocate to the mitochondria by the implantation stage, we tested the binding of mt-Dnmt1 to mtDNA by CHIP-qPCR using a non-specific anti-DNMT1 antibody and primers specific to mtDNA in E6.5 embryos, which represent a stage immediately after implantation. Results showed a significant enrichment of DNMT1 at two selected hypermethylated regions (ND5 and ND6) of mtDNA (Fig. 6E).
Discussion
It is well documented that mitochondrial functions and dynamics are of prime importance to early development. Well-organized mitochondrial modifications are thought to be essential for embryonic survival and growth during implantation, as well as for subsequent differentiation and organogenesis. Focusing on MtGs, this study provided a comprehensive and detailed understanding of the mechanisms responsible for the orchestrated mitochondrial modifications taking place during early development.
During the transition from E3.5 to E7.5, embryos undergo the differentiation of embryonic germ layers and implantation, two of the most important challenges that enable further embryonic survival and growth. On the other hand, in the period between E7.5 and E10.5, embryos initiate organogenesis following the completion of gastrulation [21]. During the transition from the blastocyst to the gastrulation stage, embryos undergo a shift in energy metabolism, in which glycolytic activity gradually decreases but OXPHOS is rapidly enhanced. Coincident with this shift is the resumption of mitochondrial biogenesis and mtDNA replication. Increased mitochondrial biogenesis and mtDNA replication in somatic cells are thought to accommodate high energy demands during physiological and pathological processes [22, 23]. In our study, we found that the expression of many key MtGs involved in mitochondrial biogenesis and mtDNA replication significantly increases during implantation. Among these, some are well-established MtGs considered responsible for the resumption of mitochondrial biogenesis in early embryos, such as Polg, Polg2, and Tfam [24,25,26]. Other MtGs that have been previously reported to play important roles in somatic cells may also be critical for this resumption. For example, Ssbp1 encodes a subunit of a single-stranded DNA-binding complex that can maintain the stability of mtDNA by removing mismatches during mtDNA replication [27] and support mitochondrial protein synthesis and morphology in human osteosarcoma and embryonic kidney cells [28, 29]. Stoml2, also known as Slp2, codes for