Abstract
Photosynthesis is a unique process that allows independent colonization of the land by plants and of the oceans by phytoplankton. Although the photosynthesis process is well understood in plants, we are still unlocking the mechanisms evolved by phytoplankton to achieve extremely efficient photosynthesis. Here, we combine biochemical, structural and in vivo physiological studies to unravel the structure of the plastid in diatoms, prominent marine eukaryotes. Biochemical and immunolocalization analyses reveal segregation of photosynthetic complexes in the loosely stacked thylakoid membranes typical of diatoms. Separation of photosystems within subdomains minimizes their physical contacts, as required for improved light utilization. Chloroplast 3D reconstruction and in vivo spectroscopy show that these subdomains are interconnected, ensuring fast equilibration of electron carriers for efficient optimum photosynthesis. Thus, diatoms and plants have converged towards a similar functional distribution of the photosystems although via different thylakoid architectures, which likely evolved independently in the land and the ocean.
Phytoplankton and plant plastids have distinct evolutionary origins and membrane organization. Here Flori et al. show that diatom photosynthetic complexes spatially segregate into interconnected subdomains within loose thylakoid stacks enabling fast diffusion of electron carriers and efficient photosynthesis
Photosynthesis is a unique process that converts sunlight energy into organic matter on Earth, feeding almost the entire food chain. Photosynthesis is accomplished on the land, which is dominated by plants, and in the ocean, which is mostly colonized by phytoplankton. In eukaryotes, this process occurs in a specialized organelle: the plastid. Plant photosynthetic plastids (chloroplasts) are derived from a cyanobacterium-like organism via primary endosymbiosis, whereas the majority of phytoplankton plastids are derived from a red eukaryotic microalga via secondary endosymbiosis. Their different phylogenetic origins have led to distinct structural plastid designs. Differences can be observed at the level of the envelope, the membrane system surrounding the stromal space and of the photosynthetic membrane network, the thylakoids. Primary plastids contain a two-membrane envelope, whereas secondary plastids generally have four envelope membranes1. Primary plastids also contain differentiated thylakoid domains that segregate the components of the photosynthetic electron flow chain: the two photosystems (PS), which perform light photochemical conversion and the cytochrome b6f, which catalyses electron exchanges between the two PSs. PSII is mostly located in the appressed grana stacks, PSI is mainly found in the non-appressed stroma lamellae whereas the cytochrome b6f is more homogeneously distributed2. The lateral heterogeneity and the consequent physical confinement of the PSs prevents energy withdrawal from PSII by PSI via the thermodynamically favourable energy transfer (energy spillover)2. However, this segregation imposes a need for long-range diffusion of intermediary electron carriers (plastoquinones, plastocyanins or soluble cytochromes) between the two domains. Restricted diffusion within the crowded thylakoid membranes and/or in the narrow luminal space are limiting the maximum rate of photosynthetic electron flow in some conditions3,4.
No thylakoid subdomains are visible in secondary plastids, where available electron micrographs show loose stacks of mostly three thylakoids (sometimes two or four) with few anastomoses in some cases5,6. Moreover, while the membrane distribution of a few complexes (PSI and the light harvesting complex, Fucoxanthin Chlorophyll Protein-FCP)6 is known, no complete picture of the arrangement of the photosynthetic machinery is available to date. Overall, the mechanisms ensuring optimum light absorption and downstream electron flow are still undetermined in secondary plastids, although the organisms containing these plastids are believe to be responsible for ∼20% of the global oxygen production7. Here, we combine functional, biochemical, immunolocalization analyses with 3D imaging in the diatom Phaeodactylum tricornutum, to reveal a sophisticated thylakoid membrane network that orchestrates photosynthetic light absorption and utilization. We show that segregation of the PSs in specific thylakoid subdomains within a functionally seamless space allows balanced light capture without restraining electron flow for optimal photosynthetic activity.
Results
Energy spillover in P. tricornutum
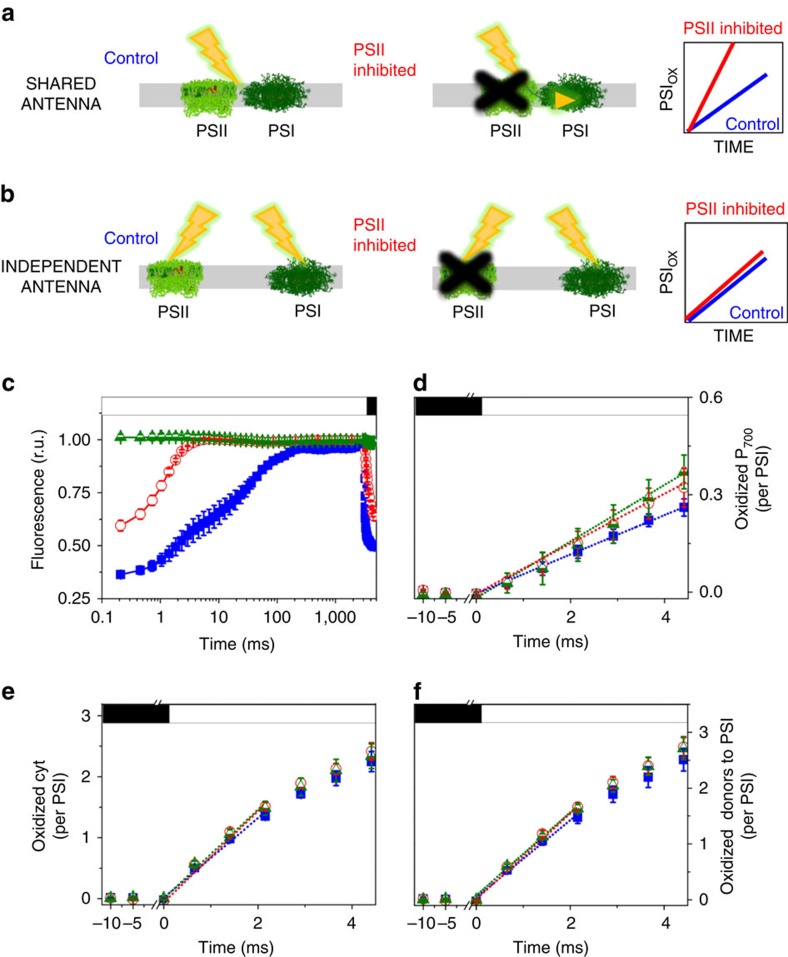
The reported loose thylakoid structure of diatoms should promote random distribution of PSI and PSII, thereby favouring PSII to PSI energy spillover via physical contacts between the complexes2. Indeed spillover has been earlier reported upon poisoning PSII (refs 8, 9) in red algae, considered to be the ancestors of secondary plastids and, more recently, in dinoflagellates (Symbiondinium)10, which are derived from secondary endosymbiosis. We tested this hypothesis by measuring changes in PSI activity upon inhibition of PSII in P. tricornutum. We reasoned that if PSI and PSII are in physical contact (Fig. 1a), inhibition of PSII photochemistry should increase the utilization of PSII-absorbed light by PSI, thus enhancing PSI activity. Conversely, no change in activity is expected if PSI and PSII are separated and do not share their excitation energy, similar to plants (Fig. 1b).
