Abstract
The methylation status of sperm DNA differs between individual bulls. However, the relationship between methylation status and bull sperm parameters is not well elucidated. The present study investigated genome-wide methylation profiles at 450,000 CpG sites in bull spermatozoa by using a human DNA methylation microarray. Semen samples from three adult Japanese Black bulls with different in vitro fertilization (IVF) results and from a young Holstein bull through sexual maturation (at ages 10, 10.5, 15, and 25 months) were used for the analysis. The heatmap displaying the results of microarray analysis shows inter- and intra-individual differences in methylation profiles. After setting a cut-off of 0.2 for differences between ages (10, 10.5 vs. 15, 25 months) or between IVF results (developed to the blastocyst-stage, > 20% vs. < 10%), different methylation levels were detected at approximately 100 CpGs. We confirmed the different DNA methylation levels of CpG sites by using combined bisulfite restriction analysis (COBRA); five of the CpG sites reflected methylation levels similar to those detected by the microarray. One of the CpG sites was thought to reflect an age-related increase in methylation levels, which was confirmed by COBRA and bisulfite sequencing. However, the relationship between methylation status and IVF results could not be shown here. In conclusion, methylation profiles of individual and age-related alterations in bull spermatozoa can be revealed using a human microarray, and methylation changes in some CpG sites can be easily visualized using COBRA. Combined analysis of DNA methylation levels and sperm parameters could be considered an effective approach for assessing bull fertility in the future.
Keywords: Bull sperm, Combined bisulfite restriction analysis (COBRA), DNA methylation, Human microarray, Sexual maturation
Evaluation of the semen obtained from breeding bulls relies on the examination of various parameters, such as ejaculate volume, sperm motility, sperm count, post-thaw motility, and sperm-membrane integrity-related tests. Although these parameters indicate the semen-producing ability of bulls and the suitability of their semen for cryopreservation, they often do not explain the underlying causes of differing fertility in bulls. It has been suggested that different features of male infertility could be related to epigenetic mechanisms occurring at different stages of spermatogenesis. Methylation in germ cells is a unique process and is necessary for proper spermatogenesis and sperm production [1]. Extensive modifications to sperm chromatin are a result of the removal of histones during spermatogenesis and their replacement with protamines, chemical modifications observed in retained histones, and methylation of the sperm DNA bound to histones [1]. The DNA methylation pattern in male germ cells does not necessarily reflect the gene expression pattern in a specific cell type, but might be involved in the germ cell-specific chromatin organization. Aberrant DNA methylation patterns have been reported to correlate with abnormal semen parameters, idiopathic male infertility, and even pregnancy failure in humans [2,3,4,5]. Sperm epigenetics may contribute to the understanding of paternal effects on embryogenesis [6].
Several methods, including microarray analysis or sequencing following methylated DNA immunoprecipitation, reduced representation bisulfite sequencing, or shotgun bisulfite sequencing, have been used to characterize the genome-wide distribution of DNA methylation [7]. Infinium Human Methylation (HM) BeadChip (Illumina, San Diego, CA, USA) is widely used for measuring genome-scale DNA methylation in humans, particularly related to epigenome-wide association studies (EWAS) [8,9,10]. However, in case of most organisms, such as mice, pigs, and cattle, equivalent tools for studying DNA methylation at a genome-scale are currently unavailable for commercial use. However, Infinium HM BeadChip arrays can be utilized for methylation profiling in non-human species. It was found that analyzing DNA methylation in the mouse genome using Infinium HM27 and HM450 platforms [11] is feasible. Recently, Kobayashi and Takeda determined the utility of Infinium HM450 BeadChips for analyzing bovine genomic DNA, using the benchmark that useful methylation data with good quality are expected to analyze approximately 50,000 CpG sites (about 10% of all probes) [12]. These results suggest that the technique can advance genome-wide screening of differentially methylated regions (DMRs) in bovine spermatozoa.
The present study aimed to investigate the methylation status of bovine sperm DNA and determine its DMRs by using Infinium HM450 platforms, and to develop a simple method for detecting differences in methylation among semen samples under different conditions by using combined bisulfite restriction analysis (COBRA).
Materials and Methods
All chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise indicated.
Semen
Fresh semen was obtained from two adult Japanese Black bulls (JA and JB) and a young Holstein bull, which were reared at the Institute of Livestock and Grassland Science, NARO (NILGS). To analyze the alteration in semen quality after puberty, semen samples were collected 1–3 times per month from a young Holstein bull through sexual maturation, from the age of 10 months until the age of 2 years (H1, 10 months old; H2, 10.5 months old; H3, 15 months old; and H4: 25 months old). Semen quality was evaluated microscopically to determine semen volume, sperm count, and motility. A 20-µl sample of fresh semen was immediately used in DNA damage and mitochondrial membrane potential assays. The remaining semen was diluted in extender (composition: 20% egg yolk, 0.13 M Tris, 0.05 M citric acid, 0.04 M lactose, 0.04 M raffinose, 0.5 mg/ml gentamicin, and 600 IU/ml penicillin G potassium), and kept at 5°C overnight. Semen diluted with 7% glycerol was frozen and stored in liquid nitrogen until use. Animal experiments were approved by the Committee for the Care and Use of Experimental Animals at NILGS (No.14111041).
Cryopreserved semen obtained from another Japanese Black bull (JC) that had 0% fertility after artificial insemination (n = 30) was also used for the following analysis.