a widely expressed mitochondrial inner membrane protein and can induce mitochondrial biogenesis in human T lymphocytes [30]. Tk2 codes for a deoxyribonucleoside kinase that localizes to the mitochondria and is required for the replication and maintenance of mtDNA in cultured human somatic cells [31]. A recent study demonstrated that Tk2 may be involved in the early development of zebrafish by modulating mtDNA metabolism [32]; however, a role for this gene in the resumption of mtDNA replication in mammalian embryos has not yet been identified.
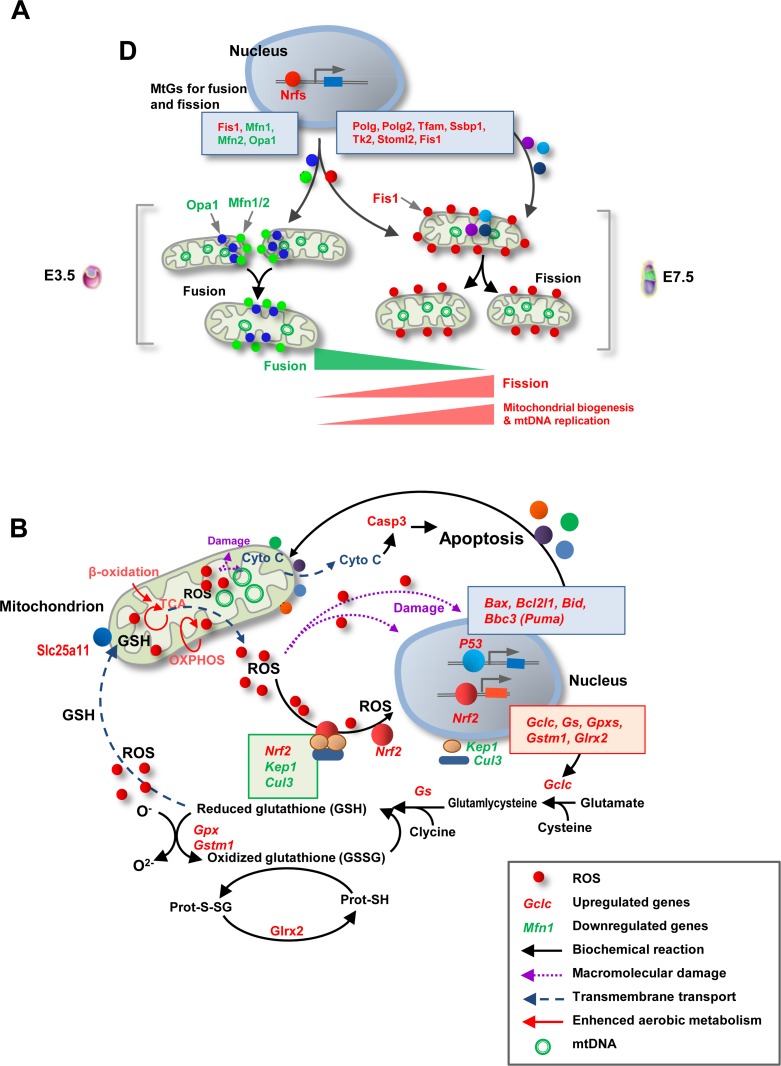
Mitochondria are dynamic organelles that are constantly undergoing fission and fusion to maintain mitochondrial homeostasis and morphology [33,34,35,36]. An interesting observation of this study was the increased level of Fis1, which is known to enable mitochondrial fission, as well as the decreased levels of Mfn1/2 and Opa1, which facilitate mitochondrial fusion. Although the role of mitochondrial fission and fusion during the period up to the implantation stage has not been well-defined, mouse embryonic fibroblasts (MEFs), as well as ES and TS cells from mouse knockout models displaying deficient fission and fusion, showed evident aberrations in mitochondrial mass and morphology (elongated or fragmented mitochondria) [37,38,39]. Thus, we predict that the resumption of mitochondrial growth by the implantation stage is not only facilitated by increased mitochondrial biogenesis and mtDNA replication, but also by the appropriate balance between fission and fusion (Fig. 7A). In addition, deficient fission and fusion result in disrupted mitochondrial functions, distribution, and apoptosis, with deficient embryos dying in midgestation [37,38,39]. These facts imply that fission and fusion dynamics may also be involved in many aspects of mitochondrial modifications during this critical period.
Fig. 7.
Schematic diagrams illustrating essential mitochondrial modifications during early development. (A) The resumption of mitochondrial biogenesis by the implantation stage appears to be facilitated not only by increased mitochondrial biogenesis and mtDNA replication but also by the skewed balance between fission and fusion. (B) Enhanced aerobic metabolism by the implantation stage, especially OXPHOS, leads to increased ROS production, which appears be the inducer of GSH biosynthesis. The apoptotic and survival signals in early embryos may be balanced by the GSH-based stress response.