Figure 1. Experimental design used to assess energy spillover in diatoms.
Expected effects of energy spillover from PSII to PSI on PSI activity: physical contact between both PSs (a), disconnected PSs (b). (c) Fluorescence emission kinetics confirm full inhibition of PSII by DCMU and HA. (d) Kinetics of P700 oxidation by light. (e) Kinetics of cyt oxidation by light. (f) Kinetics of oxidation of the entire pool of PSI electron donors by light. A cyt/PSI ratio of 3 was assumed based on the estimate shown in Supplementary Fig. 1. The light intensity was 800 μmol photons m−2 s−1. (c–f) Solid blue squares: control; empty red circles: 40 μM DCMU; green triangles: 40 μM DCMU+2 mM HA. Means±s.e.m. (n=6, from three biological samples). F0: minimum fluorescence emission (active PSII). Fm: maximum fluorescence emission (inactive PSII). Closed box: actinic light off. Open box: actinic light on. DCMU and HA were added immediately before measurements.
We found that inhibition of PSII with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) plus hydroxylamine (HA, Fig. 1c) did not appreciably accelerate PSI activity in P. tricornutum cells (Fig. 1d–f). This was revealed by the lack of significant changes in the oxidation rate of P700 (the primary donor to PSI, Fig. 1d) and of its cytochrome electron donors (Fig. 1e, see Methods), that is, of the overall pool of PSI donors (Fig. 1f). Similar results were obtained under different light intensities (Supplementary Fig. 2), indicating that if present11, spillover is of very limited amplitude in P. tricornutum. This finding is in line with earlier reports in other diatoms (Cyclotella meneghiniana)12, where absence of spillover can be deduced based on fluorescence lifetime analysis.
Segregation of photosynthetic complexes in P. tricornutum
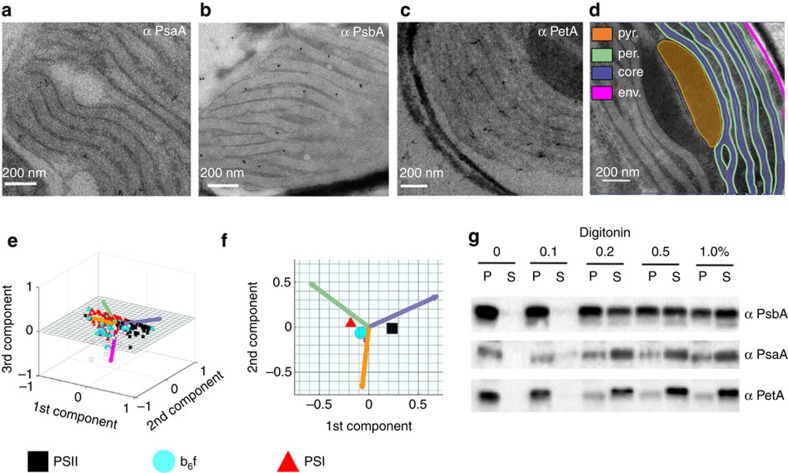
Thus, either (i) lipid or biochemical barriers prevent energy exchange between adjacent PSs or (ii) PSI and PSII are physically segregated in different thylakoid domains. To distinguish between the two possibilities, we immunolocalized the two PSs in cells prepared using the Tokuyasu protocol13, a method that ensures optimum antibody accessibility while preserving membrane structures (Supplementary Fig. 3). We localized PSI using two different antibodies against a core subunit (PsaA, Fig. 2a) and a more peripheral subunit of the complex (PsaC, Supplementary Fig. 4a). We prefentially found this complex in the external, ‘peripheral’ stromal-facing thylakoid membranes (Fig. 2d, green sectors), in agreement with earlier results6. On the other hand, we mainly located PSII in the ‘core’ thylakoid membranes (Fig. 2d, violet sectors) using two different antibodies (PsbA, Fig. 2b and PsbC, Supplementary Fig. 4b). We also immunolocalized the cytochrome b6f complex (using the PetA antibody, Fig. 2c), finding that its distribution was similar to that of PSI.
Figure 2. Immunolocalization of photosystems and of cyt b6f in the thylakoid membranes of P. tricornutum.
(a–c) TEM images of P. tricornutum labelled with antibodies directed against the PsaA subunit of PSI (a), the PsbA subunit of PSII (b) and the PetA subunit of cyt b6f (c). (d) TEM micrograph of P. tricornutum thylakoid membranes showing four distinct areas: the internal membranes (‘core’: violet); the external, peripheral membranes (‘per.’: green); the pyrenoid (‘pyr.’: orange) and the envelope (‘env.’: magenta). Bars: 200 nm. (e) Principal component analysis of PSI, cyt b6f and PSII immunolocalization with the PsbA (solid squares), PsbC (open squares), PetA (cyan circle), PsaC (solid triangles) and PsaA (open triangles) antibodies. See also Supplementary Fig. 4. A total of 258 images from four independent cultures were analysed. The first two components represent more than 91% of the variance (see Supplementary Table 1, and Methods for a more detailed explanation). Green arrow: peripheral variable; violet arrow: core variable; orange arrow: pyrenoid variable; Magenta arrow: envelope variable. (f) 2D representation of the barycentre for the PSI (α PsaA+α PsaC antibodies, black square), cyt b6f (PetA, cyan circle) and PSII (α PsbA+α PsbC antibodies, red triangle) distributions. The point size along an axis is proportional to the s.d. along the corresponding component. (g) Solubilization of P. tricornutum thylakoid membranes with increasing concentrations of digitonin (0.1%, 0.2%, 0.5%, 1%). Pellet (P) and supernatant (S) were analysed by western blotting with the same anti PSI, PSII and cyt b6f antibodies as in a–c. Representative data set of an experiment replicated on three different biological samples.
A statistical analysis of 258 micrographs (Principal Component Analysis, Fig. 2e,f, Supplementary Fig. 4d,e and Supplementary Tables 1 and 2) indicated that the barycentre of the PSI, PSII and cyt b6f complexes’ distribution do not localize in the same thylakoid compartments. This analysis confirmed the preferential ‘core’ location of PSII (black squares in Fig. 2e,f and Supplementary Fig. 4d,e) and the ‘peripheral’ location of PSI (red circles in Fig. 2e,f and Supplementary Fig. 4d,e). Conversely, while the cyt b6f complex is more concentrated in the peripheral membranes (cyan triangles in Fig. 2e,f and Supplementary Fig. 4d,e), its distribution is more homogeneous than that of PSI and PSII. Overall, this non-homogeneous distribution of the photosynthetic complexes is reminiscent of previous results in plants2 and in green algae14.
