Abstract
The expression of the cell death-inducing DNA fragmentation factor α-like effector (CIDE) family including Cidea, Cideb, and Cidec was significantly increased in mouse and human models of obesity. However, there was less information on these genes’ expression in pigs. Here, we hypothesized that different fat accumulation between lean (Duroc×Landrace×Yorkshire gilts, DLY) and obese (Lantang) pigs was attributed to porcine CIDE-modulating lipid metabolism. Our data showed that Cidea and Cidec were expressed at a high level in adipose tissue, and at a relatively high level in skeletal muscle, whereas Cideb was mainly expressed in the liver in both breeds of pig. Lantang pigs had higher white adipose and skeletal muscle Cidea and Cidec mRNA abundance, and hepatic and muscle Cideb mRNA than DLY pigs. Lipid metabolism-related genes including sterol regulatory element binding protein 1c (SREBP-1c), hepatocyte nuclear factor-4α (HNF-4α), peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), fatty acid synthase (FASN), diacylglycerol O-acyltransferase 1 (DGAT1), and DGAT2 showed a higher expression level in adipose tissue from obese pigs than in that from lean pigs. Lantang pigs exhibited higher mRNA abundance for liver SREBP-1c, HNF-4α, and PGC-1α, and higher skeletal muscle SREBP-1c, HNF-4α, PGC-1α, and DGAT2 expression, as compared with DLY pigs. However, the perlipin2 mRNA levels in adipose tissues, liver, and skeletal muscle were significantly lower in obese pigs than in their lean counterparts. Furthermore, plasma non-esterified fatty acid (NEFA), glucose, and triacylglycerol (TAG) levels were greater in obese pigs than in lean pigs. Finally, data from correlation analysis further found that CIDE mRNA expression was positively correlated with back fat thickness (BFT), abdominal fat mass (AFM), and the levels of NEFA, TAG, and glucose in the two breeds. Collectively, these data revealed that the porcine CIDEs possibly modulated lipid metabolism and contributed to the development of fat deposition and obesity in Lantang pigs.
Keywords: Cell death-inducing DNA fragmentation factor α-like effector (CIDE), Adipose tissue, Liver, Skeletal muscle, Fat deposition, Lantang pig, DLY pig
1. Introduction
Excessive fat deposition affects animal health and production efficiency, and constitutes a health risk to human consumers. Thus, modulation of fat deposition in adipose tissues of pigs is good for both animals and customers (Jiang et al., 2007). Lantang pig is a native breed (obese-type) of South China, whose carcass contains more fat content than hybrid pigs (Lu et al., 2008; Chen et al., 2010). DLY is the cross breed of three lean-type pigs, Duroc, Landrace, and Yorkshire; the lean percentage of DLY reaches 63%–65% (Lan et al., 2004). Lantang and DLY pigs show an obvious difference in total adipose mass and therefore offer an attractive comparison for studying the mechanism of obesity.
Adipose tissue, liver, and skeletal muscles play important roles in body lipid metabolism in animals (Ahima and Flier, 2000; Hulver et al., 2003; Leonhardt and Langhans, 2004). In pigs, adipose tissue is the central organ for fat synthesis and deposition (O'Hea and Leveille, 1969). Bernlohr et al. (2002) demonstrated that fat accumulation in animals depends on levels of triacylglycerol (TAG) synthesis and storage and levels of lipid mobilization and fatty acid oxidation. Excess fatty acids and glucose are converted into TAG after food intake. TAG, a major energy storage form, is stored in lipid droplets, so that other intracellular organelles can avoid lipotoxicity caused by fatty acid (Girousse and Langin, 2012). The lipid droplet is an important subcellular organelle responsible for lipid storage, and the sizes of unilocular lipid droplets reveal the lipid storage capacity and have a positive association with the development of obesity (Bell et al., 2008).
