Abstract
Chronic muscle disuse, such as that resulting from immobilization, denervation, or prolonged physical inactivity, produces atrophy and a loss of mitochondria, yet the molecular relationship between these events is not fully understood. In this review we attempt to identify the key regulatory steps mediating the loss of muscle mass and the decline in mitochondrial content and function. An understanding of common intracellular signaling pathways may provide much-needed insight into the possible therapeutic targets for treatments that will maintain aerobic energy metabolism and preserve muscle mass during disuse conditions.
Keywords: denervation, mitochondrial biogenesis, mitophagy, protein degradation, skeletal muscle
1. Introduction
Skeletal muscle is the largest tissue in the body, representing approximately 40% of the total mass of a healthy adult. This tissue is well characterized as exceptionally malleable, and a diversity of stimuli, such as prolonged inactivity,1 denervation,2 starvation,3 aging,4 or chronic disease,3, 5 can negatively impact muscle mass. Each unique stimulus yields similar, yet characteristic, molecular, functional, and phenotypic alterations in skeletal muscle. The resulting muscle atrophy is defined by an overall loss of proteins, organelles, and cytoplasm. Mitochondria are the main source of energy in skeletal muscle and they provide adenosine triphosphate by means of oxidative phosphorylation.6 It is not surprising then that mitochondria are also quite adaptable, as their cellular content can be fine-tuned to the tissue's energy requirements. Mitochondrial content is regulated by two opposing processes, mitochondrial synthesis (biogenesis)7 and mitochondrial degradation (mitophagy).8 In the context of muscle disuse, a decrement in mitochondrial abundance is one of the major adaptations observed,9 as the demand for energy is diminished. Understanding the interaction between mitochondrial biogenesis and mitophagy and their relationship to muscle atrophy during disuse is therefore invaluable for our comprehension of cellular homeostasis. The following review provides a concise summary of the mechanisms moderating muscle mass and mitochondrial content during conditions of chronic muscle disuse.
2. Atrophy signaling during muscle disuse
Skeletal muscle mass is determined by the balance between protein synthesis and degradation. One of the most significant alterations associated with prolonged inactivity of skeletal muscle is the net loss of muscle protein, which results in myofiber atrophy.10, 11, 12, 13, 14 This loss of muscle mass occurs as a consequence of enhanced activation of the cell's major proteolytic executors, the ubiquitin–proteasome system (UPS), and the autophagy–lysosome pathway (ALP), which together regulate the half-life of a majority of cellular proteins.15 The study of the molecular pathways controlling this balance is an important area of sustained research.
Muscle mass maintenance and myofiber hypertrophy are governed, in part, by insulin-like growth factor-1 signaling. This protein can promote muscle growth partially through phosphatidylinositol 3-kinase–Akt signaling,16, 17, 18 which stimulates myofibrillar protein synthesis via mammalian target of rapamycin complex 1 (mTORC1).19 Thus, the direct activation of Akt is associated with muscle hypertrophy, and interestingly is sufficient to block atrophy during muscle disuse.16 During muscle disuse, a reduction in phosphatidylinositol 3-kinase–Akt signaling is evident and could, in theory, result in reduced mammalian target of rapamycin (mTOR) activity. Notably, mTORC1 has been implicated in stimulating mitochondrial biogenesis20, 21, 22 and in the inhibition of autophagy.23 Thus, a decrease in mTOR activity would result in reduced protein synthesis and mitochondrial biogenesis, while stimulating autophagy. Both decreases in mitochondrial function and marked increases in autophagy are known to trigger cellular signaling events that contribute to muscle atrophy. However, several studies have noted a counter intuitive hyperactivation of mTOR during chronic muscle disuse, likely as a result of enhanced amino acid influx from the cellular degradative pathways, the UPS and ALP.24, 25 Therefore, although the exact role of mTOR-related signaling during muscle wasting is still contentious,24, 25, 26, 27, 28, 29 this complex appears to represent a critical rheostat in the control of an abundance of cellular processes during muscle growth and atrophy.
Central to the regulation of muscle mass during disuse are members of the forkhead box class O (FoxO) proteins. Although this family of transcription factors is involved in numerous intracellular processes,30, 31 these proteins also operate as critical mediators of myofiber atrophy during muscle disuse, as they orchestrate the induction of an atrophic gene program that is implicated in both UPS- and ALP-mediated catabolism.29, 32, 33 In this scenario Akt is able to prevent muscle atrophy by inhibitory phosphorylation of FoxO3 on multiple residues, effectively prohibiting its nuclear entry by facilitating its sequestration in the cytoplasm though interactions with 14-3-3 proteins.34 Indeed, Akt activity is reduced during muscle disuse,16 allowing FoxO3 to enter the nucleus and stimulate a gene expression program that promotes an atrophic phenotype.33 In particular, FoxO nuclear translocation has been shown to upregulate the expression of E3 ubiquitin ligases muscle RING finger 1 (MuRF1) and atrogin-1/MAFbx.33 These E3 ligases are major effectors of the UPS, as they mediate muscle loss by targeting muscle structural proteins and components related to protein translation for degradation.1, 35, 36, 37, 38
Apart from Akt-mediated phosphorylation, the activity and subcellular localization of FoxO transcription factors can be fine-tuned via post-translational modifications by other upstream factors. AMP-activated protein kinase (AMPK), a metabolic sensor, has the capacity to phosphorylate FoxO on numerous residues, which positively influences its transcriptional activity.39, 40 The activation of this AMPK–FoxO axis was documented to occur with muscle disuse41 and appears to contribute to muscle protein degradation. The increase in AMPK activation with muscle inactivity also likely inhibits protein synthesis,42, 43, 44 further accelerating muscle protein loss. AMPK activation has also been shown to promote mitochondrial biogenesis,45, 46, 47, 48, 49 however, a process that is downregulated during muscle disuse. Thus, AMPK activation at the onset of muscle disuse could serve as an early signal to mitigate energy stress, by enhancing energy substrate availability through protein breakdown and attenuating the loss of mitochondria at the early stages of muscle disuse.
FoxO3 activity can also be modulated by acetylation.50, 51, 52 In response to muscle disuse, FoxO3 can be deacetylated by Histone deacetylase 1 (HDAC1),50 promoting its nuclear translocation and activity. Conversely, recent research has also drawn attention to the role of SirT1, an NAD+-dependent deacetylase, in the inhibition of denervation-induced myofiber atrophy through the deacetylation of FoxO3.53 It appears possible that the differential acetylation of lysine residues could account for the contrasting results observed between these studies. Nonetheless, the capacity of FoxO3 to be post-translationally regulated during muscle disuse represents a critical inflection point in the control of the atrophy gene program.