We complemented the immunolocalization analyses with biochemical fractionation. In plant thylakoids15, PSI, which is located in the stromal-exposed thylakoid lamellae, is more prone to solubilization by detergents than PSII, which is buried in the appressed membranes of the grana. We investigated the detergent accessibility of PSs in chloroplasts isolated from P. tricornutum cells by exposing them to increasing concentrations of the mild detergent digitonin and analysed the solubilized supernatant and pellet fractions for the presence of the PSs and cyt b6f by immunoblotting (Supplementary Fig. 5). As shown in Fig. 2g, solubilization of PSI and cytochrome b6f requires a lower detergent concentration than for PSII, suggesting that PSI and cyt b6f are located in the stroma-accessible thylakoids while PSII is in the less accessible membranes of the diatom chloroplasts, in agreement with the immunolocalization results.
Functional consequence of photosynthetic complex segregation
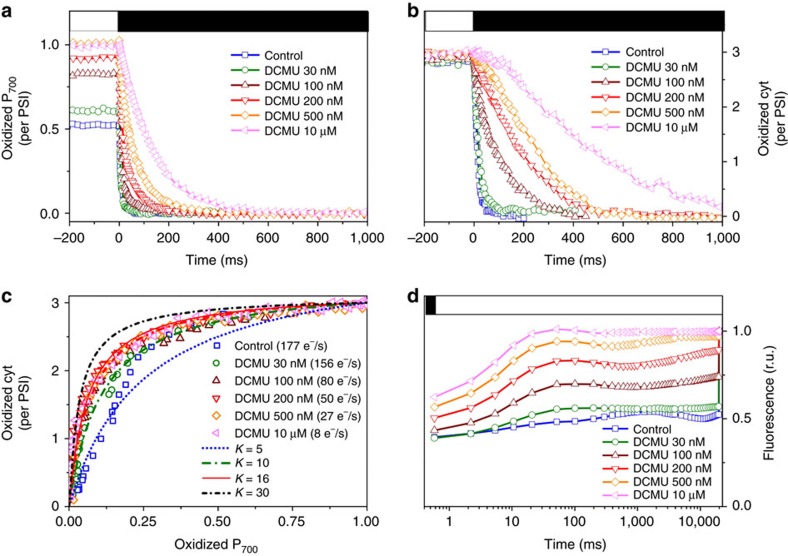
The segregation of PSI and PSII in different thylakoid sub-compartments should confine the two PSs in slow diffusion domains, as observed in plants3,4. We tested this hypothesis using a functional approach3. We compared the theoretical (Kth) and experimental (Kexp) equilibrium constants between PSI and its electron donors (cytochromes c6 and cytochrome f, see Methods). Kth was deduced from the redox potentials of cyt c6 (the soluble electron donor to PSI, 349 mV)16 and P700 (the primary electron donor to PSI, 420 mV)17. Kexp was calculated (equation (2), see Methods) from an ‘equilibration plot’ (Fig. 3), which shows the relationship between oxidized P700 (Fig. 3a) and oxidized c-type cytochromes (cyt, Fig. 3b) during dark re-reduction after illumination (Fig. 3a–c). Kexp should be equal to Kth in the absence of diffusion domains, but Kexp will be less than Kth if electron flow is limited by diffusion domains3,4. In this second case, the redox state of P700 and cyt in each domain will depend on their relative stoichiometry. During the reduction process that follows the light offset, complete reduction of P700+ and a partial reduction of cyt+ is expected in the compartments with a low P700/cyt stoichiometry. Conversely, a large fraction of P700 will still be oxidized in domains with a high P700/cyt stoichiometry. Because the equilibration plot averages the local redox states of P700 and cyt of all the different domains, the concomitant presence of P700+ (in high P700/cyt domains) and of reduced cyt c (in low P700/cyt domains) translates into a Kexp, estimate lower than the Kth value. We generated several equilibration plots (Fig. 3c) by poisoning photosynthetic electron flow (induced by saturating illumination) with increasing concentrations of DCMU (see also Fig. 3d), and found that diffusion was restricted (Kexp<Kth) when PSII generates more than 150 electrons per second (Fig. 3c, blue and green data points). However, Kexp=Kth (diffusion is no longer restricted) when the PSII rate is less than 100 electrons per second (Fig. 3c, dark red, red, orange and pink points). Thus, the compartmentalization of PSI and PSII in different thylakoid domains also generates diffusion domains in P. tricornutum, similar to plants. However, their equilibration time, 10 ms (corresponding to 100 electrons per second), is much faster than in plants (∼150 ms, corresponding to∼7 electrons per second)3.
Figure 3. Spectroscopic features of the cytochromes and P700 components of the electron transfer chain in P. tricornutum cells.
(a–b) Redox kinetics of P700 (a) and cyt (b) at the offset of a steady state illumination of 800 μmol photons m−2 s−1 in the absence and in the presence of increasing concentrations of DCMU. Closed box: actinic light off. Open box: actinic light on. (c) Equilibrium plots displaying the percentage of oxidized cyt (from b) as a function of the percentage of oxidized P700 (from a). Every point in c represents a given time after the offset of light in a,b. The dotted lines represent simulations with different values of the equilibrium constant. The rate of electron transfer (calculated as described in Methods) was modified by addition of increasing concentrations of DCMU. (d) Fluorescence induction kinetics measured for every DCMU concentration employed in a,b. The decrease in variable fluorescence indicates the progressive inhibition of PSII by DCMU. Cells were exposed to 18 μmol photons m−2 s−1 because no variable fluorescence can be observed at 800 μmol photons m−2 s−1even in the absence of DCMU (see, for example, Fig. 1c). DCMU was added immediately before measurements. Blue square: control. Green circle: DCMU 30 nM. Wine upwards triangles: DCMU 100 nM. Red downwards triangles: DCMU 200 nM. Orange losange: DCMU 500 nM. Pink leftwards triangle: DCMU 10 μM. Blue dots: simulation with an equilibrium constant of 5. Green dash and dot line: simulation with an equilibrium constant of 10. Red continuous line: simulation with an equilibrium constant of 16. Black short dash dot line: simulation with an equilibrium constant of 30.