Assessment of semen quality
DNA damage assay was performed as described previously [13]. JA and JB had already been investigated in a previous report, under the labels Bull A and Bull B, respectively [13]. Semen was diluted in Dulbecco’s phosphate-buffered saline without calcium chloride or magnesium chloride (DPBS-) to 10 or 20 × 106 spermatozoa/ml for all experiments. Sperm smears were prepared on a slide using 15–20 µl of the dilution; slides were then air dried and fixed with 2% (w/v) paraformaldehyde in DPBS- for 30 min. The level of DNA fragmentation was determined by TUNEL assay using a kit (In Situ Cell Death Detection Kit, Fluorescein, Roche, Indianapolis, IN, USA), whereby the free 3′-OH ends of DNA were labeled with fluorescein conjugated dUTP using the enzyme terminal deoxynucleotidyl transferase.
Mitochondrial membrane potential was detected using MitoTracker® Red CMXRos (MTR; Molecular Probes, Thermo Fisher Science, Eugene, OR, USA), which passively diffuses across the plasma membrane and accumulates in active mitochondria. Working concentrations of 500 nM MTR and 1 µg/ml Hoechst® 33342 (Molecular Probes) were used for staining mitochondria and nuclei in spermatozoa. Fluorescence intensity was measured using a fluorescence multi-well plate reader (Cytofluor 4000, Applied Biosystems, Foster City, CA, USA) at excitation wavelengths (nm) of 590/20 and emission wavelengths of 645/40, and excitation wavelengths of 360/40 and emission wavelengths of 460/40, respectively. The fluorescence intensity of MTR was divided by that of Hoechst, and then used to compare the mitochondrial potential among samples.
In vitro fertilization (IVF) tests were performed as previously described [14, 15]. Briefly, cumulus-oocyte complexes (COCs) were collected from slaughterhouse-derived ovaries after overnight storage in physiological saline at 15°C. Mature oocytes were collected after 24-h incubation in 199 medium (Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum at 38.5°C under 5% CO2 in air. COCs were transferred into BO medium containing 20 mg/ml crystallized BSA and 10 IU/ml Novo Heparin (Mochida Pharmaceutical, Tokyo, Japan). Each cryopreserved semen sample was thawed and washed twice in 10 ml BO medium supplemented with 10 mM caffeine sodium benzonate. Sperm suspensions were transferred to a droplet of fertilization medium (final concentration: 5 × 106 spermatozoa/ml), co-incubated for 5 h at 38.5°C under 5% CO2 in air with maximum humidity. After IVF, the zygotes were cultured in IVD 101 medium (Research Institute for Functional Peptides, Yamagata, Japan) for 7 days; IVF results were determined by counting the number of embryos at the blastocyst stage per number of embryos cultured after IVF.
Statistical analyses for sperm motility, and DNA damage by TUNEL between JA and JB were performed using Student’s t-test. IVF results among JA, JB, and JC were analyzed by one-way ANOVA. A P-value of 0.05 was considered to be significant.
DNA methylation analysis using Infinium HM450 BeadChip
Thawed semen was washed twice in DPBS(–). Genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions, with modifications; briefly, 8 µl of 2 M DTT was added to each sample before it was heated to 56°C to dissolve the spermatozoa effectively.
The Infinium HM450 BeadChip was used for genome-wide bovine DNA methylation analysis in this study. The processing of DNA for the methylation arrays (which involved bisulfite conversion, whole-genome amplification, labeling, hybridization to an Infinium HM450 BeadChip, scanning on a BeadArray scanner, and raw data processing) was performed according to Illumina protocols at G&G Science (Fukushima, Japan). The BeadChip was imaged on an Illumina iScan, and then the images were processed with Illumina GenomeStudio software and Methylation module (v1.8; Illumina). Finally, the fluorescence signal intensities of the methylated and unmethylated alleles and the total signal intensity (sum of these fluorescent signal intensities) were obtained for data analysis [16]. Similarly, the detection P-values were calculated as a proportion of 600 negative control probes with higher signal intensities than those measured for each CpG site. Incidentally, the methylation level of each CpG locus was calculated as the methylation Beta-value (signal intensity of the methylated allele / (total signal intensity + 100)).
After the methylation array analysis, the informative CpG sites were selected as described in a previous report [12]. Briefly, the CpG sites were selected for quality by checking for where detection P-values = 0 and the total signal intensities > 1000. The different methylation levels of the CpG sites were listed to compare results among ages (< 11 months old vs. > 15 months old; H1 and H2 vs. H3 and H4), or between IVF results (> 20% vs. < 10%). Spermatozoa data were also compared with previously analyzed data from other organs (liver, muscle, and brain) obtained from two Japanese Black cows [12].
DNA methylation analysis by COBRA
The BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search the highly similar sequences using human probe sequences for each quality controlled CpG site on bovine genome sequences, as previously reported [12]. Then, the CpG sites located at the restriction enzyme sites (BstUI, TaqI, and AciI) were selected from the bovine genome sequences detected by the BLAST search. Bisulfite primers were designed to amplify the regions that flanked the bovine sequences of interest using MethPrimer (http://www.urogene.org/methprimer/index.html).
Genomic DNAs were bisulfite-converted using a MethylEasy Xceed Rapid DNA Bisulphite Modification Kit (Human Genetic Signatures Pty. Ltd., New South Wales, Australia) according to the manufacturer’s instructions. The PCR of bisulfite-modified DNA was performed with a GeneAmp PCR system 9700 (PE Applied Biosystems, Foster City, CA, USA), using a TaKaRa EpiTaqTM HS kit (TaKaRa Bio, Shiga, Japan) at the following conditions: 94°C for 2 min, followed by 35 cycles at 94°C for 30 sec, 50–54°C (depending on the primer sets, see Table 1) for 30 sec, and 72°C for 30 sec. PCR products were digested with the restriction enzymes shown in Table 1. The digested fragments were subjected to electrophoresis using 3% agarose gels. The ratio of band intensity of digested and undigested fractions reflects the levels of DNA methylation at the restriction sites. The methylation status of the spermatozoa in the semen samples was compared with that of spermatozoa in the testis (TS) and epididymis (ED) samples collected from a one-month-old Japanese Black calf, and those of the ovaries (OV) collected from a slaughterhouse.