Another critical system contributing to disuse-induced muscle atrophy is the nuclear factor-κB (NF-κB) signaling pathway (Fig. 1). Extracellular factors, such as tumor necrosis factor α (TNFα), stimulate this pathway, activating the inhibitor of κB kinases (IKKα and IKKβ), which prompts the nuclear localization of NF-κB transcription factors. These transcription factors bind NF-κB response elements on atrophy genes, such as MuRF1, FoxO3, and Runx1 among others,54 and promote their transcription. Overexpression of a negative regulator of this system (inhibitor of NF-κBα) is sufficient to block disuse-induced atrophy,55, 56 and muscle-specific abolition of IKKα and IKKβ via genetic knockout (KO), or expression of a dominant negative form, is also protective of muscle mass.57, 58, 59 Attenuation of myofiber atrophy in these models appears to result from a decrease in the activity of the NF-κB transcription factors and coactivators,55, 58, 60 the most important of which are p50 and Bcl-3, which have notable roles in the expression of many genes associated with muscle atrophy.54, 61 Moreover, NF-κB signaling has also been demonstrated to impair mitochondrial biogenesis in skeletal muscle (Fig. 1).62, 63, 64 Thus, NF-κB signaling also contributes to muscle atrophy by promoting a decline in mitochondrial content and function, increasing mitochondrial reactive oxygen species (ROS) production and stimulating nuclear apoptosis.
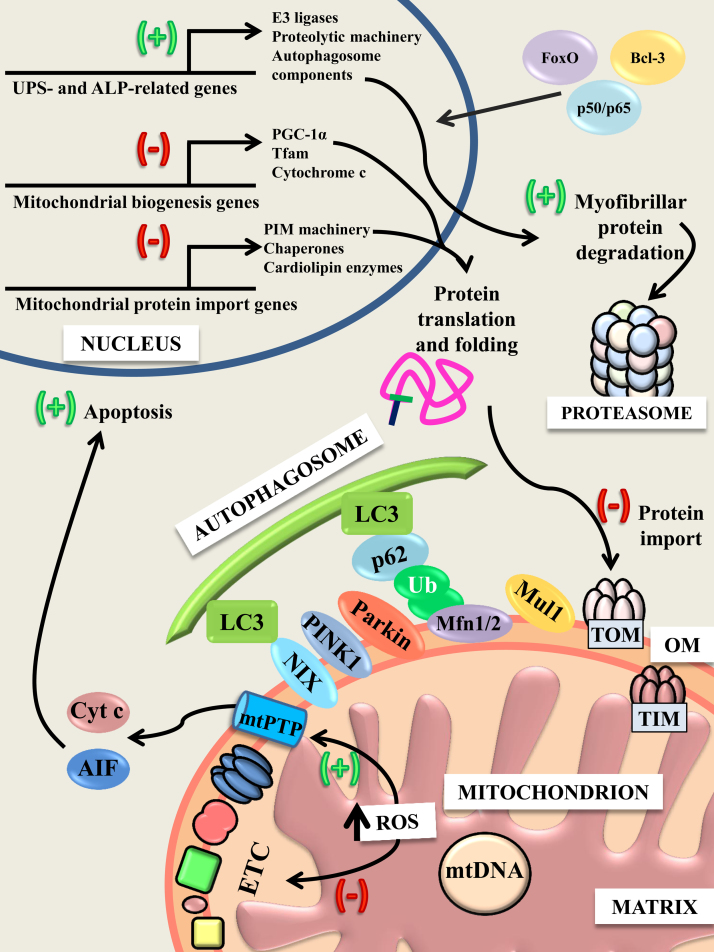
Fig. 1.
Alterations in gene expression and mitochondria during chronic muscle disuse.
During chronic muscle disuse, FoxO and NF-κB transcription factors (p50, p65) and Bcl-3 translocate to the nucleus and upregulate (+) the expression of a host of components related to proteolytic degradation pathways, resulting in increased activity of the autophagy–lysosome pathway (ALP) and the ubiquitin–proteasome system (UPS). Concurrently, the expression of factors associated with mitochondrial biogenesis and protein import are downregulated (-), and this is associated with an impairment in the import of proteins into the organelle. This may be due, in part, to a dysfunction in the electron transport chain, because respiration and mitochondrial membrane potential are impaired whereas reactive oxygen species (ROS) production surges. Dysfunctional mitochondria express PTEN-induced putative kinase 1 (PINK1) on their outer membrane (OM), which recruits Parkin. Along with Mul1, Parkin aids in the ubiquitination of OM proteins, including Mfn1/2. The adaptor factor p62 links ubiquitinated proteins to the main constituent of the autophagosomal membrane, LC3. The mitochondrial protein NIX also serves as a bridge between depolarized mitochondria and LC3. Incorporation of LC3 into the membrane continues until dysfunctional mitochondria are sequestered, at which point they can be transported to the lysosome for degradation. Mitochondrial ROS production can also promote the opening of the mitochondrial permeability transition pore (mtPTP), allowing for the release of proapoptotic factors, cytochrome c (Cyt c), and apoptosis inducing factor (AIF) into the cytosol. These factors promote the fragmentation of nuclear DNA and muscle atrophy.
Recently, the cytokine TNF-like weak inducer of apoptosis (TWEAK) and its receptor Fn14 have emerged as major effectors of disuse-induced atrophy. The expression of Fn14 is upregulated during conditions of muscle disuse,54, 65 as Fn14 promoter methylation by DNA methyltransferase 3a is decreased.66 The importance of the TWEAK–Fn14 dyad in disuse atrophy is highlighted by the TWEAK-KO mouse model, which is resistant to denervation atrophy, through impaired NF-κB signaling and MuRF1 expression.65 Downstream of the TWEAK–Fn14 system is the ubiquitin ligase TNF receptor-associated factor 6 (TRAF6). TRAF6 is a relatively unique ubiquitin ligase, in that it mediates the conjugation of Lys-63-linked polyubiquitin chains, as opposed to the more typical Lys-48-linked chains, to target proteins for degradation.67, 68 Denervation increases the expression of TRAF6, which coordinates the breakdown of muscle-specific proteins as well as mitochondrial factors.69
Recent data have also identified the transcription factors p53 and ATF4 as mediators of novel and distinct signaling pathways facilitating atrophy during disuse. ATF4 has previously been implicated in muscle atrophy, because its downstream target Gadd45a induces muscle loss during disuse through enhanced autophagy and caspase-mediated proteolysis.70 Muscle-specific deletion of either p53 or ATF4 confers partial resistance to disuse atrophy, whereas combined deletion has a synergistic impact on the preservation of myofiber size. These transcription factors have a common downstream effector, p21, which is elevated during muscle disuse and appears to be sufficient to induce muscle loss.71 Interestingly, loss of p53 also results in reduced mitochondrial content and increased ROS production.72 Thus the attenuation of muscle mass loss with p53 deletion is unlikely to involve mitochondrial pathways. Indeed, the increase in p53 protein content in muscle observed with denervation71 might be considered an important transcriptional drive that promotes muscle atrophy. Further work on this pathway is warranted.