Chloroplast structure in P. tricornutum cells
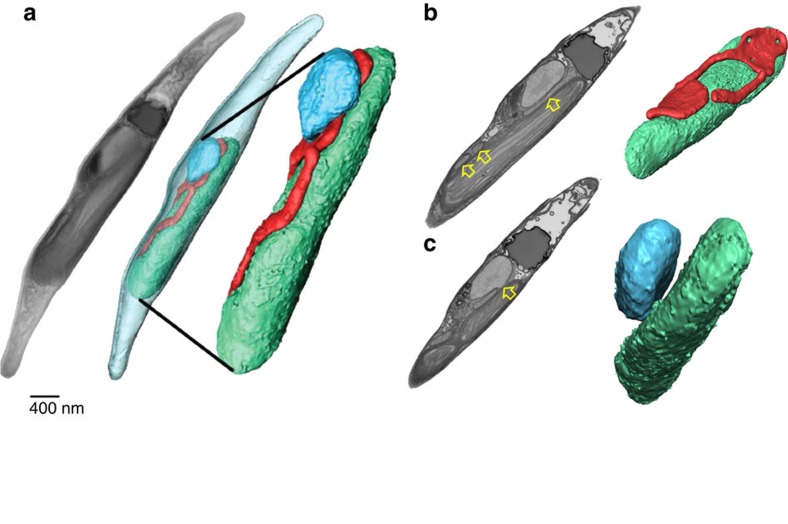
To explain the fast equilibration time of redox carriers in diatoms, we re-examined the TEM micrographs of samples prepared with the Tokuyasu protocol. By preserving the membrane structures, this technique allows observing additional features of the P. tricornutum thylakoids. We identified regions where thylakoid membranes are apparently interconnected (Supplementary Fig. 6a,b) or where they abruptly ‘disappear’ in cross-sections (Supplementary Fig. 6c, yellow circles), as if they tilt out of the micrograph plane. These features suggest the existence of a more complex 3D thylakoid network than the simple layout of three loosely juxtaposed thylakoids that is often presented in the case of secondary plastids. We collected 600 images of ultrathin sections using focused ion beam scanning electron microscopy (FIB-SEM) to reconstruct the 3D structure of a P. tricornutum cell (Supplementary Movie 1). By segmenting the 3D volume, we identified the organelles and their contacts (Fig. 4a). The mitochondrion (red) appears as a continuous network sitting on the chloroplast (green) with physical contacts between the two organelles (Fig. 4b). This mitochondrial localization likely facilitates energetic exchange between the two organelles, as recently reported18. Contact points are also seen between the chloroplast and nucleus (Fig. 4c), as expected since the outer membrane of the chloroplast envelope in secondary plastids is in connection with the nuclear ER in secondary plastids due to their evolutionary history19. These contacts could possibly mediate exchanges between the two compartments, including redox signalling20 as recently proposed in plants via the formation of transient connections between the chloroplasts and the nucleus, the stromules21.
Figure 4. Three-dimensional organization of a P. tricornutum cell.
(a) Whole cell reconstruction of an intact P. tricornutum cell based on FIB-SEM images reveals the physical contacts between the chloroplast (green), mitochondrion (red) and nucleus (blue). (b) Chloroplast–mitochondria interaction. (c) Chloroplast–nucleus interaction. Images represent frames from Supplementary movie 1. Grey pictures in a, stacks of SEM micrographs; in b,c: selected single SEM frame. Coloured pictures in a–c: 3D reconstruction. Yellow arrows highlight contacts between organelles. Bar: 400 nm.
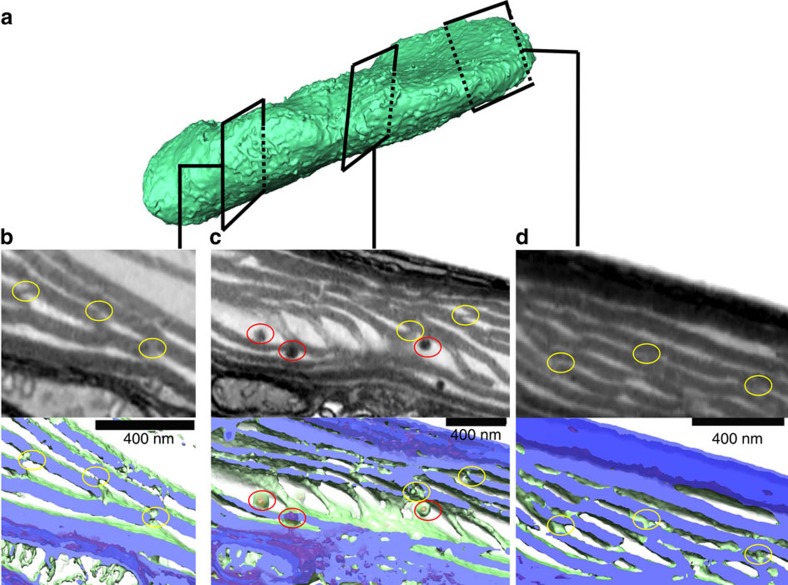
The 3D structure of the photosynthetic membranes (Fig. 5a–d) confirmed the presence of parallel layers of stacked thylakoids (purple), but also revealed the presence of connections (Fig. 5b–d, yellow circles) between them. Although the resolution of these images (4 nm pixel, see Methods) does not allow to distinguish the individual thylakoid membranes, we could nonetheless distinguish the connections from plastoglobules, chloroplast lipoprotein particles often observed between the photosynthetic membranes, which appear as globular particles in our 3D reconstruction (Fig. 5c, red circles).
Figure 5. Structural arrangement of the photosynthetic membranes in P. tricornutum.
(a) 3D image of a P. tricornutum plastid. Latitudinal (b) and longitudinal (c,d) sections reveal the 3D arrangement between the parallel photosynthetic membranes. Connections between the thylakoid layers (yellow circles) can be clearly differentiated from the plastoglobules (red circles), which appear as globular structures. (b–d): Top: representative slices of the 3D reconstruction represented as grey-levels (darker is denser). (b–d) Bottom: same areas as in top panels represented as two isosurfaces. The low density isosurface (green, thylakoid volume) and the high density isosurface (red, plastoglobules) are sliced by a semi-transparent plane (violet), to show the thylakoid stacks. Bars: 400 nm.
Discussion
Our 3D FIB-SEM reconstruction of the P. tricornutum plastid thus suggests the existence of an intricate thylakoid network, at variance with previous hypotheses suggesting that the photosynthetic membranes of secondary endosymbiotic plastids are loosely structured22. The compartmentalization of the PSs in the peripheral and core thylakoid membranes (Fig. 2) is compatible with the hypothesis that the core membranes are enriched in monogalactosyldiacylglycerol, since this lipid favours the stability and function of the dimeric PSII complex23,24. The observed organization of the PSs in the thylakoids accounts for optimum partitioning of absorbed light in low and high light conditions. Limited spillover prevents unbalanced light capture by PSI and PSII, which have similar absorption spectra in diatoms, unlike plants22. This may explain why state transitions, the migration of the light harvesting complexes between PSII and PSI to optimize low light capture in plants25, have been reported to not exist in diatoms26. Limited spillover in diatoms could also explain the high capacity of PSII to thermally dissipate excess light through non-photochemical quenching27. Indeed, non-photochemical quenching is not expected if the surplus energy in PSII were to be dissipated via spillover to PSI, as in red algae9.
Our results suggest that viridiplantae (including plants and green algae) and diatoms have achieved a similar functional topology of the PSs to optimize photosynthetic light utilization. However, this functional equivalence is achieved with different thylakoid architectures, which likely evolved independently in primary and secondary plastids, and differently affect the electron flow capacity. While PSs confinement constrains electron flow in plants, possibly limiting photosynthesis, no such limitation is observed in diatoms, where the less structured thylakoids allow very fast redox equilibration between the two PSs. Indeed, the presence of connections between thylakoid layers should facilitate diffusion of cyt c6 between the cyt b6f complexes in the core membranes and the PSI in the peripheral ones, and the diffusion of plastoquinones from PSII in the core membranes towards the cyt b6f complexes in the peripheral regions. Overall, the faster diffusion of the soluble electron carriers would promote fast redox equilibration between the photosynthetic complexes in the diatom even at very high electron flux, unlike plants.