Table 1. Primer sequences for COBRA.
| Forward and reverse primer sequences | Product length (bp) |
Annealing (°C) |
Restriction enzyme |
Bovine features of the sequences including the target CpG | Accession No., location | Target ID of array | |
| CpG1 | 5'GTTTTATTTTATTTTGTTTTTTTT3' | 181 | 50 | TaqaI | mitochondrial glutamate carrier 1 isoform | AC_000186.1, 50835888 | cg27100471 |
| 5'ACCATTCTACTAATTCTACAACCT3' | |||||||
| CpG2 | 5'GGGTTTTAGGTAGATTAATTAGGTT3' | 112 | 54 | AciI | ubiquitin-conjugating enzyme E2 D2, CXXC-type zinc finger protein 5 isoform | AC_000162.1, 52499000 | cg26301389 |
| 5'CTCTAAAACACAAACCCCAAAAC3' | |||||||
| CpG3 | 5'GGGATTTGGTTAATATGATAGGTTT3' | 294 | 54 | TaqaI | leucine-rich repeat and fibronectin typ-III domain-containing protein 3 precursor | AC_000175.1, 46821245 | cg15658249 |
| 5'CCTCCCAAAACAACTACTCCA3' | |||||||
| CpG4 | 5'TGTTATTTGTTGGTTAGAGGGTGTAT3' | 203 | 52 | BstUI | myosin-9 | AC_000162.1, 75099407 | cg01816775 |
| 5'ATAATAATACCAAACCTCTCCTCCC3' | |||||||
| CpG5 | 5'GTTGGTGGAGGATAGTAGGGTTA3' | 191 | 52 | BstUI | Transmembrane protein 125 | AC_000160.1, 103338929 | cg14088090 |
| 5'CCAAATCAACCATATCTAAAAAAAA3' |
DNA methylation analysis by bisulfite sequencing
The methylation status of one of the CpGs (CpG1) was confirmed by bisulfite sequencing. The PCR of bisulfite-modified DNA was performed using the forward primer 5′-TGGTTTGGAAATATTTTTGAAAGTAG-3′ and reverse primer 5′-AAAAACCAAAAATCAACCAAAATC-3′ at the following conditions: 94°C for 2 min, followed by 35 cycles at 94°C for 30 sec, 54°C for 30 sec, and 72°C for 45 sec. The PCR products (446 bp) were electrophoresed on 2% agarose gels and purified using an illustraTM GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare, Buckinghamshire, UK). The products were then cloned into the pMD18-T vector (TaKaRa Bio). The colony-PCR was performed with an EmeraldAmpR PCR system (TaKaRa Bio) using the universal primers (M13 Primer M4 and M13 Primer RV). The PCR products were then purified (FastGene Gel/PCR Extraction Kit, Nippon Genetics, Tokyo, Japan) and used for sequencing.
Verification of age-related DMRs detected by COBRA
To verify age-related DMRs including CpG1, we performed COBRA using cryopreserved semen samples obtained from three Japanese Black bulls at different ages (JD: 14 months, 19 months, 28 months, 54 months, and 162 months, JE: 15 months, 20 months, and 26 months, and JF: 14 months, 18 months, 21 months). To examine the methylation changes among samples, the intensity of DNA bands in each lane on 3% agarose gels was measured by densitometry using ImageJ 1.50i (http://imagej.nih.gov/ij; National Institutes of Health, Bethesda, MD).
Results
Semen quality
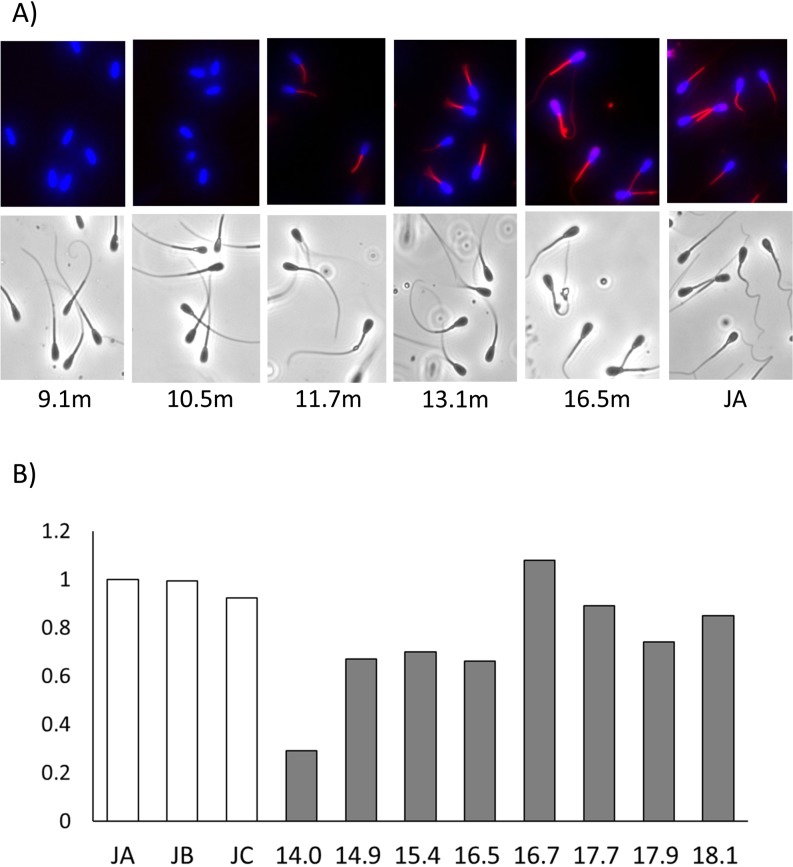
The quality of semen obtained from a young Holstein bull (H1–4) is shown in Table 2. Motility, sperm count, and IVF results improved during the process of sexual maturation, and at the age of 15 months, the parameters reached levels almost similar to those of adult bulls. Mitochondrial activity was not observed in spermatozoa immediately after puberty (9–10 months age), but, at the age of 16.7 months, it was almost equal to that observed in adult bulls (Fig. 1).