ROS are elevated in skeletal muscle during disuse.2, 73, 74, 75, 76 Although mitochondria are not the sole site of ROS production in the cell, the increase in mitochondrial-specific ROS during muscle inactivity is notable. In particular, ROS are implicated in the damage of mitochondrial membranes, proteins, and DNA,77, 78, 79, 80 as well as the opening of the mitochondrial permeability transition pore,81 an event that reduces mitochondrial membrane potential, decreases ATP production, and promotes the release of proapoptotic proteins that induce myonuclear decay. Several studies have noted increases in apoptotic protein expression, DNA fragmentation,2, 82, 83, 84, 85 and heightened mitochondrial susceptibility to permeability transition pore opening with denervation.86 Although controversial,87 this potential loss of myonuclei could impair the capacity of the cell to maintain a sufficient transcription of genes required for the maintenance of muscle mass and mitochondrial content. In support for a role in mitochondrially-mediated apoptosis, deletion of the proapoptotic protein Bax, alone or in combination with Bak, attenuates apoptotic signaling, oxidative stress, and loss of muscle mass, illustrating at least a partial role for apoptotic cell death in disuse atrophy.88, 89 In addition, the pharmacological inhibition of calpain and caspase-3, two proteases implicated in the activation of the UPS and apoptosis, has been shown to attenuate myofiber atrophy during muscle disuse.90 These data suggest that an elevated incidence of apoptotic signaling during skeletal muscle disuse is a likely contributor to muscle atrophy.
Taken together, it is evident that the regulation of muscle mass during muscle inactivity largely depends on the interplay between the regulation of protein turnover, the activity of apoptotic signaling, and the balance between cellular energy supply and demand mediated by mitochondrial form and function.
3. Expression of nuclear genes encoding mitochondrial proteins during muscle disuse
During muscle disuse, the loss of muscle mass is accompanied, or preceded, by decrements in mitochondrial content and function. This is likely due to a reduced drive for mitochondrial biogenesis, and an increased removal of dysfunctional organelles. Research delving into the alterations in the pathways governing mitochondrial biogenesis and degradation in the context of muscle disuse has gained significant ground in recent years, and the resolution of these mechanisms will provide formidable insight into the maintenance of skeletal muscle health during periods of prolonged inactivity.
Mitochondrial biogenesis is a product of the integrated contributions of both the nuclear and mitochondrial genomes, and the coordinated expression of these genomes is required for optimal mitochondrial function. Although the nuclear genome encodes >99% of the proteins within mitochondria, the mitochondrial genome contributes 13 proteins critical to electron transport chain (ETC) function, which is embedded within the inner mitochondrial membrane (IMM).91, 92 Mitochondrial biogenesis is a complex process that synchronizes the transcription, translation, and subsequent import of nuclear encoded-proteins into the mitochondrion, along with the replication and expression of mitochondrial DNA .7, 92 Newly-synthesized mitochondrial proteins and lipids are incorporated into the existing mitochondrial reticulum and contribute to its expansion. The peroxisome proliferator activated receptor γ coactivator-1 (PGC-1) family of transcriptional coactivators is critical for the coordinated expression of these genomes.93, 94 A member of this family, PGC-1α1, is well established as an orchestrator of mitochondrial biogenesis, as it can bind a number of nuclear transcription factors, including nuclear respiratory factors (NRF)-1 and -2, thereby promoting the expression of nuclear genes encoding mitochondrial proteins.92, 95 One of these is mitochondrial transcription factor A (Tfam), a vital factor for mitochondrial DNA replication and transcription.96, 97, 98, 99
Because PGC-1α1 plays a fundamental role in the regulation of mitochondrial biogenesis, the study of this coactivator and its downstream effectors is of interest in the context of muscle disuse. The reduced expression of this family of coactivators with denervation-induced disuse occurs early after the cessation of muscle activity.32, 100 This is followed by decreased expression of estrogen-related receptor α (ERR α), NRF-1/2, and Tfam, in addition to the mitochondrial markers cytochrome c and COX IV at the mRNA and protein level.2, 100, 101, 102, 103 Interestingly, an overexpression of either PGC-1α1 or -β attenuates protein degradation and muscle atrophy in denervated muscle,104 implying a role for these coactivators in the maintenance of muscle mass. The attenuation of muscle atrophy by PGC-1α during disuse appears to be mediated via a suppression of FoxO3-mediated transcription of E3 ligases MuRF1 and atrogin-1.32 The protection conferred by PGC-1α1 overabundance is also observed in the sarcopenic muscle of aged mice, because overexpression of this coactivator mitigates the age-associated reductions in muscular function, mitochondrial content, antioxidant response, and protein synthesis.105 Because muscle inactivity promotes a strong reduction in PGC-1α expression, it is likely that the PGC-1α-induced suppression of FoxO3 signaling is released, promoting myofiber atrophy.
In addition to a dampening of FoxO activity, a recent study has demonstrated a role for PGC-1α in the neutralization of TWEAK-mediated muscle atrophy.62 PGC-1α overexpression is sufficient to attenuate the loss of muscle mass during denervation by restraining Fn14 expression and TWEAK-mediated NF-κB signaling, further highlighting the importance of this transcriptional coactivator in the protection of muscle mass.
The PGC-1 family of coactivators also facilitates the expression of genes involved in mitochondrial morphology. In particular, the interaction between PGC-1α/β and ERRα stimulates the transcription of mitofusin (Mfn) 2,106, 107 which regulates outer mitochondrial membrane (OMM) fusion.108, 109 Because these factors are downregulated during muscle disuse, it is not surprising that the integrity of the mitochondrial reticulum is compromised. Muscle inactivity induces a substantial decrease in the expression of the OMM fusion proteins Mfn2 and Opa1, presumably shifting the cellular balance in favor of mitochondrial fission.9, 103, 110 An increase in fission facilitates mitochondrial-specific degradation processes, such as mitophagy,111, 112 resulting in a net loss of mitochondria from muscle.
PGC-1α also plays a regulatory role in the biosynthesis and remodeling of cardiolipin (CL), a mitochondria-specific phospholipid implicated in the physical stabilization of the ETC, and in the assembly of the protein import machinery (PIM).113, 114 Chronic muscle disuse is associated with a decrease in CL content within muscle,115, 116 which is linked with mitochondrial dysfunction. This impairment in mitochondrial function drives a vicious cycle, because the production of mitochondrial ROS is enhanced, likely driving the oxidation of CL. This facilitates the involvement of CL in proapoptotic events,117, 118, 119 which mediate a portion of the muscle atrophy observed with muscle disuse.
4. Protein import into the mitochondrion
Mitochondria rely heavily on nuclear transcription for a majority of the genes necessary for their proper function.120 Since mitochondria are not formed de novo, proteins that are translated from nuclear-derived transcripts must translocate into the mitochondria via the PIM.121 The mechanisms of this protein import process are highly specialized and complex,122, 123 and the process represents a critical step at which mitochondrial content can be regulated.
It is well known that chronic exercise or contractile activity is accompanied by mitochondrial biogenesis that is closely associated with the elevated expression of PIM components124, 125, 126 as well as mitochondrial CL content.115, 127 Chronic contractile activity also augments the expression of factors important for the assembly of the translocase of the outer mitochondrial membrane complex, which serves as the main channel for protein import.124 Furthermore, contractile activity elicits increases in the expression of the cytosolic and mitochondrial chaperone proteins Hsp60, mtHsp70, and Hsp70,128 which are integral to the stabilization, import, and refolding of precursor proteins into mitochondria. Indeed, increases in the PIM following contractile activity correlate well with the accelerated rates of import of proteins such as mitochondrial Tfam into the mitochondrial matrix.125, 129 Taken together, the increase in mitochondrial protein import with contractile activity is critical for mitochondrial biogenesis, and it promotes the expansion of the mitochondrial reticulum.