We propose that these features, along with the tight interactions between organelles for efficient energetic exchange18, provide the most adapted framework for high photosynthetic efficiency and acclimation capacity to the ever-changing ocean environment. Indeed, the less ‘rigid’ structure of secondary plastid could allow the establishment of physical contacts between PSs possible under conditions where substantial protection of PSII is needed. Consistent with this idea, red microalgae can develop sustained spillover to protect PSII in high light9, while the symbiotic alga Symbiodinium triggers PSII spillover in response to temperature stress10. In the latter case, occurrence of topological changes favouring physical contacts between the two PSs has been proposed to account for the enhancement of spillover10. On the other hand, accumulation of PSI in specific thylakoids domains has been reported in P. tricornutum cells exposed to a particular light regime (prolonged far red light illumination), possibly to segregate it from PSII (ref. 28).
Similar structural features have been reported in green algae. In Chlamydomonas, where the number of stacks can vary from 2 to 15, with a median of 3 thylakoids29,30, connections between thylakoids also appear in cryo-tomograms29. While no spillover exists between PSII and PSI in this alga31, recent data have shown that exposure to different light qualities induces major structural changes in the thylakoids (as revealed by SANS, Small Angle Neutron Scattering), triggering changes in the harvesting capacity of PSII (ref. 32).
Methods
Phaeodactylum tricornutum cultivation
The P. tricornutum Pt1 strain (CCAP 1055/3) was obtained from the Culture Collection of Algae and Protozoa, Scottish Marine institute, UK. Cells were grown in the ESAW (Enriched Seawater Artificial Water) medium33, in 50 ml flasks in a growth cabinet (Certomat BS-1, Sartorius Stedim, Germany), at 19 °C, a light intensity of 20 μmol photon m−2 s−1, a 12-h light/12-h dark photoperiod and shaking at 100 r.p.m. Cells were collected in exponential phase, concentrated to a density of 2 × 107 cells per ml and used for experimental characterization.
Spectroscopic measurements
Spectroscopic analysis was performed on intact cells at 20 °C, using a JTS-10 spectrophotometer (Biologic, France). To assess energy spillover from PSII to PSI, redox changes of P700 and of its electron donor pool were monitored. Because of the high equilibrium constant between P700 and its electron donor pool, one needs to estimate the redox states of both P700 and of this pool to quantify the whole amount of electrons that is delivered to PSI. In diatoms, a c-type cytochrome acts as the electron donor to PSI, equivalent to plastocyanin in plants. This cytochrome will be referred as to cytochrome c6 (ref. 34), instead of cytochrome cx (ref. 35) or cytochrome c6A (ref. 36) as used in other publications. Cyt c6 and cytochrome f of the cyt b6f complex have very similar absorption features. It is thus not possible to distinguish them spectroscopically. We define therefore as ‘cyt’, the pool of cyt c6+cyt f. Cyt redox changes were calculated as [554]−0.4 × [520]−0.4 × [566], where [554], [520] and [566] are the absorption difference signals at 554, 520 and 566 nm, respectively18. P700 redox changes were measured at 705 nm. To rule out any possible contribution of fluorescence emission at this wavelength, experiments were repeated at 820 nm (where P700+ is still detected but chlorophyll fluorescence is not measured). Similar results were obtained at both wavelengths, indicating that the interference between P700 redox changes and chlorophyll fluorescence emission was negligible.
Kinetics of oxidation of P700, cyt and of the total donors to PSI oxidation result from concomitant electron injection by PSII and withdrawal by PSI. Inhibiting PSII activity with DCMU also modifies the rate of electron injection into cyt+ and P700+. This translates into an increase of the net oxidation rate of P700 and of cyt, which could be misinterpreted as an increase of the PSI activity. Therefore, to calculate the true PSI oxidation rates, we evaluate the reduction rate of this electron donor pool as the slope (SD) of signal relaxation upon switching the light off (Supplementary Fig. 1b). This rate was added to the net oxidation rate, which we estimate from the slope in the light (SL). The sum (SL+SD) provides the absolute oxidation rate (see Supplementary Fig. 1 for an example in the case of the total PSI electron donor pool).
Inhibition of PSII activity by DCMU and HA was probed following changes in chlorophyll emission from F0 (minimum fluorescence level in which QA, the primary quinone acceptor of PSII, is oxidized) to the Fm level, in which QA is fully reduced because PSII is blocked. As shown in Fig. 1c and Supplementary Fig. 2a–c, light reduces QA in DCMU poisoned samples but this inhibitor alone is not sufficient to fully reduce QA in the short time (4 ms) employed in our tests to measure oxidation of P700 and of cyt. This is particularly evident in low light (for example, Supplementary Fig. 2a, red circles), because at low photon flux the rate of QA reduction is diminished. Since full reduction of QA is needed to induce spillover, DCMU alone could not be sufficient to probe the occurrence of energy spillover in our experimental conditions. On the other hand, a complete reduction of QA is observed in the presence of HA, because this inhibitor prevents re-oxidation of reduced QA in PSII (ref. 37). By ensuring QA reduction (Fm level) at the beginning of illumination (Fig. 1c, Supplementary Fig. 2a–c green triangles), HA and DCMU ensure optimum conditions to test the occurrence of spillover.
To assess the existence of restricted diffusion domains, we compared the theoretical equilibrium constant (Kth) between PSI and its electron donors (cyt) with the experimental one (Kexp), following previous approaches in plants3 and bacteria38. In P. tricornutum we calculate a value of 16 for Kth, based on the redox potentials of cyt c6 (349 mV)16 and of P700 (420 mV)17. To evaluate Kexp the following equation was used to relate redox changes of P700 and of cyt in an ‘equilibration plot’ (Fig. 3c).
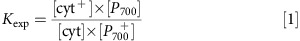 |
where [cyt], [cyt+], [P700] and [P700+] represent the concentration of the oxidized and reduced form of the cyt and P700 pools.
From equation (1) the relationship between the relative amount of oxidized P700 and of cyt can be derived as:
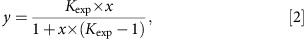 |
with
 |
and
 |
Experiments were performed under light saturated conditions, in which photosynthesis is limited by electron flow itself rather than by other factors (that is, light harvesting by PSI and PSII). In these conditions, it is possible to quantify the rate of equilibration between the diffusion domains from measurements of photosynthetic electron flow.
Finally, the P700/cyt stoichiometry was calculated in intact cells exposed to a saturating single turnover laser flash. This flash generates 1 turnover per PSI, leading to oxidation of 1 cyt per PSI. The amount of oxidized cytochrome was quantified 300 μs after the flash (that is, when P700 is fully re-reduced by the cytochromes), and compared to the amount of cyt oxidized in continuous light in the presence of DCMU (40 μM). Because the flash (which generates one positive charge per PSI) oxidizes 33% of the total oxidable cyt pool (per PSI), we conclude that the c-type cytochromes (cyt c6+cyt f)/PSI ratio is ∼3.