Enhanced mitochondrial functions not only provide more ATP but also produce more ROS. Energy metabolism in mitochondria, including TCA, β-oxidation, and (especially) OXPHOS, is the major source of ROS. Our results regarding the GSH/GSSG ratio suggested that embryos face increased oxidative stress soon after implantation. Mitochondria are both a major source and a target of ROS [40]. Mitochondrial glutathione (mGSH), which is synthesized in the cytosol and then imported into the mitochondrial matrix, is thought to be the key survival antioxidant for maintaining the appropriate mitochondrial redox environment [41]. Interestingly, the temporal pattern of GSH biosynthesis is similar to the dynamics of the resumption of mitochondrial biogenesis by the implantation stage [42]. Thus, increased GSH biosynthesis may be a physiological response to enhanced mitochondrial functions and the consequent increase in ROS levels. Indeed, we observed an evident activation of many key MtGs involved in GSH biosynthesis and metabolism. The expression of Gclc and Gs (Glul), which code for the enzymes that catalyze the first (rate-limiting) and second steps of glutathione biosynthesis, increased during implantation. We also found that Nrf1 and Nrf2 were upregulated; these well-known transcription factors can activate GSH-based antioxidative genes, including Gclc, Gss, Glrxs, and Gpxs [43, 44]. Consistently, Keap1 and Cul3, which code for proteins that inactivate Nrf2 by forming a KAP1/CUL3/NRF2 complex in the cytosol, were downregulated during the transition from E3.5 blastocysts to E7.5 epiblasts. Previous studies in normal somatic cells and cancer cells indicated that the inhibition of either Keap1 or Cul3 increases the nuclear accumulation of Nrf2, leading to the activation of Nrf2-dependent antioxidative gene expression [45, 46]. Although the role of ROS in uncoupling NRF2 from the KAP1/CUL3 complex and thereby upregulating the transcription of GSH-based antioxidative genes has been well documented in many somatic cell types [47], the involvement of ROS in the Nrf2-induced activation of the GSH system has never been demonstrated in early embryogenesis. Our data suggest that the enhancement of mitochondrial functions in embryos by the implantation stage may increase the production of cellular ROS, activating the expression of the GSH-based antioxidative system via the Nrf2-Keap1 signaling pathway to protect mitochondria against oxidative stress (Fig. 7B). The transition from E3.5 blastocysts to E7.5 epiblasts was also characterized by a marked increase in the expression of many Gpxs, which are the genes coding for proteins that catalyze the reduction of hydrogen peroxide and lipid peroxides by GSH. Among these, Gpx3 and Gpx4 are well-known mitochondrial Gpx genes that are essential for oxidative homeostasis in mitochondria [48, 49].
We also found many apoptotic genes undergoing significant changes during early development. TUNEL analysis showed that E7.5 epiblasts displayed a high rate of apoptosis compared to E3.5 blastocysts, consistently to previously reported observations in hESCs undergoing differentiation [50]. Compared with MtGs associated with ‘Mitochondrial biogenesis and mtDNA replication’ and ‘GSH-based antioxidative response’, the gene expression dynamics of ‘Apoptotic mitochondrial change’ showed a complicated pattern. Indeed, properly controlled mitochondrial apoptotic signaling is not harmful to early embryos; in contrast, it is considered significant for the survival and differentiation of early embryos [50, 51]. A PPI analysis showed that the term ‘Apoptotic mitochondrial change’ was highly associated with ‘GSH-based antioxidative response’ and ‘Mitochondrial biogenesis’. Similarly, REVIGO analysis showed that ‘Apoptotic mitochondrial change’ interacted significantly with ‘Oxoacid metabolism’. These observations imply that the apoptotic and survival signals may be balanced by the GSH-based stress response (Fig. 7B).
Our data suggest that, in addition to successful implantation, mitochondrial functions or dynamics may be also involved in early organogenesis. Using the MGI database, we found that many of the MtGs whose expression was significantly altered (P < 0.05, FC > 2) were associated with embryonic death or aberrant development. At E7.5, when embryos are in an advanced stage of the gastrulation process and have a formed neural plate, many of these MtGs were annotated with phenotypes such as ‘Decreased neuron number’, ‘Abnormal neural crest cell apoptosis’, and ‘Abnormal neural tube morphology/development’. From E7.5 to E10.5, the neural plate folds to form the neural tube and the brain, while structures and organs such as somites, the heart, and the limb buds start developing. MtGs at this stage were annotated with phenotypes that included ‘Abnormal nervous system morphology’, ‘Abnormal eye development’, ‘Abnormal heart morphology’, and ‘Abnormal limb morphology’. It is noteworthy that the MAPK and Wnt signaling pathways were tightly clustered with MtGs that are responsible for mitochondrial functions. Previous studies in mouse and human ES cells indicated that mitochondrial apoptotic signals might contribute to differentiation programs, probably via the MAPK and Wnt signaling pathways [50, 51].