In direct contrast to contractile activity, chronic muscle disuse results in substantial reductions in mitochondrial content,2, 76, 116 which correlate well with diminished protein import into the mitochondrion.76 This suggests that the reduction in mitochondrial content is mediated, at least in part, by reductions in protein import. This is attributable to a number of factors, including changes in the content and distribution of CL, reductions in the expression of PIM components, and an increase in the production of ROS.115, 116 With respect to CL, reductions in the content of this phospholipid can result in destabilization of ETC complexes III and IV130, 131, 132 and a loss of membrane potential, which impairs the import of proteins into the organelle.133 Furthermore, muscle disuse may alter the membrane localization of CL. A majority of mitochondrial CL typically resides in the IMM.134, 135 However, denervation has been shown to increase the expression of phospholipid scramblase-3, which serves to transport CL from the IMM to the OMM.115 Since the location of CL can have an impact on its cellular role in the context of apoptosis,117, 136, 137 the probable increase in OMM CL with denervation may contribute to the increased mitochondrial apoptosis observed with denervation.2
In addition to CL, several PIM components have been shown to be significantly reduced in response to muscle disuse, including Tim23, Tom20, and mtHsp70. These effects were observed to occur as early as 3 days following denervation, and likely precede the loss in mitochondria and muscle mass observed with denervation.76 In addition to the reduction in the expression of PIM components, the increase in mitochondrial ROS noted with denervation also appears to underlie deficits in mitochondrial matrix-destined import during muscle disuse. Elevated ROS levels inhibit the import of matrix-destined proteins and increase their susceptibility to proteasome-mediated degradation.76, 138 The reduction in import is likely related to the redox modulation of the cysteine- or thiol-rich residues of the import components. In the future, investigations should be conducted to identify the principal mechanisms that enable ROS to inhibit protein import.
5. Autophagy and mitophagy during muscle disuse
Macroautophagy (hereafter referred to as autophagy) is a conserved cellular process that is responsible for the elimination of long-lived proteins, as well as dysfunctional organelles. The ALP is critical for the turnover and maintenance of organelles in skeletal muscle in particular, because this tissue is postmitotic. This process involves the formation of double membrane vesicles, known as autophagosomes, which sequester cytoplasmic materials that are destined for degradation and deliver them to the lysosome. Upon autophagosome–lysosome fusion, the acidic pH and hydrolases native to the lysosome digest the cargo into its constituent parts, which can subsequently be utilized for energy provision. Given its catabolic nature, it is not surprising that autophagy has been implicated in muscle atrophy. The expression of multiple autophagic factors was found to be upregulated under various atrophic conditions, including denervation.3, 29, 139 These findings have raised interest in the pharmacological inhibition of autophagy as a potential target during muscle-wasting conditions. Studies involving animals with a deficient ALP, specifically in skeletal muscle, however, have revealed that an intact pathway is actually required for the maintenance of muscle mass and function.140, 141 Moreover, the absence of a functional autophagy program during muscle disuse only exacerbates muscle wasting.140 Thus, aberrant autophagy underlies several myopathies and muscular dystrophies,141, 142, 143, 144, 145 and interventions restoring ALP functionality ameliorate some of the myopathic phenotypes.141, 142, 143, 144 Conversely, excessive autophagy can also be detrimental by inducing a net catabolic effect, culminating in loss of muscle mass as demonstrated by overactive autophagy.146 Nonetheless, it appears that the maintenance of a basal level of autophagy is required for the appropriate turnover of long-lived proteins and organelles in skeletal muscle, and that constrained activity of the ALP is essential for muscle mass maintenance and health.
Interestingly, many of the myopathies characterized by deficient autophagy also exhibit the accumulation of dysfunctional and swollen mitochondria, which certainly contribute to disease pathogenesis.140, 142 This is likely due to the insufficient turnover of mitochondria in these conditions, highlighting the integral relationship between rigorous mitochondrial quality control and muscle health. Although mitochondrial content can be regulated in a variety of ways, mitochondria-specific autophagy, known as mitophagy, is currently the only known mechanism for the wholesale removal of mitochondria from postmitotic tissues, such as skeletal muscle. This process is required for the proper progression of the mitochondrial life cycle and is therefore vital for mitochondrial vigor.147
Mitophagy is a constitutively active housekeeping mechanism in all cells, and can be further induced under conditions of energetic distress. During an energy deficit, AMPK is activated whereas mTOR is inhibited, leading to the induction of autophagosome formation by the activation and lipidation of microtubule-associated protein light chain 3 (LC3). This is achieved through a coordinated effort by various AuTophaGy (ATG) related genes-dependent conjugation steps. Continuous incorporation of membrane-bound LC3 leads to the formation and elongation of the autophagosome around the tagged cargo until it is completely encapsulated. The specific targeting of mitochondria for elimination by mitophagy is thought to be mediated by two generally distinct pathways: one being indirect and requiring the recruitment of an E3 ubiquitin ligase; and the other necessitating a mitophagy-specific receptor. Current evidence suggests that mitochondrial dysfunction often results in the activation of the indirect pathway, whereas removal of mitochondria under a programed biological event is carried out by the receptor-mediated pathway.148, 149
With respect to the indirect pathway, loss of mitochondrial membrane potential or a surge in ROS production prompt the stabilization of PTEN-induced putative kinase 1 (PINK1) on the mitochondrial membrane. PINK1 subsequently recruits Parkin, which ubiquitinates various OMM proteins.148 Interestingly, mice that lack Parkin are partially protected against loss of muscle mass and mitochondria in denervated slow-twitch but not fast-twitch muscle, suggesting a fiber type-specific preference for Parkin-mediated mitophagy.150 In addition to Parkin, several mitochondria-associated E3 ubiquitin ligases have been implicated in mitophagy, including Mul1 and Gp78.151, 152, 153 These ligases preferentially ubiquitinate the OMM proteins of dysfunctional mitochondria, allowing for their recognition by autophagy adaptor proteins such as p62 and NBR1,154, 155 which effectively link the autophagosome to the malfunctioning mitochondrion, allowing it to be sequestered. Interestingly, Parkin and Mul1 have both been implicated in disuse atrophy. Parkin KO mice showed resistance to denervation in slow-twitch muscle, likely due to reduced denervation-induced proteasomal activation via Nuclear Factor Erythroid 2-Like 1 (NFE2L1/Nrf1).150 Moreover, Lokireddy et al152 demonstrated that Mul1 expression is induced by denervation through a mechanism mediated by FoxO1/3 transcription factors, and that Mul1 appears to be both sufficient and required for mitophagy in skeletal muscle.152 Interestingly, E3 ligases appear to compensate for one another, because Mul1 was recently demonstrated to play a role in maintaining mitochondrial integrity in light of a loss in PINK1/Parkin.156
Mitochondria can also be directly targeted for degradation by the receptor-mediated pathway involving BNIP3 and the BNIP3-like protein NIX. These factors are regulated by FoxO3 and are able to localize to the mitochondrial membrane, anchoring it to the autophagosome directly via their LC3 interacting region. BNIP3 and NIX were both found to be sufficient to induce muscle wasting, and appear to mediate, in part, FoxO3-induced muscle atrophy.157 BNIP3 has also been documented to have a role in mitochondrial fragmentation, a prerequisite for mitophagy.158 Indeed, mitochondrial morphology and dynamics play a key role in sealing mitochondrial fate, and organelle fission alone is sufficient to activate mitophagy.159 In skeletal muscle, mitochondrial fragmentation induced by Fis1 enhances muscle wasting by leading to energetic stress and the activation of the AMPK–FoxO3 axis. Pharmacological inhibition of mitochondrial fragmentation was sufficient to dampen autophagy, as well as muscle atrophy.41, 160 Of note, the profusion proteins mitofusin1 and 2 are well-characterized targets of both Parkin and Mul1.152, 156, 161 Thus, mitochondrial dynamics appear to play a key role in muscle atrophy and this effect may be mediated, at least in part, by the activation of mitophagy.