Table 2. Changes in semen quality after puberty in a young Holstein bull.
| Semen | Age (months) | Motility | Concentration (/ml) |
DNA damage by TUNEL * (%) |
IVF {%, (no. of embryos, r)} | |
| H1 | 10 | 3 | 1.1 × 108 | 0.2 | 0.0 | (0/95, 2) |
| H2 | 10.5 | 5 | 2.4 × 108 | 2.9 | 0.0 | (0/121, 2) |
| H3 | 15 | 80 | 5.2 × 108 | 2.1 | 29.5 | (51/173, 3) |
| H4 | 25 | 70 | 2.7 × 108 | - | 31.3 | (21/67, 2) |
No. of embryos: number of blastocyst-stage embryos for a certain number of oocytes cultured after in vitro fertilization. r: repeat number for IVF experiment. * Takeda et al., 2015 [13].
Fig. 1.
Mitochondrial membrane potential of spermatozoa after puberty in a young Holstein bull. (A) Active mitochondria (red) and nuclei (blue) in spermatozoa were observed in samples obtained from bulls aged from 9.1 to 16.5 months. (B) The mitochondrial membrane potential of spermatozoa was measured at age from 14.0 to 18.1 months. The MTR fluorescence intensity was divided by the Hoechst® 33342 fluorescence intensity and each value was calculated to determine the control value of JA as 1.0. JA–JC: Japanese Black bulls.
The quality of semen obtained from three Japanese Black bulls (JA, JB, and JC) is shown in Table 3. IVF results differed among these bulls (P < 0.05). The different methylation levels analyzed using human microarray were compared (H4 and JA vs. JB and JC); these results showed above 20% and below 10% methylation, respectively.
Table 3. Semen quality of three Japanese Black bulls.
| Bull | Age (months) | Motility | Concentration (/ml) |
DNA damage by TUNEL * (%) |
IVF {%, (no. of embryos, r)} | |
| JA | 159.1 | 73 a | 1.2 × 109 | 2.3 a* | 22.8 a | (34/149, 3) |
| JB | 75.3 | 45 b | 1.0 × 109 | 6.9 b* | 8.9 b | (24/284, 6) |
| JC | 19.9 | 95 | 0.4 × 109 | 6.9 | 2.0 c | (4/177, 4) |
No. of embryos: number of blastocyst-stage embryos for a certain number of oocytes cultured after in vitro fertilization. r: repeat number for IVF experiment. a, b, c The values with different superscripts within the same column are significantly different (P < 0.05). a*, b* Takeda, et al., 2015 [13].
Comparison of DNA methylation status among different semen samples by using Infinium HM450 BeadChip
According to DNA methylation status analysis of the bovine spermatozoa in the semen samples using Infinium HM450 BeadChips, the mean number of CpG sites in the examined samples with a detection P-value of 0 was 75,067 (15.5%; 43,315–91,703 sites). There was no difference in the distribution in each sample. In total, 40,171 CpG sites were associated with a detection P-value of 0 in all examined samples (8.3%). In total, 37,224 CpG sites fulfilled the second criterion; a mean total signal intensity greater than 1000 was observed for all examined samples (7.7%).
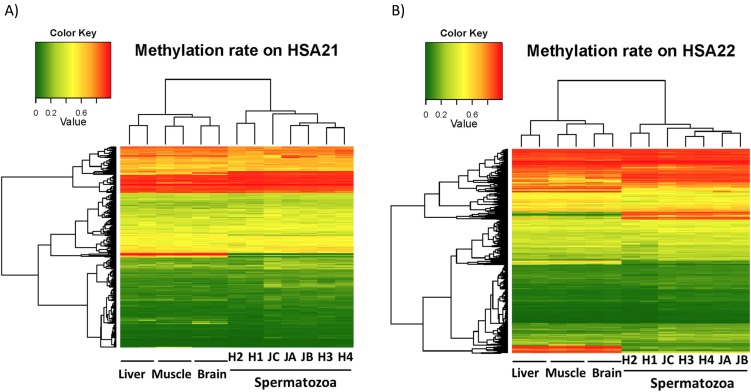
Unsupervised hierarchical clustering of samples using this subset of the probes suggests that distinct DNA methylation profiles of bovine spermatozoa can be developed using the HM450 BeadChip, compared to those of samples from three other tissues (brain, liver, and muscle) (Kobayashi & Takeda, in preparation). As shown in Fig. 2, for example, there are major differences between the methylation levels of these tissues and those of spermatozoa in the CpG sites on human chromosomes 21 and 22. The heatmap also showed inter- and intra-individual differences in methylation profiles among spermatozoa in the semen samples.
Fig. 2.
Heatmap displaying the methylation status of (A) CpG loci (n = 265) mapping on the human chromosome 21, and (B) CpG loci (n = 757) mapping on the human chromosome 22. The dendrograms above the heatmap show hierarchical clustering based on the methylation data. Methylation levels are represented in the scale on the upper side of the heatmap and are arranged from lowest to highest as green (0.0) to red (1.0).