Changes of lipid homeostasis by over-expression or deletion of specific genes often result in obesity. Genetically modified animal models have highlighted that expression of several adipogenic and lipogenic genes, including fatty acid synthase (FASN), diacylglycerol O-acyltransferase (DGAT), sterol regulatory element binding protein 1c (SREBP-1c), and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), played important roles in fatty acid synthesis, TAG synthesis, and lipid storage (Liu et al., 2008; Malaguarnera et al., 2009). Over the past decade, it has been reported that the cell death-inducing DNA fragmentation factor α-like effector (CIDE) family plays an important role in lipid and fat metabolism (Zhou et al., 2003; Gong et al., 2009; Yonezawa et al., 2011). Previous studies have reported that animals with a deficiency in Cidea, Cideb, or Cidec exhibited a typical lean phenotype with high energy expenditure, high levels of plasma TAG and non-esterified fatty acid (NEFA) reduction (Zhou et al., 2003; Li et al., 2007; Nishino et al., 2008), and altered genes’ expression in various metabolic and signaling networks (Li et al., 2010). These studies suggest that the CIDE family plays an important role in TAG synthesis, lipid storage, and the development of obesity (Gong et al., 2009). However, all the studies on CIDEs have been carried out in mice and humans; there was far less information on the expression of these genes in pigs.
In the present study, we evaluated mRNA abundance of the adipose, liver, and skeletal muscle tissues for CIDEs and several lipid metabolism genes in both genetically obese and lean pigs. Our aim was to explain the relationship between porcine CIDE gene expression and lipid accumulation. We think that this basic molecular information might be useful for further investigation of the functions of CIDEs in pig models.
2. Materials and methods
2.1. Animals and sample collection
Eight castrated male Duroc×Landrace×Yorkshire pigs of similar liveweight ((20.21±0.57) kg) as well as eight Lantang pigs, also castrated males of similar liveweight ((16.12±0.63) kg), were used in this study. All pigs were raised in the pig farm of the animal facilities of the Institute of Animal Science in the Guangdong Academy of Agricultural Sciences, China and were provided feed and watered ad libitum. The diets for the two breeds were made up of different proportions of ingredients because the growth rates of the two breeds were different (Tables 1 and 2). The trials were terminated when the pigs of DLY and Lantang reached 100 and 70 kg body weight, respectively. An ultrasonic instrument (Renco, USA) was used to determine back fat thickness (BFT) at the 1st, 10th, and last ribs before slaughter. Blood sample was obtained from the anterior vena cava using vacuum tubes (using ethylenediaminetetra-acetic acid (EDTA) as anticoagulant) and centrifuged for 5 min at 3000g at 4 °C, and the plasma was separated and stored at −20 °C following slaughter. The left-side carcass was dissected into abdominal fat following the procedure of Walstra and Merkus (1995). Subcutaneous adipose tissue (subcutaneous back fat) and liver and skeletal muscle (longissimus muscle) samples were immediately removed and frozen in liquid nitrogen, and stored at −80 °C.
Table 1.
Composition of experimental diets fed to DLY and Lantang pigs
| Group | Corn (%) | Soybean meal (%) | Wheat bran (%) | Fish meal (%) | Rice bran (%) | Zeolite powder (%) | Methionine (%) | Calcium hydrophosphate (%) | Salt (%) | Premix* (%) | Total (%) |
| DLY | 67.38 | 23 | 4 | 0 | 0 | 0.96 | 0 | 0.6 | 0.06 | 4 | 100 |
| Lantang | 56.00 | 18 | 15 | 1 | 5 | 0.90 | 0.1 | 0 | 0 | 4 | 100 |
Premix provided the following nutrients per kilogram of diet: vitamin A 1300 IU, vitamin D3 150 IU, vitamin E 11 mg, vitamin K3 0.5 mg, thiamin 1 mg, riboflavin 2 mg, pyridoxine 1 mg, vitamin B12 6 µg, niacin 7.5 mg, pantothenic acid 7 mg, biotin 0.05 mg, folacin 0.3 mg, choline chloride 300 mg, Fe 50 mg, Zn 50 mg, Mn 2 mg, Cu 3.5 mg, and I 0.14 mg
Table 2.