Increases in autophagy and mitophagy have been noted during various models of muscle atrophy. We have previously demonstrated an upregulation in the expression of numerous autophagy proteins and transcripts in response to denervation. Furthermore, denervation resulted in the increased localization of LC3-II, p62, and Parkin to mitochondrial membranes, indicating an enhanced targeting of mitochondria for degradation by autophagy.88, 139, 162 This may be due to the decrements in mitochondrial oxygen consumption, which prompt an increase in ROS production observed during muscle disuse.2, 76 ROS promote skeletal muscle autophagy and this effect is mediated, in part, through activation of the AMPK and inhibition of Akt, because the scavenging of ROS significantly decreased autophagic flux in mouse skeletal muscles.163
Thus, autophagy and mitophagy are activated during a variety of muscle-wasting conditions and participate in muscle atrophy. However, these pathways act as a double-edged sword whereby suppression of mitophagy may temporarily attenuate muscle loss, but if left unchecked can result in mitochondrial dysfunction and cellular oxidative stress, exacerbating myofiber atrophy. It is still unclear whether the activation of autophagy and mitophagy in skeletal muscle during disuse atrophy is a byproduct of the loss of energetic homeostasis, or whether these processes themselves instigate muscle wasting.
6. Conclusion
The regulation of skeletal muscle mass during chronic disuse largely depends on the balance between protein synthesis and protein degradation, with cellular energetic status playing a key role in the signals that fine-tune protein turnover and expression. Mitochondrial quality control is at the epicenter of cellular metabolic regulation and contributes meaningfully to muscle mass maintenance. During periods of muscle disuse, decreases in mitochondrial biogenesis, protein import, and oxidative phosphorylation, coupled with enhanced fragmentation and removal of organelles by mitophagy, lead to a net loss of mitochondrial content and an increase in mitochondrially-mediated nuclear apoptosis. All of these greatly contribute to the observed atrophy. Thus, resolving energetic distress by improving mitochondrial function and enhancing mitochondrial network formation appear to be protective of muscle mass in this scenario. With this in mind, few studies have examined the early signaling events that occur upon the onset of muscle disuse, making it difficult to predict the kinetics or infer causality between mitochondrial alterations and muscle wasting during disuse. Further studies delineating these early signaling events responsible for mitochondrial malfunction and its relation to muscle wasting are warranted.
Conflicts of interest
All contributing authors declare no conflicts of interest.
References
- 1.Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 2.Adhihetty P.J., O’Leary M.F.N., Chabi B., Wicks K.L., Hood D.A. Effect of denervation on mitochondrially mediated apoptosis in skeletal muscle. J Appl Physiol. 2007;3:1143–1151. doi: 10.1152/japplphysiol.00768.2006. [DOI] [PubMed] [Google Scholar]
- 3.Lecker S.H., Jagoe R.T., Gilbert A., Gomes M., Baracos V., Bailey J. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 4.Chabi B., Ljubicic V., Menzies K.J., Huang J.H., Saleem A., Hood D.A. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell. 2008;7:2–12. doi: 10.1111/j.1474-9726.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 5.Fearon K.C.H. The mechanisms and treatment of weight loss in cancer. Proc Nutr Soc. 1992;51:251–265. doi: 10.1079/pns19920036. [DOI] [PubMed] [Google Scholar]
- 6.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood D.A. Invited Review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 8.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iqbal S., Ostojic O., Singh K., Joseph A.M., Hood D.A. Expression of mitochondrial fission and fusion regulatory proteins in skeletal muscle during chronic use and disuse. Muscle Nerve. 2013;48:963–970. doi: 10.1002/mus.23838. [DOI] [PubMed] [Google Scholar]
- 10.Argadine H.M., Hellyer N.J., Mantilla C.B., Zhan W., Sieck G.C. The effect of denervation on protein synthesis and degradation in adult rat diaphragm muscle. J Appl Physiol. 2009;107:438–444. doi: 10.1152/japplphysiol.91247.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eley H.L., Tisdale M.J. Skeletal muscle atrophy, a link between depression of protein synthesis and increase in degradation. J Biol Chem. 2007;282:7087–7097. doi: 10.1074/jbc.M610378200. [DOI] [PubMed] [Google Scholar]
- 12.Furuno K., Goodman M., Goldberg A.L. Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J Biol Chem. 1990;265:8550–8557. [PubMed] [Google Scholar]
- 13.Tawa N.E., Odessey R., Goldberg A.L. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wing S.S., Haas A.L., Goldberg A.L. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochem J. 1995;307:639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masiero E., Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy. 2010;6:307–309. doi: 10.4161/auto.6.2.11137. [DOI] [PubMed] [Google Scholar]
- 16.Bodine S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R.Z. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 17.Coleman M.E., DeMayo F., Yin K.C., Lee H.M., Geske R., Montgomery C. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem. 1995;270:12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- 18.Rommel C., Bodine S.C., Clarke B.A., Rossman R., Nunez L., Stitt T.N. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 19.Lai K.M., Gonzalez M., Poueymirou W.T., Kline W.O., Na E., Zlotchenko E. Conditional activation of Akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blättler S.M., Verdeguer F., Liesa M., Cunningham J.T., Vogel R.O., Chim H. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor yin yang 1 in skeletal muscle. Mol Cell Biol. 2012;32:3333–3346. doi: 10.1128/MCB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter H.N., Hood D.A. Contractile activity-induced mitochondrial biogenesis and mTORC1. Am J Physiol Cell Physiol. 2012;303:C540–C547. doi: 10.1152/ajpcell.00156.2012. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 23.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quy P.N., Kuma A., Pierre P., Mizushima N. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J Biol Chem. 2013;288:1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H., Inoki K., Lee M., Wright E., Khuong A., Khuong A. mTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Sci Signal. 2014;7:ra18. doi: 10.1126/scisignal.2004809. [DOI] [PubMed] [Google Scholar]
- 26.Argadine H.M., Mantilla C.B., Zhan W., Sieck G.C. Intracellular signaling pathways regulating net protein balance following diaphragm muscle denervation. Am J Physiol Cell Physiol. 2011;300:C318–C327. doi: 10.1152/ajpcell.00172.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentzinger C.F., Lin S., Romanino K., Castets P., Guridi M., Summermatter S. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle. 2013;3:6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machida M., Takeda K., Yokono H., Ikemune S., Taniguchi Y., Kiyosawa H. Reduction of ribosome biogenesis with activation of the mTOR pathway in denervated atrophic muscle. J Cell Physiol. 2012;227:1569–1576. doi: 10.1002/jcp.22871. [DOI] [PubMed] [Google Scholar]