In case of the 37,224 probes studied to analyze DNA methylation in bovine samples using the HM450 BeadChip, the standard deviation of the Beta-values of each probe among the semen samples was calculated (Supplementary Table 1: online only). After setting a cut-off of 0.2 for differences between ages (H1 and H2 vs. H3 and H4), it was found that 107 CpG sites demonstrated greater differences in Beta-values across the samples analyzed (Supplementary Table 2: online only). Comparison between IVF results (JA and H4 vs. JB and JC) revealed 94 CpG sites with different levels of methylation (Supplementary Table 3: online only).
Alteration in DNA methylation levels revealed by COBRA and bisulfite sequencing
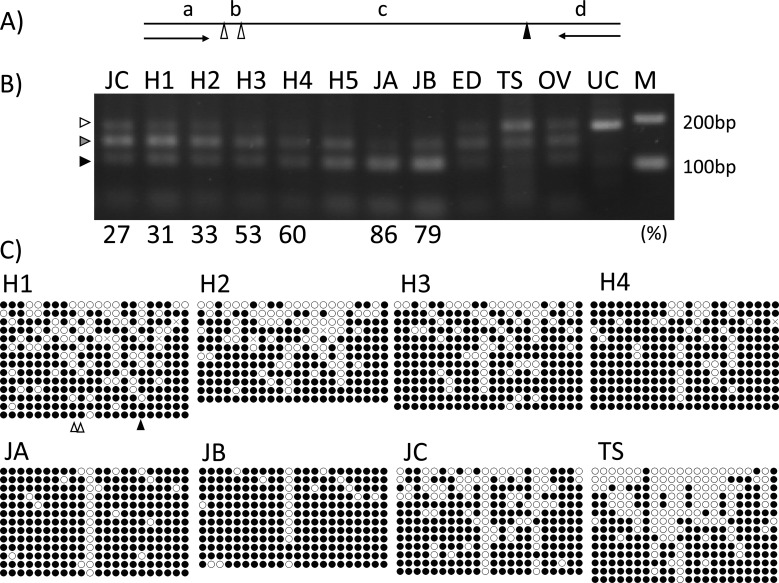
Comparison between samples obtained from bulls of different ages revealed 107 CpG sites with different methylation levels; furthermore, 35 matching bovine DNA sequences were picked up after BLAST analysis (32.7%). Twelve CpG sites, including restriction enzyme sites, were selected and primer sets were designed for ten of these. The changes in DNA methylation levels were further analyzed, and CpG1 (Table 1) was found to reflect an age-related change in methylation levels (Fig. 3, and Table 4). Unfortunately, the three methylated CpG sites, including CpG1, were digested by TaqI (Fig. 3A), such that only four DNA bands dependent on the methylation patterns could be visualized at 181 bp (0% methylation, indicated by a white arrow in Fig. 3B), 140–151 bp (indicated by a gray arrow in Fig. 3B), 101–112 bp (indicated by a black arrow in Fig. 3B), and at 11–41 bp on the gel. Bisulfite sequencing of the 446-bp fragment, including CpG1, revealed both age-related and individual differences (Fig. 3C).
Fig. 3.
Age-related and individual differences in methylation levels of CpG1 revealed by COBRA and bisulfite sequencing. (A) Positions of the restriction enzyme TaqI of the PCR product (181 bp) amplified by CpG1 primers (arrows) for COBRA. Three methylated CpG sites indicated by triangles, including CpG1 (indicated by a black triangle), were digested by TaqI. The DNA bands, 151 bp (b + c + d), 142 bp (a + b + c), 140 bp (c + d), 112 bp (b + c), 101 bp (c), 41 bp (a + b), 39 bp (d), 30 bp (a), and 11 bp (b) can be obtained by TaqI digestion. (B) The percentage indicated below the gels are the CpG1 methylation levels measured using a human microarray. H1–H4, JA, JB, JC: spermatozoa; H5: spermatozoa in the semen samples collected from the same Holstein bull at the age of 35 months; ED: epididymis; TS: testis; OV: ovary; UC: un-cut by restriction enzyme; M: 100-bp ladder DNA size marker. DNA bands dependent on the methylated patterns were visualized at 181 bp (0% methylated, indicated by a white arrow), 140–151 bp (indicated by a gray arrow), 101–112 bp (indicated by a black arrow), and a small band at 11–41 bp on the gel. (C) The data for each methylation level are shown in Table 4. The location of CpG1 is indicated by a black triangle and the other locations of restriction enzyme sites (TaqI) are indicated by white triangles below the panel H1.
Table 4. Methylation levels (%) of CpG1 and the CpG1 area in samples analyzed using different methods.
| Sample | H1 | H2 | H3 | H4 | H5 | JA | JB | JC | TS |
| CpG-Array | 31.3 | 33.4 | 53.1 | 59.6 | - | 86.1 | 78.5 | 26.6 | - |
| CpG-BiSeq | 52.4 | 25.0 | 38.5 | 46.2 | 68.4 | 84.6 | 91.7 | 15.4 | 50.0 |
| Area-BiSeq | 77.9 | 71.8 | 79.0 | 87.0 | 86.8 | 90.6 | 92.1 | 72.7 | 62.0 |
CpG-Array: The methylation level of the CpG1 analyzed using human microarray. CpG-BiSeq: The methylation level of the CpG1 analyzed using bisulfate sequencing. Area-BiSeq: The average methylation level of all 22 CpG sites including CpG1 in the area (446 bp) analyzed using bisulfate sequencing.