Our next step was to investigate whether the well-orchestrated mitochondrial functions and dynamics taking place by the implantation stage are regulated by epigenetic modifications. Coinciding with the dramatic mitochondrial changes during early development, embryos undergo de novo DNA methylation. Aberrant de novo DNA methylation caused by deleting or mutating Dnmts in mice or ES cells results in embryonic lethality and failed differentiation [52, 53]. Thus, we decided to investigate the possible functional association of these two early developmental events. Our data showed that most MtGs (clusters 1 and 2) underwent de novo methylation in their promoter regions during early development. These observations were very similar to those reported by Borgel et al. [19] and Smith et al. [54], who demonstrated that embryos undergo global promoter de novo methylation in the period between the preimplantation and the postimplantation stages. In our study, the integrated analysis of MtGs that underwent promoter de novo methylation revealed that approximately half of them displayed reduced expression levels. Many of these MtGs are significantly associated with mitochondrial functions and diseases. These results demonstrate that de novo DNA methylation, one of the most important epigenetic events during mammalian embryogenesis, may have considerable functional consequences for the mitochondrial modifications that are essential for embryonic survival and growth.
Another finding was that the mtDNA of both E7.5 and E10.5 embryos showed significant enrichment of MeDIP reads. This novel finding suggests that mtDNA methylation, which has been reported in many somatic cell types [20, 55, 56], can be observed as early as immediately after implantation. We also showed that this enrichment in early embryos was not limited to regions that coded for enzymes essential for OXPHOS (ND5, ND6), but also in noncoding regions (12S rRNA) and a control region (D-loop). In addition, in our study mtDNA methylation appeared to be independent of CpG density, as it was detected in both high- and low-CpG regions. This result is consistent with the findings of Bellizzi et al., who reported that both CpG and non-CpG regions of mtDNA can be methylated in human blood and cultured cells [56]. To the best of our knowledge, this is the first study to show that mtDNA becomes methylated in early embryos and that mt-DNMT1 can translocate into the mitochondria matrix and bind to mtDNA for maintaining mtDNA methylation by the implantation stage. Indeed, our unpublished data indicate that mtDNA undergoes a process similar to the de novo DNA methylation of the nuclear genome, thereby demonstrating that highly dynamic DNA methylation modifications are critical not only for the nuclear genome but also for the mitochondrial genome.
Overall, our study provided high-resolution dynamic expression profiles of MtGs during early embryogenesis. Our data indicate that orchestrated MtG expression is critical for the ‘embryonic shift’ in energy metabolism and the resumption of mitochondrial biogenesis, which is of prime importance for successful embryonic implantation and survival. In addition, the mitochondria-mediated balance between apoptosis and the stress response plays a significant role in the differentiation and organogenesis of postimplantation embryos. Our data also suggest that promoter de novo methylation of nDNA-encoded MtGs may have considerable functional consequences for mitochondrial modifications. Finally, mtDNA methylation in somatic cells can be observed as early as immediately after implantation. Our study can serve as a comprehensive reference for the further study of mitochondrial modulations during early mammalian embryogenesis and provides a new insight into the epigenetic mechanisms modulating mitochondrial functions and dynamics at this stage.
Supplementary
Acknowledgments
This work was supported by grants from the National High-Tech R&D Program (2013AA102506) and the National Natural Science Foundation of China (No. 4933167246 and 31472092).