- 29.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Calnan D.R., Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 31.Eijkelenboom A., Burgering B.M.T. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 32.Sandri M., Lin J., Handschin C., Yang W., Arany Z.P., Lecker S.H. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A. FoxO transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S. Akt promotes cell survival by phosphorylating and inhibiting forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 35.Cohen S., Brault J.J., Gygi S.P., Glass D.J., Valenzuela D.M., Gartner C. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fielitz J., Kim M., Shelton J.M., Latif S., Spencer J.A., Glass D.J. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagirand-Cantaloube J., Offner N., Csibi A., Leibovitch M.P., Batonnet-Pichon S., Tintignac L.A. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lokireddy S., Wijesoma I.W., Sze S.K., McFarlane C., Kambadur R., Sharma M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am J Physiol Cell Physiol. 2012;303:C512–C529. doi: 10.1152/ajpcell.00402.2011. [DOI] [PubMed] [Google Scholar]
- 39.Greer E.L., Oskoui P.R., Banko M.R., Maniar J.M., Gygi M.P., Gygi S.P. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez A.M.J., Csibi A., Raibon A., Cornille K., Gay S., Bernardi H. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 41.Romanello V., Guadagnin E., Gomes L., Roder I., Sandri C., Petersen Y. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolster D.R., Crozier S.J., Kimball S.R., Jefferson L.S. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 43.Chan A.Y.M., Soltys C-LM, Young M.E., Proud C.G., Dyck J.R.B. Activation of AMP-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J Biol Chem. 2004;279:32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 44.Horman S., Browne G., Krause U., Patel J., Vertommen D., Bertrand L. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr Biol. 2002;12:1419–1423. doi: 10.1016/s0960-9822(02)01077-1. [DOI] [PubMed] [Google Scholar]
- 45.Bergeron R., Ren J.M., Cadman K.S., Moore I.K., Perret P., Pypaert M. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–E1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 46.Irrcher I., Adhihetty P.J., Sheehan T., Joseph A.M., Hood D.A. PPARγ coactivator-1α expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. Am J Physiol Cell Physiol. 2003;284:C1669–C1677. doi: 10.1152/ajpcell.00409.2002. [DOI] [PubMed] [Google Scholar]
- 47.Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winder W.W., Holmes B.F., Rubink D.S., Jensen E.B., Chen M., Holloszy J.O. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol. 2000;88:2219–2226. doi: 10.1152/jappl.2000.88.6.2219. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Uguccioni G., Ljubicic V., Irrcher I., Iqbal S., Singh K. Multiple signaling pathways regulate contractile activity-mediated PGC-1α gene expression and activity in skeletal muscle cells. Physiol Rep. 2014;2:1–12. doi: 10.14814/phy2.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beharry A.W., Sandesara P.B., Roberts B.M., Ferreira L.F., Senf S.M., Judge A.R. HDAC1 activates FoxO and is both sufficient and required for skeletal muscle atrophy. J Cell Sci. 2014;127:1441–1453. doi: 10.1242/jcs.136390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertaggia E., Coletto L., Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302:C587–C596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 52.Senf S.M., Sandesara P.B., Reed S.A., Judge A.R. p300 acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am J Physiol Cell Physiol. 2011;300:C1490–C1501. doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee D., Goldberg A.L. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J Biol Chem. 2013;288:30515–30526. doi: 10.1074/jbc.M113.489716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C.L., Kandarian S.C., Jackman R.W. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS One. 2011;6:e16171. doi: 10.1371/journal.pone.0016171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Judge A.R., Koncarevic A., Hunter R.B., Liou H.C., Jackman R.W., Kandarian S.C. Role for IkappaBalpha, but not c-Rel, in skeletal muscle atrophy. Am J Physiol Cell Physiol. 2007;292:C372–C382. doi: 10.1152/ajpcell.00293.2006. [DOI] [PubMed] [Google Scholar]
- 56.Mourkioti F., Kratsios P., Luedde T., Song Y.H., Delafontaine P., Adami R. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116:2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai D., Frantz J.D., Tawa N.E., Melendez P.A., Oh B.C., Lidov H.G.W. IKKB/NF-κB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 58.Van Gammeren D., Damrauer J.S., Jackman R.W., Kandarian S.C. The IκB kinases IKKα and IKKB are necessary and sufficient for skeletal muscle atrophy. FASEB J. 2009;23:362–370. doi: 10.1096/fj.08-114249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed S.A., Senf S.M., Cornwell E.W., Kandarian S.C., Judge A.R. Inhibition of IkappaB kinase alpha (IKKα) or IKKbeta (IKKβ) plus forkhead box O (Foxo) abolishes skeletal muscle atrophy. Biochem Biophys Res Commun. 2011;405:491–496. doi: 10.1016/j.bbrc.2011.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Senf S.M., Dodd S.L., McClung J.M., Judge A.R. Hsp70 overexpression inhibits NF-κB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB J. 2008;22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunter R.B., Kandarian S.C. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J Clin Invest. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hindi S.M., Mishra V., Bhatnagar S., Tajrishi M.M., Ogura Y., Yan Z. Regulatory circuitry of TWEAK-Fn14 system and PGC-1α in skeletal muscle atrophy program. FASEB J. 2014;28:1398–1411. doi: 10.1096/fj.13-242123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Remels A.H.V., Gosker H.R., Bakker J., Guttridge D.C., Schols A.M.W.J., Langen R.C.J. Regulation of skeletal muscle oxidative phenotype by classical NF-κB signalling. Biochim Biophys Acta. 2013;1832:1313–1325. doi: 10.1016/j.bbadis.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 64.Remels A.H.V., Gosker H.R., Langen R.C., Polkey M., Sliwinski P., Galdiz J. Classical NF-κB activation impairs skeletal muscle oxidative phenotype by reducing IKK-α expression. Biochim Biophys Acta. 2014;1842:175–185. doi: 10.1016/j.bbadis.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Mittal A., Bhatnagar S., Kumar A., Lach-Trifilieff E., Wauters S., Li H. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol. 2010;188:833–849. doi: 10.1083/jcb.200909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tajrishi M.M., Shin J., Hetman M., Kumar A. DNA methyltransferase 3a and mitogen-activated protein kinase signaling regulate the expression of fibroblast growth factor-inducible 14 (Fn14) during denervation-induced skeletal muscle atrophy. J Biol Chem. 2014;289:19985–19999. doi: 10.1074/jbc.M114.568626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng L., Wang C., Spencer E., Yang L., Braun A., You J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 68.Lamothe B., Besse A., Campos A.D., Webster W.K., Wu H., Darnay B.G. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paul P.K., Gupta S.K., Bhatnagar S., Panguluri S.K., Darnay B.G., Choi Y. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bongers K.S., Fox D.K., Ebert S.M., Kunkel S.D., Dyle M.C., Bullard S.A. Skeletal muscle denervation causes skeletal muscle atrophy through a pathway that involves both Gadd45a and HDAC4. Am J Physiol Endocrinol Metab. 