Nutrient contents of experimental diets fed to DLY and Lantang pigs
| Group | Digestive energy (MJ/kg) | Crude protein (%) | Available phosphorus (%) | Lys (%) | Met+cysteine (%) |
| DLY | 13.38 | 16.0 | 0.25 | 0.76 | 0.58 |
| Lantang | 12.04 | 15.1 | 0.17 | 0.72 | 0.63 |
2.2. Real-time quantitative PCR analysis
Real-time quantitative polymerase chain reaction (PCR) was done as we described previously (Tian et al., 2016). Briefly total RNA was isolated from subcutaneous adipose and liver tissue samples using TRIzol reagent (Invitrogen Co., Carlsbad, CA, USA) according to the manufacturer’s instructions. All complementary DNAs (cDNAs) were synthesized from 1 μg of total RNA using a reverse transcription kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s recommendations. Then the synthesized cDNA was diluted (1:10, v/v) and real-time quantitative PCR amplification was performed with SYBR green (TaKaRa, Tokyo, Japan) and specific primers for pig messenger RNA (mRNA) sequences (Table 3). Conditions for real-time PCR were an initial denaturation at 95 °C for 180 s, followed by 40 cycles at 95 °C for 15 s and 58 °C for 30 s, with a final elongation at 72 °C for 30 s. Each sample for each gene was amplified three times, in three independent wells, in order to have technical replicates. To normalize expression data, we used multiple internal control genes as described by Vandesompele et al. (2002). The expression stability was evaluated by the M value and pairwise variations of geNorm (Version 3.5; PrimerDesign Ltd., Southampton, Hampshire, UK). We found that β-actin had a lower M value than glyceraldehyde-3-phosphate dehydrogenase (GAPDH), both below 1.5. Thus, β-actin was ranked as the most stably expressed gene. Furthermore, the pairwise variations of β-actin and GAPDH from subcutaneous adipose and liver tissue samples of Lantang and DLY pigs were below the threshold (0.150) that required the inclusion of an additional normalization gene. Therefore, β-actin and GAPDH could be used for normalization. In the present study, the expression of the target genes relative to the β-actin was analyzed by the 2−ΔΔCt method. ΔCt=Ct (target gene)−Ct (β-actin) and ΔΔCt=ΔCt (Lantang pigs)−ΔCt (DLY pigs), where Ct is cycle threshold.
Table 3.
Primer sequences used in this study
| Gene | Sequence (5'→3') | Product size (bp) | GenBank accession |
| Cidea | Forward: CACCGTGGTAGATACAGAGG | 292 | NM_001112696.1 |
| Reverse: GGACAGGAACCGCAACA | |||
| Cideb | Forward: TGGGGACTCTGATGCTGAA | 284 | NM_001112688.1 |
| Reverse: CCCGTAGAATGTGGCTTTG | |||
| Cidec | Forward: CGGTGCCTACTCCCTTTCCT | 184 | NM_001112689.1 |
| Reverse: TGGGTCTTTGCCCTTGGT | |||
| Perilipin2 | Forward: GCTGGCGACATCTACTCA | 250 | NM_214200.2 |
| Reverse: AAGTCCACAACAGAACCCTA | |||
| DGAT1 | Forward: AGGACGGACACGACGAT | 287 | NM_214051.1 |
| Reverse: GAACGCAGTCACAGCAAA | |||
| DGAT2 | Forward: TCCTGTCTTTCCTCGTGC | 131 | NM_001160080.1 |
| Reverse: ACCTTTCTTGGGCGTGT | |||
| HNF-4α | Forward: ATCGCCACCATCGTCAA | 200 | NM_001044571.1 |
| Reverse: CCTCACCCTTTCCACTACCA | |||
| SREBP-1c | Forward: AAGCGGACGGCTCACAA | 121 | NM_214157.1 |
| Reverse: GCAAGACGGCGGATTTATT | |||
| PGC-1α | Forward: TCACCACCCAAATCCTTAT | 295 | NM_213963.2 |
| Reverse: ATTCTTCCCTCTTCAGCCT | |||
| FASN | Forward: CCTGGGAAGAGTGTAAGCA | 108 | NM_001099930.1 |
| Reverse: GGAACTCGGACATAGCG | |||
| β-actin | Forward: CATCGTCCACCGCAAAT | 210 | NC_010445 |
| Reverse: TGTCACCTTCACCGTTCC |
2.3. Analysis of biochemical variables in plasma
Plasma concentrations of high density lipoprotein (HDL), low density lipoprotein (LDL), glucose, cholesterol, NEFA, and TAG were measured using commercial kits purchased from the Nanjing Jiancheng Institute of Bioengineering, China.