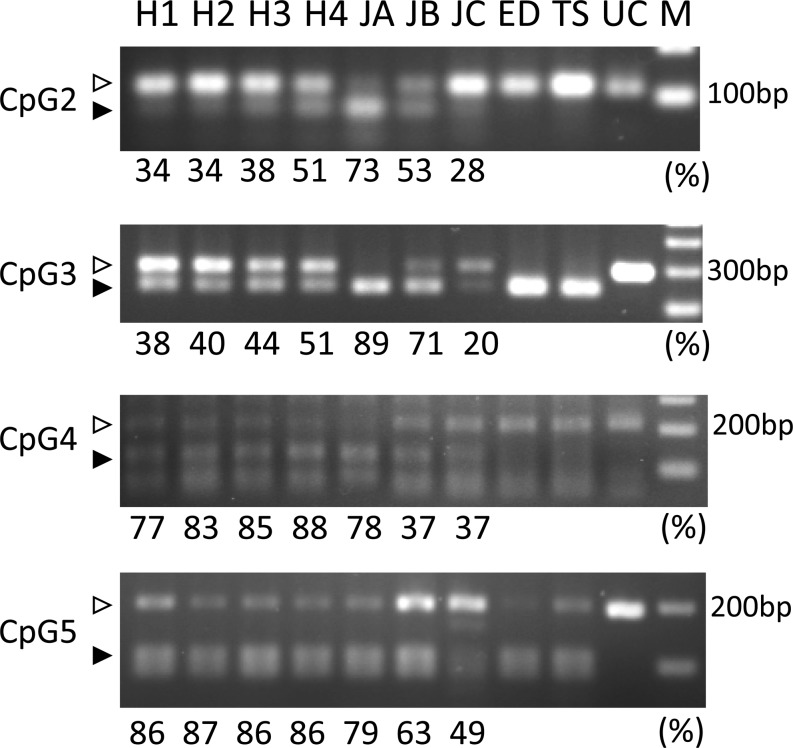
Comparison between IVF results revealed 94 CpG sites with different methylation levels; 63 matching bovine DNA sequences were picked up after BLAST analysis (67.0%). Nineteen CpG sites, including restriction enzyme sites, were selected and primer sets were designed for 16 of these. CpG1 was again selected as a differential methylation site. We performed COBRA for 12 CpG sites, and four sites (Table 1) reflected methylation levels similar to those observed in a human array (Fig. 4). However, from the results of COBRA of H1–H3, these sites were noted to be unrelated to those found in the IVF results (Table 2, Fig. 4).
Fig. 4.
Different methylation levels of the spermatozoa revealed by COBRA. The percentages indicated below the gels are the methylation levels measured using a human microarray. COBRA results reflected that CpG2–5 showed different methylation levels among samples analyzed by human microarray. H1–H4, JA, JB, JC: spermatozoa; ED: epididymis; TS: testis; UC: un-cut by restriction enzyme; M: 100-bp ladder DNA size marker. White arrows indicate hypomethylation, whereas black arrows indicate hypermethylation.
Verification of age-related DMRs detected by COBRA
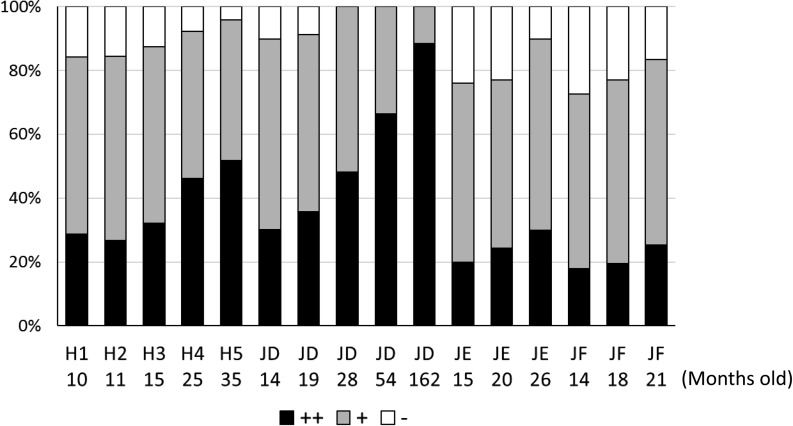
All analyzed bulls showed an age-related increasing trend in levels of the CpG1 DMR, as examined by COBRA (Fig. 5). One of the bulls (JD) showed an age-dependent increase until the age of 162 months (13.5 years).
Fig. 5.
Age-related increase in the levels of methylated CpG1, examined by COBRA, in spermatozoa obtained from a Holstein bull (H1–H5, Fig. 3) and three Japanese Black bulls (JD, JE, and JF). Each lane of the three DNA bands was measured by densitometry and is shown as percentages. ++: 101–112 bp (indicated by a black arrow in Fig. 3A), +: 140–151 bp (indicated by a gray arrow in Fig. 3A), –: 181 bp (indicated by a white arrow in Fig. 3A). Age (months) of bulls at the time of semen collection is indicated below each bull number.
Discussion
The current study was undertaken to explore the differences in genome-wide methylation at about 37,000 CpG sites in bull sperm DNA. The heatmap reflected the different methylation statuses between spermatozoa and organs (liver, muscle, brain) as revealed by a human microarray (Kobayashi & Takeda, in preparation). The heatmap displaying the results of the human DNA methylation microarray analysis showed inter- and intra-individual differences. Thus, to the best of our knowledge, this is the first study establishing a method for detecting the differentially methylated CpG sites/areas in bull spermatozoa by using the convenient and inexpensive COBRA method.