2013;305:E907–E915. doi: 10.1152/ajpendo.00380.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fox D.K., Ebert S.M., Bongers K.S., Dyle M.C., Bullard S.A., Dierdorff J.M. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am J Physiol Endocrinol Metab. 2014;307:E245–E261. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saleem A., Adhihetty P.J., Hood D.A. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 73.Kavazis A.N., Talbert E.E., Smuder A.J., Hudson M.B., Nelson W.B., Powers S.K. Mechanical ventilation induces diaphragmatic mitochondrial dysfunction and increased oxidant production. Free Radic Biol Med. 2009;46:842–850. doi: 10.1016/j.freeradbiomed.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Min K., Smuder A.J., Kwon O.S., Kavazis A.N., Szeto H.H., Powers S.K. Mitochondrial-targeted antioxidants protect skeletal muscle against immobilization-induced muscle atrophy. J Appl Physiol. 2011;111:1459–1466. doi: 10.1152/japplphysiol.00591.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller F.L., Song W., Jang Y.C., Liu Y., Sabia M., Richardson A. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1159–R1168. doi: 10.1152/ajpregu.00767.2006. [DOI] [PubMed] [Google Scholar]
- 76.Singh K., Hood D.A. Effect of denervation-induced muscle disuse on mitochondrial protein import. Am J Physiol Cell Physiol. 2011;300:C138–C145. doi: 10.1152/ajpcell.00181.2010. [DOI] [PubMed] [Google Scholar]
- 77.Davies K.J.A. Protein damage and degradation by oxygen radicals. J Biol Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 78.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 79.Powers S.K., Wiggs M.P., Duarte J.A., Zergeroglu A.M., Demirel H.A. Mitochondrial signaling contributes to disuse muscle atrophy. Am J Physiol Endocrinol Metab. 2012;303:E31–E39. doi: 10.1152/ajpendo.00609.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turrens J.F. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lemasters J.J., Nieminen A.L., Qian T., Trost L.C., Elmore S.P., Nishimura Y. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 82.Borisov A.B., Carlson B.M. Cell death in denervated skeletal muscle is distinct from classical apoptosis. Anat Rec. 2000;258:305–318. doi: 10.1002/(SICI)1097-0185(20000301)258:3<305::AID-AR10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 83.Ferreira R., Neuparth M.J., Vitorino R., Appell H.J., Amado F., Duarte J.A. Evidences of apoptosis during the early phases of soleus muscle atrophy in hindlimb suspended mice. Physiol Res. 2008;57:601–611. doi: 10.33549/physiolres.931272. [DOI] [PubMed] [Google Scholar]
- 84.O’Leary M.F.N., Hood D.A. Effect of prior chronic contractile activity on mitochondrial function and apoptotic protein expression in denervated muscle. J Appl Physiol. 2008;105:114–120. doi: 10.1152/japplphysiol.00724.2007. [DOI] [PubMed] [Google Scholar]
- 85.Siu P.M., Alway S.E. Mitochondria-associated apoptotic signalling in denervated rat skeletal muscle. J Physiol. 2005;565:309–323. doi: 10.1113/jphysiol.2004.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Csukly K., Ascah A., Matas J., Gardiner P.F., Fontaine E., Burelle Y. Muscle denervation promotes opening of the permeability transition pore and increases the expression of cyclophilin D. J Physiol. 2006;574:319–327. doi: 10.1113/jphysiol.2006.109702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bruusgaard J.C., Gundersen K. In vivo time-lapse microscopy reveals no loss of murine myonuclei during weeks of muscle atrophy. J Clin Invest. 2008;118:1450–1457. doi: 10.1172/JCI34022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Leary M.F.N., Vainshtein A., Carter H.N., Zhang Y., Hood D.A. Denervation-induced mitochondrial dysfunction and autophagy in skeletal muscle of apoptosis-deficient animals. Am J Physiol Cell Physiol. 2012;303:C447–C454. doi: 10.1152/ajpcell.00451.2011. [DOI] [PubMed] [Google Scholar]
- 89.Siu P.M., Alway S.E. Deficiency of the Bax gene attenuates denervation-induced apoptosis. Apoptosis. 2006;11:967–981. doi: 10.1007/s10495-006-6315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Talbert E.E., Smuder A.J., Min K., Kwon O.S., Powers S.K. Calpain and caspase-3 play required roles in immobilization-induced limb muscle atrophy. J Appl Physiol. 2013;114:1482–1489. doi: 10.1152/japplphysiol.00925.2012. [DOI] [PubMed] [Google Scholar]
- 91.Hock M.B., Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 92.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 93.Scarpulla R.C., Vega R.B., Kelly D.P. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uguccioni G., D'souza D., Hood D.A. Regulation of PPARγ coactivator-1α function and expression in muscle: effect of exercise. PPAR Res. 2010 doi: 10.1155/2010/937123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 97.Ngo H.B., Lovely G.A., Phillips R., Chan D.C. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi Y., Dierckx A., Wanrooij P.H., Wanrooij S., Larsson N.G., Wilhelmsson L.M. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci USA. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 100.Sacheck J.M., Hyatt J.P., Raffaello A., Jagoe R.T., Roy R.R., Edgerton V.R. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 101.Booth F.W., Lou W., Hamilton M.T., Yan Z. Cytochrome c mRNA in skeletal muscles of immobilized limbs. J Appl Physiol. 1996;81:1941–1945. doi: 10.1152/jappl.1996.81.5.1941. [DOI] [PubMed] [Google Scholar]
- 102.Kang C., Ji L.L. Muscle immobilization and remobilization downregulates PGC-1α signaling and the mitochondrial biogenesis pathway. J Appl Physiol. 2013;115:1618–1625. doi: 10.1152/japplphysiol.01354.2012. [DOI] [PubMed] [Google Scholar]
- 103.Wagatsuma A., Kotake N., Mabuchi K., Yamada S. Expression of nuclear-encoded genes involved in mitochondrial biogenesis and dynamics in experimentally denervated muscle. J Physiol Biochem. 2011;67:359–370. doi: 10.1007/s13105-011-0083-5. [DOI] [PubMed] [Google Scholar]
- 104.Brault J.J., Jespersen J.G., Goldberg A.L. Peroxisome proliferator-activated receptor γ coactivator 1α or 1B overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wenz T., Rossi S.G., Rotundo R.L., Spiegelman B.M., Moraes C.T. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Liesa M., Borda-d’Agua B., Medina-Gómez G., Lelliott C.J., Paz J.C., Rojo M. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS One. 2008;3:e3613. doi: 10.1371/journal.pone.0003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Soriano F.X., Liesa M., Bach D., Chan D.C., Palacín M., Zorzano A. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-γ coactivator-1α, estrogen-related receptor-α, and mitofusin 2. Diabetes. 2006;55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 108.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song Z., Ghochani M., Mccaffery J.M., Frey T.G., Chan D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagatsuma A., Kotake N., Kawachi T., Shiozuka M., Yamada S., Matsuda R. Mitochondrial adaptations in skeletal muscle to hindlimb unloading. Mol Cell Biochem. 2011;350:1–11. doi: 10.1007/s11010-010-0677-1. [DOI] [PubMed] [Google Scholar]