Chloroplast purification
An original protocol was developed to purify intact chloroplasts from P. tricornutum. Cells were collected by centrifugation at 5,000g, 10 min, 4 °C. The pellet was resuspended gently in 10 ml of isolation buffer (0.5 M Sorbitol; 50 mM Hepes-KOH; 6 mM EDTA; 5 mM MgCl2; 10 mM KCl; 1 mM MnCl2; 1% (w/v) Poly Vinyl Pyrrolidone 40 [K30]; 0.5% BSA; 0.1% cysteine, pH 7.2–7.5) and passed slowly through a French Press at 90 MPa. Ten millilitres of the isolation buffer were added to the mixture of broken cells on ice in the dark before centrifugation at 300g for 8 min to remove intact cells and cell debris. The supernatant was collected and subjected to centrifugation at 2,000g for 10 min at 4 °C. The pellet containing the chloroplasts was gently resuspended with a soft paint-brush in 2 ml of washing buffer (0.5 M Sorbitol; 30 mM Hepes-KOH; 6 mM EDTA; 5 mM MgCl2; 10 mM KCl; 1 mM MnCl2; 1% PVP 40 [K30]; 0.1% BSA, pH 7.2–7.5) and loaded on a discontinuous Percoll gradient (10, 20, 30%) in the same buffer. After centrifugation (SW41Ti rotor) at 10,000g for 35 min, the chloroplast fraction was recovered in the 20% Percoll layer of the gradient, diluted in the washing buffer (without BSA) and subjected to centrifugation at 14,000g for 10 min at 4 °C. Chloroplasts were resuspended in washing buffer and intactness was tested with a Clark electrode (Hansatek, UK) using sodium ferricyanide (1.5 mM) as an electron acceptor. Oxygen evolution in saturating light was measured before and after an osmotic shock (induced by incubation for 5 min in the washing buffer without sorbitol). The ratio between the two rates was used to evaluate intactness, which was approximately 70% in our case.
Membrane solubilization and immunoblot analysis
To differentially solubilize the two thylakoids compartments (core and peripheral), chloroplasts were incubated at a final chlorophyll concentration of 0.2 mg ml−1 for 10 min at 4 °C with digitonin (C56H92O29, Sigma Aldrich) at increasing final concentrations (0.1, 0.2, 0.5 and 1%). Samples were subjected to centrifugation at 100,000g for 5 min (rotor TLA-100), supernatants were collected and pellets were resuspended in the same volume of washing buffer without sorbitol. Samples (1.4 μg chlorophyll) were loaded onto 4–20% polyacrylamide SDS gels and blotted onto nitrocellulose membranes. Antisera against PSI (PsaA and PsaC, subunits of photosystem I, Agrisera, Se, catalogue numbers: AS06172 and AS10939, respectively), PSII (PsbA and PsbC core subunits of PSII, Agrisera, Se, catalogue numbers: AS05084 and AS111787, respectively) and cytochrome b6f (PetA, Agrisera, Se, catalogue number: AS06119) were detected by ECL using a CCD (charge-coupled device) imager (Chemidock MP Imaging, Bio-Rad, USA). Antibodies were used at a dilution of 1/10,000 (PsaA, PsaC, PsbA and PsbC) or 1/2,000 (PetA) (Supplementary Fig. 5).
Sample preparation for immunolocalization
Cells of P. tricornutum were fixed in a double-strength fixative (4% (w/v) formaldehyde, 0.4% (v/v) glutaraldehyde) in PHEM buffer (PIPES 60 mM, HEPES 25 mM, EGTA 10 mM, MgCl2 2 mM; pH 7.0) in an equal volume to the culture medium (ESAW), and then diluted into a standard strength fixative (2% (w/v) formaldehyde (EMS, USA) and 0.2% (v/v) glutaraldehyde (EMS, USA)). After 15 min, fresh standard strength fixative was replaced and fixation proceeded for 30 min at 20 °C, under agitation. Cells were washed three times with 50 mM glycine in PHEM buffer and after centrifugation were embedded in 12% gelatin in PHEM. The gelatin-embedded blocks were cryo-protected in 2.3 M sucrose in rotating vials at 4 °C (16 h). Samples vitrification was obtained in liquid nitrogen following the plunge and freezing technique13. Thin sections (80 nm) were prepared at −110 °C with a diamond knife (Diatome, Switzerland). Ribbons were picked-up with a drop of 1% (w/v) methylcellulose/1.15 M sucrose in PHEM buffer. Sections were thawed and transferred to Formvar carbon-coated nickel grids.
Immunolabelling was performed using an automated system (Leica microsystems EM IGL). Samples were post-fixed with 2% glutaraldehyde in PBS, pH 7.4, for 5 min and finally washed (three times in PBS, pH 7.4, for 2 min and six times with deionized water for 2 min). Six nanometers gold conjugate goat anti-rabbit secondary antibodies (Aurion, Wageningen, the Netherlands, catalogue number: 806.011, dilution 1/5) were used to detect PsaA and PetA. Goat anti-rabbit gold ultra-small 1.4 nm secondary antibodies (Aurion, Wageningen, the Netherlands, catalogue number 800.011, dilution 1/20), were used to dectect PsbA, PsaC and PsbC and sections were enhanced with silver (Aurion R-Gent SE-EM) for 25 min and again washed on deionized water (six times for 2 min). For observation, grids were incubated 5 min on 2% uranyl acetate (pH 7.0) and transferred to a mixture of 1.6% methyl cellulose and 0.4% uranyl acetate on ice, the excess of the viscous solution was drained away and the grids were let to dry. Grids were imaged in an electron Tecnai 12 electron microscope (FEI, USA), using an Orius CCD camera (Gatan, USA). Primary antibodies were used at a dilution of 1/50. Gold particle counting for statistical analysis was done manually. First, the total number of labels (11,932) was assessed and then particles were attributed to various compartments. If gold particles were uncertainly located (3,995), they were not considered for further analysis.
Principal components analysis
The principal components analysis (PCA) allows reducing the dimensionality39 detecting possible groupings in a given data set40. We performed PCA on our immunolabelling data considering four possible subcellular compartments for the antibodies against PSI, PSII and the cytochrome b6f complex: the internal (core) and external (peripheral) thylakoid membranes, as well as the pyrenoid and the envelope, to account for possible aspecific labelling. This led to a 4-dimension localization space (core, peripheral, pyrenoid and envelope) of 258 images from four independent cultures, where values represent the number of immunolabelling in a given localization. For data analysis, we first normalized the localization space of each of the 258 images. To do so, for each localization, we calculated the number of gold particles for a given image minus the average number of particles in that localization (for example, for a x(i,j), we obtained x1(i,j)=x(i,j)−mean(column j). This value was then normalized by the s.d. in the same localization (for example, x1(i,j)/s.d.(column j) in the above considered case).
To represent the distribution of these normalized dimensional data for the 258 images, the direction (a four-dimensional vector) giving the largest possible variance of the distribution (that is, accounts for as much of the variability in the data as possible) was selected as the direction for the first principal component. Then, the direction (another four-dimensional vector) orthogonal to the previous one(s) giving the largest possible variance of the distribution was selected as the direction for the second principal component. The repetition of this procedure automatically selects vectors representing the scatter of the distribution from major ones to minor ones. Based on singular value decomposition, PCA is a principal axis rotation of the original variables that preserves the variation in the data. Therefore, the total variance of the original variables is equal to the total variance of the principal components. The principal component coefficients correspond to the percentage of explained variance. All statistical analysis was done with the R software41.