Recently, numerous studies have explored the relationship between DNA methylation levels and male fertility in humans [2,3,4,5, 8]. The sperm chromatin, owing to the replacement of approximately 85% DNA-binding histones with protamines during spermiogenesis, shows higher levels of DNA packaging and silent gene expression [1]. DNA methylation patterns are largely established in spermatogonia via both demethylation and de novo methylation events [9]. Epigenetic alterations can involve histone-to-protamine transition and histone removal and degradation, inducing protamine replacement errors [17]. From analysis using Infinium HM Human Methylation 450K arrays (Illumina), aberrant DNA methylation patterns were observed in the spermatozoa of infertile men [10, 18]. For farm animals, several studies also showed altered methylation in infertile buffalo bulls and boars [19, 20]. Verma et al. (2014) [20] used a custom-designed 180K buffalo (Bubalus bubalis)-specific CpG island/promoter microarray, and found differential methylation in 96 individual genes covered under CpG islands in the sperm of high-fertile and sub-fertile buffalo bulls (with conception rates in the ranges of 54–58% and 31–36%, respectively). They found that differentially methylated genes in high-fertile versus sub-fertile buffalo bull sperm have a role in sperm functions and embryogenesis. In the current study, the heatmap displaying the methylation status of bovine sperm DNA did not show the differences between the samples grouped by IVF results (Fig. 2). Approximately 100 CpG sites were detected to have differential methylation levels above 0.2 between groups, and they would be useful for detecting DMRs among samples. To confirm a correlation between DMRs and male fertility, more samples should be analyzed. We believe it will be useful to establish and analyze the differences in methylation among many bull semen samples using an easy and low cost methodology, such as COBRA.
The microarray was able to detect differences at about 200 CpG sites, and five of them could be clearly visualized by COBRA. Some of the CpG sites did not reflect their exact methylation levels after analysis with microarray. The selected areas, including each target CpG site, showed more than 80% homology between human and bovine sequences. However, it seems necessary to confirm the methylation levels of the target CpG sites by other methods. In the case of the CpG1 site, it was difficult to accurately demonstrate the methylation level by COBRA, because the PCR products were digested by three CpG sites, including CpG1. Thus, it was difficult to detect the small differences revealed among semen samples on agarose gel using COBRA. Bisulfite sequencing is more effective for detecting detailed differences in DNA methylation levels in several CpG sites located in DMRs. Nonetheless, the DMR that includes CpG1 will be a useful marker for detecting age-related alterations in methylation levels of cryopreserved bull sperm that can be verified by COBRA (Fig. 5). CpG1 is located at one of the CpG islands in the mitochondrial glutamate carrier 1 gene. The mitochondrial aspartate/glutamate carrier (Aralar/AGC1/Slc25a12) is a member of the mitochondrial carrier family, plays a central role in several important ubiquitous processes, and is linked to the regulatory role of Ca2+ in mitochondrial function [21, 22]. Indeed, an age-related increase in mitochondrial activity was observed in the spermatozoa of a young bull through sexual maturation (Fig. 1). However, it seemed there is no relationship between the methylation status of CpG islands, including CpG1 (CpG1 area), and mitochondrial activity among individual bulls (Fig. 1 and Fig. 3). There have been no reports on the role of the mitochondrial carrier in spermatozoa function to date, except one study which indicated that post-meiotic male germ cells might benefit from the high potential for mitochondrial pyruvate carrier (MPC) activity provided by the expression of the elevated MPC subunits (MPC1L and MPC2) [23]. It is interesting that CpG1 is detected as a differentially methylated CpG site when comparing results against both age and IVF status. CpG2 is located in the ubiquitin-conjugating enzyme E2 D2 gene, which plays an important role in initiating the DNA damage response [24]. TUNEL results could not reflect the methylation status of CpG2 (Table 2, Fig. 4). It is unknown whether the methylation status of CpG2 is related to sperm DNA damage. The function of DMRs (including CpG1–5) detected by a human microarray in spermatozoa remains unknown. Although there is not sufficient information for constructing a bovine epigenome yet and the process is still very expensive, whole-genome bisulfite sequencing using next generation sequencer will be beneficial for future methylome analysis.
Age-related differences were observed in a young bull throughout sexual maturation (Fig. 2), and the differences could be visualized with both COBRA and bisulfite sequencing (Fig. 3, Table 4). Not only CpG1, but also CpG2 and CpG3 showed age-related changes as revealed by COBRA. Semen quality showed remarkable age-related differences throughout sexual maturation (Fig. 1, and Table 2). The altered methylation status of these sites may be reflected in the altered DNA packaging status of spermatozoa during sexual maturation. The results of bisulfite sequencing of the CpG1 area reflected the variable methylation patterns of each individual spermatozoon (Fig. 3C, horizontal lines). DNA methylation changes can also accumulate with increasing paternal age in spermatozoa, as is the case for multiple tissues, including the gametes; exposure of male mice to toxins can also induce changes in the epigenome of germ cells [25]. Puberty is typically defined as the age at which reproductive function is initiated in animals. More specifically, puberty can be defined in bulls as the age at which the animal is first able to produce ejaculate containing 50 million sperms with a minimum of 10% motility [26]. Age at first freezable semen is defined as the age at which a bull first produces ejaculate containing at least 500 million sperms with > 50% progressive motility [27]. It is known that the semen obtained around puberty is not suitable for cryopreservation, as its motility after thawing is often poor. The reproductive development and performance of young bulls has gained attention as beef breeders attempt to accelerate improvement of economic traits in cattle and reduce costs by using younger sires. The age-dependent increase in these DMRs in spermatozoa samples may change throughout the bull’s lifetime (Fig. 5). Such age-related DMRs may be useful markers of sperm DNA methylation status in bulls. Further study will be needed to confirm the relationship between aging and methylation status.
In conclusion, we detected different methylation statuses in spermatozoa by using a human microarray, and found DMRs that could be easily detected using COBRA. The combined analysis of DNA methylation levels and sperm parameters can be considered an effective approach for assessing bull aging or fertility in the future.