- 111.Dagda R.K., Cherra S.J., Kulich S.M., Tandon A., Park D., Chu C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chicco A.J., Sparagna G.C. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 114.Gebert N., Joshi A.S., Kutik S., Becker T., McKenzie M., Guan X.L. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ostojic O., O’Leary M.F.N., Singh K., Menzies K.J., Vainshtein A., Hood D.A. The effects of chronic muscle use and disuse on cardiolipin metabolism. J Appl Physiol. 2013;114:444–452. doi: 10.1152/japplphysiol.01312.2012. [DOI] [PubMed] [Google Scholar]
- 116.Wicks K.L., Hood D.A. Mitochondrial adaptations in denervated muscle: relationship to muscle performance. Am J Physiol. 1991;260:C841–C850. doi: 10.1152/ajpcell.1991.260.4.C841. [DOI] [PubMed] [Google Scholar]
- 117.Garcia Fernandez M., Troiano L., Moretti L., Nasi M., Pinti M., Salvioli S. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ. 2002;13:449–455. [PubMed] [Google Scholar]
- 118.Kagan V.E., Tyurin V.A., Jiang J., Tyurina Y.Y., Ritov V.B., Amoscato A.A. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 119.Petrosillo G., Ruggiero F.M., Paradies G. Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria. FASEB J. 2003;17:2202–2208. doi: 10.1096/fj.03-0012com. [DOI] [PubMed] [Google Scholar]
- 120.Scarpulla R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 121.Baker M.J., Frazier A.E., Gulbis J.M., Ryan M.T. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 122.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 124.Joseph A.M., Hood D.A. Plasticity of TOM complex assembly in skeletal muscle mitochondria in response to chronic contractile activity. Mitochondrion. 2012;12:305–312. doi: 10.1016/j.mito.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 125.Takahashi M., Chesley A., Freyssenet D., Hood D.A. Contractile activity-induced adaptations in the mitochondrial protein import system. Am J Physiol. 1998;274:C1380–C1387. doi: 10.1152/ajpcell.1998.274.5.C1380. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Y., Iqbal S., O’Leary M.F.N., Menzies K.J., Saleem A., Ding S. Altered mitochondrial morphology and defective protein import reveal novel roles for Bax and/or Bak in skeletal muscle. Am J Physiol Cell Physiol. 2013;305:C502–C511. doi: 10.1152/ajpcell.00058.2013. [DOI] [PubMed] [Google Scholar]
- 127.Takahashi M., Hood D.A. Chronic stimulation-induced changes in mitochondria and performance in rat skeletal muscle. J Appl Physiol. 1993;74:934–941. doi: 10.1152/jappl.1993.74.2.934. [DOI] [PubMed] [Google Scholar]
- 128.Ornatsky O.I., Connor M.K., Hood D.A. Expression of stress proteins and mitochondrial chaperonins in chronically stimulated skeletal muscle. Biochem J. 1995;311:119–123. doi: 10.1042/bj3110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gordon J.W., Rungi A.A., Inagaki H., Hood D.A. Effects of contractile activity on mitochondrial transcription factor A expression in skeletal muscle. J Appl Physiol. 2001;90:389–396. doi: 10.1152/jappl.2001.90.1.389. [DOI] [PubMed] [Google Scholar]
- 130.Lange C., Nett J.H., Trumpower B.L., Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pfeiffer K., Gohil V., Stuart R.A., Hunte C., Brandt U., Greenberg M.L. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 132.Zhang M., Mileykovskaya E., Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 133.Jiang F., Ryan M.T., Schlame M., Zhao M., Gu Z., Klingenberg M. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 134.Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerma F., Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem. 1990;265:18797–18802. [PubMed] [Google Scholar]
- 135.Mejia E.M., Nguyen H., Hatch G.M. Mammalian cardiolipin biosynthesis. Chem Phys Lipids. 2014;179:11–16. doi: 10.1016/j.chemphyslip.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 136.Kuwana T., Mackey M.R., Perkins G., Ellisman M.H., Latterich M., Schneiter R. Bid. Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 137.Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat Cell Biol. 2000;2:754–761. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 138.Wright G., Terada K., Yano M., Sergeev I., Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 139.O’Leary M.F.N., Hood D.A. Denervation-induced oxidative stress and autophagy signaling in muscle. Autophagy. 2009;5:230–231. doi: 10.4161/auto.5.2.7391. [DOI] [PubMed] [Google Scholar]
- 140.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 141.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grumati P., Coletto L., Sabatelli P., Cescon M., Angelin A., Bertaggia E. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 143.De Palma C., Morisi F., Cheli S., Pambianco S., Cappello V., Vezzoli M. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pauly M., Daussin F., Burelle Y., Li T., Godin R., Fauconnier J. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 145.Sugie K., Noguchi S., Kozuka Y., Arikawa-Hirasawa E., Tanaka M., Yan C. Autophagic vacuoles with sarcolemmal features delineate Danon disease and related myopathies. J Neuropathol Exp Neurol. 2005;64:513–522. doi: 10.1093/jnen/64.6.513. [DOI] [PubMed] [Google Scholar]
- 146.Carmignac V., Svensson M., Körner Z., Elowsson L., Matsumura C., Gawlik K.I. Autophagy is increased in laminin α2 chain-deficient muscle and its inhibition improves muscle morphology in a mouse model of MDC1A. Hum Mol Genet. 2011;20:4891–4902. doi: 10.1093/hmg/ddr427. [DOI] [PubMed] [Google Scholar]
- 147.Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Narendra D.P., Jin S.M., Tanaka A., Suen D., Gautier C.A., Shen J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Furuya N., Ikeda S.I., Sato S., Soma S., Ezaki J., Oliva Trejo J.A. PARK2/Parkin-mediated mitochondrial clearance contributes to proteasome activation during slow-twitch muscle atrophy via NFE2L1 nuclear translocation. Autophagy. 2014;10:631–641. doi: 10.4161/auto.27785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fu M., St-Pierre P., Shankar J., Wang P.T.C., Joshi B., Nabi I.R. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–1162. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lokireddy S., Wijesoma I.W., Teng S., Bonala S., Gluckman P.D., McFarlane C. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 153.Narendra D., Tanaka A., Suen D., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 155.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yun J., Puri R., Yang H., Lizzio M.A., Wu C., Sheng Z.H. Mul1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 158.Kubli D.A., Ycaza J.E., Gustafsson A.B. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Benard G., Karbowski M. Mitochondrial fusion and division: regulation and role in cell viability. Semin Cell Dev Bio. 2009;20:365–374. doi: 10.1016/j.semcdb.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Romanello V., Sandri M. Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr Hypertens Rep. 2010;12:433–439. doi: 10.1007/s11906-010-0157-8. [DOI] [PubMed] [Google Scholar]
- 161.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D., Karbowski M. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O’Leary M.F., Vainshtein A., Iqbal S., Ostojic O., Hood D.A. Adaptive plasticity of autophagic proteins to denervation in aging skeletal muscle. Am J Physiol Cell Physiol. 2013;304:C422–C430. doi: 10.1152/ajpcell.00240.2012. [DOI] [PubMed] [Google Scholar]
- 163.Rahman M., Mofarrahi M., Kristof A.S., Nkengtac B., Harel S., Hussain S.N. Reactive oxygen species regulation of autophagy in skeletal muscles. Antioxid Redox Signal. 2014;20:443–459. doi: 10.1089/ars.2013.5410. [DOI] [PubMed] [Google Scholar]