Logistic regression
Logistic regression was used to describe data and to explain the relationship between one dependent binary variable and one or more independent variables. The two major assumptions are: (i) that the outcome must be discrete, that is, the dependent variable should be dichotomous in nature and (ii) there should be no high intercorrelations (as demonstrated42, the assumption is met for values less than 0.9) among the predictors.
We use a dose–response relationship model where the predictors are the multiple continuous variables, that is, the number of immunolabelling in the different localizations (core, peripheral, pyrenoid and envelope). Since probabilities have a limited range and regression models could predict off-scale values below zero or above 1, it makes more sense to model the probabilities of getting a given antibody on a transformed scale; this is what is done in logistic regression analysis43. A linear model for transformed probabilities can be set up as 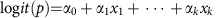 in which
in which 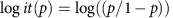 is the log odds. Each xi is the number of gold beads in the localization i and statistics about the coefficients αi will provide insight about the impact of the localization I on the probability to get a given antibody. The analysis of deviance table and the Akaike information criterion allows the identification of the relevant predictors44.
is the log odds. Each xi is the number of gold beads in the localization i and statistics about the coefficients αi will provide insight about the impact of the localization I on the probability to get a given antibody. The analysis of deviance table and the Akaike information criterion allows the identification of the relevant predictors44.
The table of correlations shows that there are no strong intercorrelations between the variables (Supplementary Note 1). Starting from a complete model (Supplementary Note 1) and based on the variable coefficients P values (Pr(>|z|)), we see that we can recursively delete the two variables envelope, env. and pyrenoid, pyr. without significantly reducing the Akaike information criterion45, which is a common measure of the relative quality of a statistical model for a given set of data. The final model demonstrates that the relevant variables to predict the antibody are the number of immunolabelling in core (P value=2 e−03) and peripheral, per. (P value<8 e−8) area. Bootstrap procedure allows the evaluation of the average percentage of wrong prediction (18%, Supplementary Table 2).
FIB-SEM and 3D reconstruction
P. tricornutum cells were fixed in 0.1 M cacodylate buffer (Sigma-Aldrich), pH 7.4, containing 2.5% glutaraldehyde (TAAB), 2% formaldehyde (Polysciences) for 1 h at 20 °C and prepared according to a modified protocol from (https://ncmir.ucsd.edu/sbem-protocol). FIB tomography was performed with a Zeiss NVision 40 dual-beam microscope. In this technique, the Durcupan (Sigma-Aldrich) resin-embedded cells of P. tricornutum were cut in cross-section, slice by slice, with a Ga+ ion beam (of 700 nA at 30 kV). After a thin slice was removed with the ion beam, the newly exposed surface was imaged in SEM at 5 kV using the in-column EsB backscatter detector. For each slice, a thickness of 4 nm was removed, and the SEM images were recorded with a pixel size of 4 nm. The image stack was then registered by cross-correlation using the StackReg plugin in the Fiji software.
For 3D reconstruction, a stack of 600 images was analysed with FIJI ImageJ software and projected in three-dimension (x,y,z axis) using the AVIZO (FEI, USA) and CHIMERA softwares (https://www.cgl.ucsf.edu/chimera/, UCSF, USA). Experiments were also performed at higher resolution (voxel size 2 nm, Supplementary movie 2). However, no significant improvement of the resolution was observed in these conditions. This likely stems from the fact that under these imaging conditions, the recorded backscatter signals emerge primarily from an area <5 nm across and <10 nm thick, setting an empirical lower limit for pixel size and slicethickness46. Moreover, the higher electron dose per surface unit might also enhance electron beam fluctuations during the acquisition and/or thermal damages to the sample, thus further limiting the imaging resolution.
Data availability
The authors declare that all data supporting the findings of this study are available within the manuscript and its supplementary files or are available from the corresponding authors on request.
Additional information
How to cite this article: Flori, S. et al. Plastid thylakoid architecture optimizes photosynthesis in diatoms. Nat. Commun. 8, 15885 doi: 10.1038/ncomms15885 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors are grateful to Pierre Joliot (Institut de Biologie Physico Chimique, Paris, France), Arthur Grossman (The Carnegie Institution, Stanford, USA) and Chris Bowler (Ecole Normale Supérieure, Paris, France) for critically reading the manuscript. We thank the Marie Curie Initial Training Network Accliphot (FP7-PEOPLE-2012-ITN; 316427 to S.F., G.F.), the HFSP (HFSP0052 to G.F.), GRAL (ANR-10-Labx-49-01 to B.G., C.M., L.F.E., D.P., C.B., G.S.), the DRF impulsion FIB-Bio program (to P.-H.J., B.G., C.M., L.F.E., D.F., G.S., G.F.), the University of Konstanz (to A.S., C.R.B., P.G.K.) and a stipend from the Graduate School of Chemical Biology (KoRS-CB to A.S.). This work used platforms from ScopeM (ETH Zurich) and the Grenoble Instruct Centre (ISBG: UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) within the Grenoble Partnership for Structural Biology (PSB).
Footnotes
The authors declare no competing financial interests.
Author contributions B.B., A.S., C.R.B., E.M., P.G.K., D.P., S.Z., C.B., G.S., D.F. and G.F. designed the study. S.F., P.-H.J., B.B., B.G., L.F.E., C.M., O.B., S.E., C.B., D.F., and G.F. performed the experiments (S.F.: chloroplast purification, biochemical analyses, spectroscopy, immunolabelling and tomography; P.-H.J.: tomography; B.B.: spectroscopy; B.G.: immunolabelling; L.F.E. tomography; C.M.: immunolabelling; O.B.: PCA analysis; S.E.: tomography; C.B.: biochemical analyses; D.F. immunolabelling and tomography; G.F.: spectroscopy). S.F. B.B., B.G., L.F.E. C.M., O.B. E.M., D.P., S.Z., C.B., G.S., D.F. and G.F. analysed the data. C.B., S.Z., D.F. and G.F. wrote the manuscript, and all authors revised and approved the manuscript.