Supplementary
Acknowledgments
This work was supported by the Ito Foundation Grant (H27-Ken157, H28-Ken42) and JSPS KAKENHI Grant Number JP16K07999. The authors are grateful to the members of the Livestock Research Support Center in NARO for their help with semen collection.
References
- 1.Carrell DT. Epigenetics of the male gamete. Fertil Steril 2012; 97: 267–274. [DOI] [PubMed] [Google Scholar]
- 2.Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, Guérin JF. Influence of global sperm DNA methylation on IVF results. Hum Reprod 2005; 20: 768–773. [DOI] [PubMed] [Google Scholar]
- 3.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE 2007; 2: e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boissonnas CC, Jouannet P, Jammes H. Epigenetic disorders and male subfertility. Fertil Steril 2013; 99: 624–631. [DOI] [PubMed] [Google Scholar]
- 5.Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, Carrell DT. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 2015; 104: 1388–1397.e1: 5. [DOI] [PubMed] [Google Scholar]
- 6.Stuppia L, Franzago M, Ballerini P, Gatta V, Antonucci I. Epigenetics and male reproduction: the consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin Epigenetics 2015; 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammoud SS, Cairns BR, Carrell DT. Analysis of gene-specific and genome-wide sperm DNA methylation. Methods Mol Biol 2013; 927: 451–458. [DOI] [PubMed] [Google Scholar]
- 8.Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, Boekelheide K. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS ONE 2011; 6: e20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci USA 2007; 104: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dere E, Huse S, Hwang K, Sigman M, Boekelheide K. Intra- and inter-individual differences in human sperm DNA methylation. Andrology 2016; 4: 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong NC, Ng J, Hall NE, Lunke S, Salmanidis M, Brumatti G, Ekert PG, Craig JM, Saffery R. Exploring the utility of human DNA methylation arrays for profiling mouse genomic DNA. Genomics 2013; 102: 38–46. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi E, Takeda K. A data driven approach to utilizing Human Methylation arrays in genome-wide study for bovine DNA methylation. J Anim Genet 2016; 44: 45–52. [Google Scholar]
- 13.Takeda K, Uchiyama K, Kinukawa M, Tagami T, Kaneda M, Watanabe S. Evaluation of sperm DNA damage in bulls by TUNEL assay as a parameter of semen quality. J Reprod Dev 2015; 61: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akagi S, Hosoe M, Matsukawa K, Ichikawa A, Tanikawa T, Takahashi S. Culture of bovine embryos on a polydimethylsiloxane (PDMS) microwell plate. J Reprod Dev 2010; 56: 475–479. [DOI] [PubMed] [Google Scholar]
- 15.Takeda K, Tasai M, Akagi S, Matsukawa K, Takahashi S, Iwamoto M, Srirattana K, Onishi A, Tagami T, Nirasawa K, Hanada H, Pinkert CA. Microinjection of serum-starved mitochondria derived from somatic cells affects parthenogenetic development of bovine and murine oocytes. Mitochondrion 2010; 10: 137–142. [DOI] [PubMed] [Google Scholar]
- 16.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics 2011; 98: 288–295. [DOI] [PubMed] [Google Scholar]
- 17.Dada R, Kumar M, Jesudasan R, Fernández JL, Gosálvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet 2012; 29: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urdinguio RG, Bayón GF, Dmitrijeva M, Toraño EG, Bravo C, Fraga MF, Bassas L, Larriba S, Fernández AF. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod 2015; 30: 1014–1028. [DOI] [PubMed] [Google Scholar]
- 19.Congras A, Yerle-Bouissou M, Pinton A, Vignoles F, Liaubet L, Ferchaud S, Acloque H. Sperm DNA methylation analysis in swine reveals conserved and species-specific methylation patterns and highlights an altered methylation at the GNAS locus in infertile boars. Biol Reprod 2014; 91: 137. [DOI] [PubMed] [Google Scholar]
- 20.Verma A, Rajput S, De S, Kumar R, Chakravarty AK, Datta TK. Genome-wide profiling of sperm DNA methylation in relation to buffalo (Bubalus bubalis) bull fertility. Theriogenology 2014; 82: 750–759.e1. [DOI] [PubMed] [Google Scholar]
- 21.Thangaratnarajah C, Ruprecht JJ, Kunji ER. Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat Commun 2014; 5: 5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rueda CB, Llorente-Folch I, Traba J, Amigo I, Gonzalez-Sanchez P, Contreras L, Juaristi I, Martinez-Valero P, Pardo B, Del Arco A, Satrustegui J. Glutamate excitotoxicity and Ca2+-regulation of respiration: Role of the Ca2+ activated mitochondrial transporters (CaMCs). Biochim Biophys Acta 2016; 1857: 1158–1166. [DOI] [PubMed] [Google Scholar]
- 23.Vanderperre B, Cermakova K, Escoffier J, Kaba M, Bender T, Nef S, Martinou JC. MPC1-like is a placental mammal-specific mitochondrial pyruvate carrier subunit expressed in postmeiotic male germ cells. J Biol Chem 2016; 291: 16448–16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Schwacke R, Kunze R. DNA Damage-Induced Transcription of Transposable Elements and Long Non-coding RNAs in Arabidopsis Is Rare and ATM-Dependent. Mol Plant 2016; 9: 1142–1155. [DOI] [PubMed] [Google Scholar]
- 25.Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav 2011; 59: 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lunstra DD, Echternkamp SE. Puberty in beef bulls: acrosome morphology and semen quality in bulls of different breeds. J Anim Sci 1982; 55: 638–648. [DOI] [PubMed] [Google Scholar]
- 27.Lunstra DD, Cundiff LV. Growth and pubertal development in Brahman-, Boran-, Tuli-, Belgian Blue-, Hereford- and Angus-sired F1 bulls. J Anim Sci 2003; 81: 1414–1426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.