2.4. Statistical analysis
Data were presented as mean±standard error of the mean (SEM). Analysis was performed using GraphPad Prism Version 5 (GraphPad Software Inc., San Diego, CA, USA). Significance was predominantly established using a two-tailed Student’s t-test. P<0.05 was considered a statistically significant difference. Correlation analysis between CIDE mRNA expression levels and apparent index (BFT, abdominal fat mass (AFM), NEFA, TAG, and glucose) was calculated using the Pearson’s correlation coefficient of the IBM SPSS Statistics 22 software (IBM Corp., Armonk, NY, USA). P<0.05 and P<0.01 were considered as significant and highly significant correlations, respectively.
3. Results
3.1. Animal performance
As shown in Table 4, Lantang pigs exhibited significantly greater BFT and AFM than DLY pigs.
Table 4.
Growth performance of DLY and Lantang pigs
| Group | Initial body weight (kg) | Final body weight (kg) | Average back fat thickness (cm) | Abdominal fat mass (kg) |
| DLY | 20.21±0.57 | 101.60±2.40 | 2.10±0.24 | 0.61±0.06 |
| Lantang | 16.12±0.63 | 68.90±1.83 | 4.05±0.03* | 1.57±0.06* |
Data are expressed as mean±SEM, with n=8.
P<0.05 vs. DLY pigs
3.2. Expression of CIDEs in adipose, liver, and skeletal muscle tissues
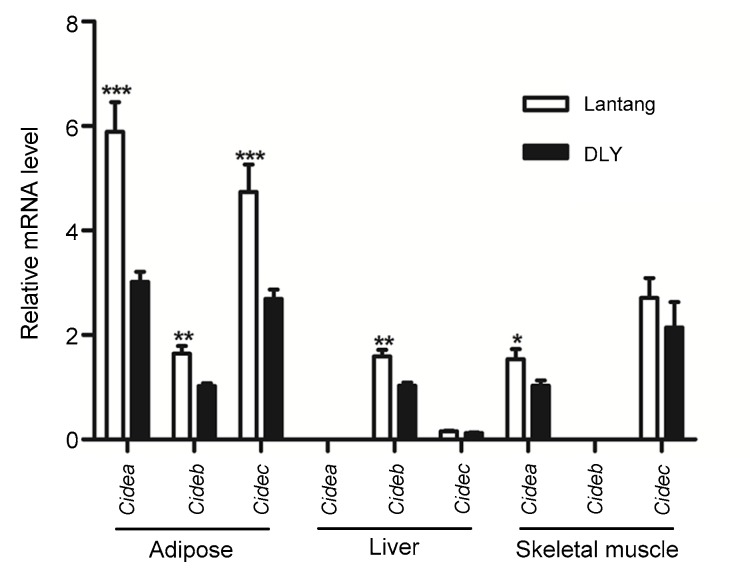
As shown in Fig. 1, porcine Cidea and Cidec mRNAs were highly expressed in white adipose tissue. Both were expressed at a relatively high level in skeletal muscle. However, Cidea was not expressed in liver, and Cidec mRNA was expressed at a much lower level in liver. Cideb mRNA was mainly expressed in porcine liver tissue, at a lower level in adipose tissue, and was not detected in skeletal muscle. Moreover, obese pigs (Lantang) had a significantly higher Cidea and Cidec mRNA levels in adipose and skeletal muscle tissues, and a higher Cideb in adipose and liver than lean breeds (DLY).