References
- Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote–eukaryote chimaeras (meta-algae). Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 109–133 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. P. & Boekema E. J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 (2005). [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Schöttler M.-A., Maurer J. & Weis E. Plastocyanin redox kinetics in spinach chloroplasts: evidence for disequilibrium in the high potential chain. Biochim. Biophys. Acta 1659, 63–72 (2004). [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. et al. Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl Acad. Sci. USA 108, 20248–20253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoshvili Y. D., Popkova T. P. & Likhoshway Y. V. Chloroplast structure of diatoms of different classes. Cell Tissue Biol. 3, 297–310 (2009). [Google Scholar]
- Pyszniak A. M. & Gibbs S. P. Immunocytochemical localization of photosystem I and the fucoxanthin-chlorophyll a/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma 166, 208–217 (1992). [Google Scholar]
- Field C., Behrenfeld M., Randerson J. & Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- Ley A. C. & Butler W. L. Energy transfer from photosystem II to photosystem I in Porphyridium cruentum. Biochim. Biophys. Acta 462, 290–294 (1977). [DOI] [PubMed] [Google Scholar]
- Kowalczyk N. et al. Photosynthesis in Chondrus crispus: the contribution of energy spill-over in the regulation of excitonic flux. Biochim. Biophys. Acta 1827, 834–842 (2013). [DOI] [PubMed] [Google Scholar]
- Slavov C. et al. ‘Super-quenching’ state protects Symbiodinium from thermal stress—implications for coral bleaching. Biochim. Biophys. Acta 1857, 840–847 (2016). [DOI] [PubMed] [Google Scholar]
- Yokono M., Nagao R., Tomo T. & Akimoto S. Regulation of excitation energy transfer in diatom PSII dimer: how does it change the destination of excitation energy? Biochim. Biophys. Acta 1847, 1274–1282 (2015). [DOI] [PubMed] [Google Scholar]
- Chukhutsina W. U., Büchel C. & van Amerongen H. Variations in the first steps of photosynthesis for the diatom Cyclotella meneghiniana grown under different light conditions. Biochim. Biophys. Acta 1827, 10–18 (2013). [DOI] [PubMed] [Google Scholar]
- Tokuyasu K. T. A technique for ultracryotomy of cell suspensions and tissues. J. cell biol. 57, 551–565 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Wollman F. A. & Olive J. Lateral distribution of the main protein complexes of the photosynthetic apparatus in Chlamydomonas reinhardtii and in spinach—an immunocytochemical study using intact thylakoid membranes and a PS-II enriched membrane preparation. Photobiochem. Photobiophys. 12, 203–220 (1986). [Google Scholar]
- Berthold D. A., Babcock G. T. & Yocum C. F. A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes. FEBS lett. 134, 231–234 (1981). [Google Scholar]
- Akazaki H. et al. Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 Å resolution. Biosci. Biotechnol. Biochem. 73, 189–191 (2009). [DOI] [PubMed] [Google Scholar]
- Witt H. et al. Species-specific differences of the spectroscopic properties of P700: analysis of the influence of non-conserved amino acid residues by site-directed mutagenesis of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 278, 46760–46771 (2003). [DOI] [PubMed] [Google Scholar]
- Bailleul B. et al. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369 (2015). [DOI] [PubMed] [Google Scholar]
- Gibbs S. P. The chloroplast endoplasmic reticulum: structure, function, and evolutionary significance. Int. Rev. Cytol. 72, 49–99 (1981). [Google Scholar]
- Lepetit B. et al. High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant physiol 161, 853–865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K. J., Turkan I. & Krieger-Liszkay A. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant physiol. 171, 1541–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel C. Evolution and function of light harvesting proteins. J. Plant Physiol. 172, 62–75 (2015). [DOI] [PubMed] [Google Scholar]
- Kansy M., Wilhelm C. & Goss R. Influence of thylakoid membrane lipids on the structure and function of the plant photosystem II core complex. Planta 240, 781–796 (2014). [DOI] [PubMed] [Google Scholar]
- Lepetit B., Goss R., Jakob T. & Wilhelm C. Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth. res. 111, 245–257 (2012). [DOI] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G. & Wollman F. A. The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515 (2008). [DOI] [PubMed] [Google Scholar]
- Owens T. G. Light-harvesting function in the diatom Phaeodactylum tricornutum: II. distribution of excitation energy between the photosystems. Plant physiol. 80, 739–746 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavaud J., Rousseau B., van Gorkom H. J. & Etienne A.-L. Influence of the diadinoxanthin pool size on photoprotection in the marine planktonic diatom Phaeodactylum tricornutum. Plant physiol. 129, 1398–1406 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bína D., Herbstová M., Gardian Z., Vácha F. & Litvín R. Novel structural aspect of the diatom thylakoid membrane: lateral segregation of photosystem I under red-enhanced illumination. Sci. Rep. 6, 25583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B. D. et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polukhina I., Fristedt R., Dinc E., Cardol P. & Croce R. Carbon supply and photoacclimation cross talk int the green alga Chlamydomonas reinhardtii. Plant. Physiol. 172, 1494–1505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F. A. & Diner B. A. Cation control of fluorescence emission, light scatter, and membrane stacking in pigment mutants of Chlamydomonas reinhardtii. Arch. biochem. biophys. 201, 646–659 (1983). [DOI] [PubMed] [Google Scholar]
- Nagy G. et al. Chloroplast remodeling during state transitions in Chlamydomonas reinhardtii as revealed by noninvasive techniques in vivo. Proc. Natl Acad. Sci. USA 111, 5042–5047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges J. A., Franklin P. J. & Harrison P. J. evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 37, 1138–1145 (2001). [Google Scholar]
- Inda L. A., Erdner D. L., Peleato M. L. & Anderson D. M. Cytochrome c6 isolated from the marine diatom Thalassiosira weissflogii. Phytochemistry 51, 1–4 (1999). [Google Scholar]
- Weigel M., Pesaresi P. & Leister D. Tracking the function of the cytochrome c6-like protein in higher plants. Trends Plant Sci. 8, 513–517 (2003). [DOI] [PubMed] [Google Scholar]
- Howe C. J., Schlarb-Ridley B. G., Wastl J., Purton S. & Bendall D. S. The novel cytochrome c6 of chloroplasts: a case of evolutionary bricolage? J. Exp. Bot. 57, 13–22 (2006). [DOI] [PubMed] [Google Scholar]
- Cheniae G. M. & Martin I. F. Effects of hydroxylamine on photosystem II: I. factors affecting the decay of O2 evolution. Plant physiol. 47, 568–575 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B. et al. The thermodynamics and kinetics of electron transfer between cytochrome b6f and photosystem I In the chlorophyll d-dominated cyanobacterium, Acaryochloris marina. J. Biol. Chem. 283, 25218–25226 (2008). [DOI] [PubMed] [Google Scholar]
- Lebart L., Morineau A. & Piron M. Statistique exploratoire multidimensionnelle 3rd edn Dunod (2000). [Google Scholar]
- Takeuchi F., Futamura Y., Yoshikura H. & Yamamoto K. Statistics of trinucleotides in coding sequences and evolution. J. theor. biol. 222, 139–149 (2003). [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing http://www.R-project.org (2008). [Google Scholar]
- Tabachnick B. G. & Fidell L. S. Using Multivariate Statistics 6th edn Pearson (2012). [Google Scholar]
- Hosmer D. W. Jr, Lemeshow S. & Sturdivant R. X. Applied logistic regression Vol 398, (John Wiley & Sons (2013). [Google Scholar]
- Dalgaard P. Introductory Statistics with R Springer-Verlag (2002). [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE trans. automat. control 19, 716–723 (1974). [Google Scholar]
- Narayan K. & Subramaniam S. Focused ion beams in biology. Nat. Methods 12, 1021–1031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the manuscript and its supplementary files or are available from the corresponding authors on request.