References:
- 1.Harvey A, Gibson T, Lonergan T, Brenner C. Dynamic regulation of mitochondrial function in preimplantation embryos and embryonic stem cells. Mitochondrion 2011; 11: 829–838. [DOI] [PubMed] [Google Scholar]
- 2.Ramalho-Santos J, Varum S, Amaral S, Mota PC, Sousa AP, Amaral A. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum Reprod Update 2009; 15: 553–572. [DOI] [PubMed] [Google Scholar]
- 3.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003; 115: 629–640. [DOI] [PubMed] [Google Scholar]
- 4.Jian B, Yang S, Chen D, Zou L, Chatham JC, Chaudry I, Raju R. Aging influences cardiac mitochondrial gene expression and cardiovascular function following hemorrhage injury. Mol Med 2011; 17: 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiao CP, Wang D, Kaushal A, Saligan L. Mitochondria-related gene expression changes are associated with fatigue in patients with nonmetastatic prostate cancer receiving external beam radiation therapy. Cancer Nurs 2013; 36: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren L, Wang Z, An L, Zhang Z, Tan K, Miao K, Tao L, Cheng L, Zhang Z, Yang M, Wu Z, Tian J. Dynamic comparisons of high-resolution expression profiles highlighting mitochondria-related genes between in vivo and in vitro fertilized early mouse embryos. Hum Reprod 2015; 30: 2892–2911. [DOI] [PubMed] [Google Scholar]
- 7.Takasugi M, Yagi S, Hirabayashi K, Shiota K. DNA methylation status of nuclear-encoded mitochondrial genes underlies the tissue-dependent mitochondrial functions. BMC Genomics 2010; 11: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly RD, Mahmud A, McKenzie M, Trounce IA, St John JC. Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res 2012; 40: 10124–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashemi M, Rezaei H, Kaykhaei MA, Taheri M. Association of the Promoter Methylation of Mitochondrial Transcription Factor A With Susceptibility to Metabolic Syndrome. Gene Cell Tissue 2014; 1: e18308. [Google Scholar]
- 10.Gemma C, Sookoian S, Dieuzeide G, García SI, Gianotti TF, González CD, Pirola CJ. Methylation of TFAM gene promoter in peripheral white blood cells is associated with insulin resistance in adolescents. Mol Genet Metab 2010; 100: 83–87. [DOI] [PubMed] [Google Scholar]
- 11.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 2009; 10: 189–198. [DOI] [PubMed] [Google Scholar]
- 12.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development 1993; 118: 1255–1266. [DOI] [PubMed] [Google Scholar]
- 13.Sui L, An L, Tan K, Wang Z, Wang S, Miao K, Ren L, Tao L, He S, Yu Y, Nie J, Liu Q, Xing L, Wu Z, Hou Z, Tian J. Dynamic proteomic profiles of in vivo- and in vitro-produced mouse postimplantation extraembryonic tissues and placentas. Biol Reprod 2014; 91: 155. [DOI] [PubMed] [Google Scholar]
- 14.Giritharan G, Talbi S, Donjacour A, Di Sebastiano F, Dobson AT, Rinaudo PF. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction 2007; 134: 63–72. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Khatib H. Comparison of transcriptomic landscapes of bovine embryos using RNA-Seq. BMC Genomics 2010; 11: 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kepkova KV, Vodicka P, Toralova T, Lopatarova M, Cech S, Dolezel R, Havlicek V, Besenfelder U, Kuzmany A, Sirard MA, Laurincik J, Kanka J. Transcriptomic analysis of in vivo and in vitro produced bovine embryos revealed a developmental change in cullin 1 expression during maternal-to-embryonic transition. Theriogenology 2011; 75: 1582–1595. [DOI] [PubMed] [Google Scholar]
- 17.Nie J, An L, Miao K, Hou Z, Yu Y, Tan K, Sui L, He S, Liu Q, Lei X, Wu Z, Tian J. Comparative analysis of dynamic proteomic profiles between in vivo and in vitro produced mouse embryos during postimplantation period. J Proteome Res 2013; 12: 3843–3856. [DOI] [PubMed] [Google Scholar]
- 18.Zitka O, Skalickova S, Gumulec J, Masarik M, Adam V, Hubalek J, Trnkova L, Kruseova J, Eckschlager T, Kizek R. Redox status expressed as GSH:GSSG ratio as a marker for oxidative stress in paediatric tumour patients. Oncol Lett 2012; 4: 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 2010; 42: 1093–1100. [DOI] [PubMed] [Google Scholar]
- 20.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA 2011; 108: 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng X, Oliver G. Elucidating the molecular characteristics of organogenesis in human embryos. Genome Biol 2010; 11: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem 2010; 47: 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol 2005; 37: 822–834. [DOI] [PubMed] [Google Scholar]
- 24.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet 2005; 14: 1775–1783. [DOI] [PubMed] [Google Scholar]
- 25.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 1998; 18: 231–236. [DOI] [PubMed] [Google Scholar]