Fig. 1.
CIDE gene expression patterns Relative mRNA levels of Cidea, Cideb, and Cidec in adipose tissue, liver, and skeletal muscle of Langtang and DLY pigs. Data were expressed as mean±SEM, with n=8. * P<0.05, ** P<0.01, *** P<0.001 vs. DLY pigs
3.3. Expression levels of genes responsible for lipid metabolism
The mRNA levels of PGC-1ɑ and SREBP-1c in adipose tissue from Lantang pigs increased 3.67-fold (P<0.01) and 10-fold (P<0.001), respectively, in comparison with DLY pigs. mRNA abundances for HNF-4α, FASN, DGAT1, and DGAT2 mRNAs were significantly higher in adipose tissue from the Lantang breed than in that from DLY pigs. We observed that Lantang pigs exhibited higher PGC-1ɑ, SREBP-1c, and HNF-4α mRNA expression in liver tissue than the DLY breed (P<0.05, P<0.01, and P<0.05, respectively). FASN, DGAT1, and DGAT2 mRNA expression tended to be greater in liver tissue from obese pigs than in that from lean pigs, although at a non-significant level by the Student’s t-test. In skeletal muscle, the amounts of PGC-1ɑ, SREBP-1c, and DGAT2 mRNAs in Lantang pigs were higher than those in DLY pigs. However, the mRNA levels of perlipin2 in adipose, liver, and skeletal muscle from obese pigs were significantly decreased, as compared with their lean counterparts (Fig. 2).
Fig. 2.
Relative mRNA levels of SREBP-1c, HNF-4α, PGC-1α, FASN, DGAT1, DGAT2, and perlipin2 in adipose tissue, liver, and skeletal muscle from Lantang and DLY pigs Data were expressed as mean±SEM, with n=8. * P<0.05, ** P<0.01, *** P<0.001 vs. DLY pigs
3.4. Assessment of biochemical variables in different swine breeds
No difference in the level of HDL, LDL, or cholesterol was observed between Lantang and DLY breeds (Figs. 3d‒3f). However, plasma NEFA and TAG as well as glucose levels in Lantang pigs were significantly higher than those in DLY breeds (Figs. 3a‒3c).
Fig. 3.
Plasma levels of NEFA (a), TAG (b), glucose (c), total cholesterol (d), LDL-cholesterol (e), and HDL-cholesterol (f) in Lantang and DLY pigs
The results show higher plasma levels of NEFA, TAG, and glucose in Lantang pigs than in DLY pigs. Data were expressed as mean±SEM, with n=8. * P<0.05, ** P<0.01, *** P<0.001. NS: not statistical significant; NEFA: non-esterified fatty acid; TAG: triacylglycerol; LDL: low density lipoproteins; HDL: high density lipoproteins
3.5. Relationship between CIDE mRNA expression and BFT, AFM, NEFA, TAG, and glucose
The results of correlation analysis between CIDE mRNA expression and BFT, AFM, NEFA, TAG, and glucose are presented in Table 5. Cidea mRNA expression in adipose is positively correlated with BFT, AFM, TAG, and glucose (P<0.01) in the two breeds of pig. Cidea mRNA expression in skeletal muscle had a positively significant correlation with BFT, AFM, and NEFA (P<0.05). The correlations of Cidec mRNA expression level in adipose but not in liver or skeletal muscle, and BFT, AFM, NEFA, TAG, and glucose were positive and highly significant (P<0.01) in the two breeds. Expression of Cideb mRNA in both adipose and liver tissues correlated positively with BFT, AFM, NEFA, TAG, and glucose in two porcine breeds (P<0.01).