- 26.Humble MM, Young MJ, Foley JF, Pandiri AR, Travlos GS, Copeland WC. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet 2013; 22: 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazak L, Reyes A, Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol 2012; 13: 659–671. [DOI] [PubMed] [Google Scholar]
- 28.He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, Walker JE, Holt IJ. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res 2012; 40: 6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arakaki N, Nishihama T, Kohda A, Owaki H, Kuramoto Y, Abe R, Kita T, Suenaga M, Himeda T, Kuwajima M, Shibata H, Higuti T. Regulation of mitochondrial morphology and cell survival by Mitogenin I and mitochondrial single-stranded DNA binding protein. Biochim Biophys Acta 2006; 1760: 1364–1372. [DOI] [PubMed] [Google Scholar]
- 30.Christie DA, Lemke CD, Elias IM, Chau LA, Kirchhof MG, Li B, Ball EH, Dunn SD, Hatch GM, Madrenas J. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol Cell Biol 2011; 31: 3845–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rampazzo C, Fabris S, Franzolin E, Crovatto K, Frangini M, Bianchi V. Mitochondrial thymidine kinase and the enzymatic network regulating thymidine triphosphate pools in cultured human cells. J Biol Chem 2007; 282: 34758–34769. [DOI] [PubMed] [Google Scholar]
- 32.Artuso L, Romano A, Verri T, Domenichini A, Argenton F, Santorelli FM, Petruzzella V. Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim Biophys Acta 2012; 1817: 1002–1011. [DOI] [PubMed] [Google Scholar]
- 33.Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr Opin Cell Biol 2003; 15: 706–716. [DOI] [PubMed] [Google Scholar]
- 34.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci 2001; 114: 867–874. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Liu Y, Dorn GW., 2nd. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res 2011; 109: 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 2006; 25: 2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 2003; 160: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 2005; 280: 26185–26192. [DOI] [PubMed] [Google Scholar]
- 39.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 2009; 11: 958–966. [DOI] [PubMed] [Google Scholar]
- 40.Reinecke F, Smeitink JA, van der Westhuizen FH. OXPHOS gene expression and control in mitochondrial disorders. Biochim Biophys Acta 2009; 1792: 1113–1121. [DOI] [PubMed] [Google Scholar]
- 41.Marí M, Morales A, Colell A, García-Ruiz C, Fernández-Checa JC. Mitochondrial glutathione, a key survival antioxidant. Antioxid Redox Signal 2009; 11: 2685–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen JM, Harris C. Glutathione during embryonic development. Biochim Biophys Acta 2015; 1850: 1527–1542. [DOI] [PubMed] [Google Scholar]
- 43.Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1. Mol Cell Biol 2005; 25: 5933–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chepelev NL, Zhang H, Liu H, McBride S, Seal AJ, Morgan TE, Finch CE, Willmore WG, Davies KJ, Forman HJ. Competition of nuclear factor-erythroid 2 factors related transcription factor isoforms, Nrf1 and Nrf2, in antioxidant enzyme induction. Redox Biol 2013; 1: 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 2004; 24: 8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 2008; 373: 151–154. [DOI] [PubMed] [Google Scholar]
- 47.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 2014; 39: 199–218. [DOI] [PubMed] [Google Scholar]
- 48.Passaia G, Fonini LS, Caverzan A, Jardim-Messeder D, Christoff AP, Gaeta ML, de Araujo Mariath JE, Margis R, Margis-Pinheiro M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci 2013; 208: 93–101. [DOI] [PubMed] [Google Scholar]
- 49.Savaskan NE, Ufer C, Kühn H, Borchert A. Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol Chem 2007; 388: 1007–1017. [DOI] [PubMed] [Google Scholar]
- 50.Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem 2007; 282: 5842–5852. [DOI] [PubMed] [Google Scholar]
- 51.Duval D, Trouillas M, Thibault C, Dembelé D, Diemunsch F, Reinhardt B, Mertz AL, Dierich A, Boeuf H. Apoptosis and differentiation commitment: novel insights revealed by gene profiling studies in mouse embryonic stem cells. Cell Death Differ 2006; 13: 564–575. [DOI] [PubMed] [Google Scholar]
- 52.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257. [DOI] [PubMed] [Google Scholar]
- 53.Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 2015; 47: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012; 484: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging 2012; 33: 2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bellizzi D, DAquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res 2013; 20: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.