Table 5.
Correlation analysis between CIDE mRNA expression and BFT, AFM, NEFA, TAG, and glucose in adipose, liver, and skeletal muscle
| Parameter |
Cidea mRNA expression |
Cideb mRNA expression |
Cidec mRNA expression |
||||
| Adipose | Muscle | Adipose | Liver | Adipose | Liver | Muscle | |
| BFT | 0.788** | 0.580* | 0.707** | 0.751** | 0.743** | 0.120 | 0.214 |
| AFM | 0.818** | 0.570* | 0.668** | 0.614** | 0.668** | 0.246 | 0.170 |
| NEFA | 0.460 | 0.514* | 0.636** | 0.549** | 0.618** | −0.265 | 0.383 |
| TAG | 0.685** | 0.488 | 0.665** | 0.659** | 0.675** | 0.123 | 0.237 |
| Glucose | 0.652** | 0.385 | 0.810** | 0.583** | 0.582** | 0.103 | 0.067 |
BFT: back fat thickness; AFM: abdominal fat mass; NEFA: non-esterified fatty acid; TAG: triacylglycerol.
P<0.05,
P<0.01
4. Discussion
In the present study, we have observed several interesting findings regarding obese Lantang pigs exhibiting higher CIDE gene expression than lean DLY pigs. Some expressions of lipid metabolism genes-related CIDEs also were up-regulated or down-regulated in Lantang pigs, as compared with DLY pigs, and the correlation analysis data found that porcine CIDE mRNA abundances were positively associated with BFT, AFM, NEFA, TAG, and glucose in the two breeds. These observations were considered novel as there were no reports, to our knowledge, comprehensively evaluating the difference of porcine adipose, liver, or skeletal muscle CIDE mRNA expression between obese and lean pigs, or elucidating the relationship between the roles of CIDEs and fat accumulation.
Earlier studies have reported that Cidea was mainly expressed in the heart, and at a lower level in the brain, skeletal muscle, thymus, appendix, lymph nodes, and bone marrow, but neither in normal adult human nor mouse liver tissue (Inohara et al., 1998; Zhou et al., 2003). Our study found that porcine Cidea was highly expressed in white adipose and at a relatively high level in skeletal muscle tissue, but was not detected in liver, although Li et al. (2009) showed that porcine Cidea can be detected in the liver of Tongcheng and Large White pigs. Danesch et al. (1992) demonstrated that Cidec was an adipocyte-specific marker gene. Consistently, we found that porcine Cidec gene mRNA was expressed at a high level in white adipose tissue, at a relatively high level in skeletal muscle, and at a lower level in liver. It has been reported that Cideb mRNA was expressed in many tissues with the highest levels in the liver (Inohara et al., 1998). Our result agreed with the report that porcine Cideb was mainly expressed in the liver, but was not detected in skeletal muscle. The differential tissue distribution patterns of the three CIDE genes can imply that they may have different functions.
Previous studies reported that with CIDE deficiency mice showed a typical lean phenotype and a markedly lower adiposity index. Plasma TAG and NEFA levels, and the size of white adipocytes in CIDE-deficient mice were significantly reduced, compared with those in wild-type mice (Li et al., 2007; Nishino et al., 2008; Toh et al., 2008; Gong et al., 2009). In addition, it has been shown that over-expressed Cidec can increase lipid droplet size and enhance the accumulation of lipids (Keller et al., 2008), whereas deletion of Cidea by RNA interference in human adipocyte stimulated lipolysis (Nordström et al., 2005). In our results, we found that mRNA expressions of Cidea and Cidec in adipose tissue and skeletal muscle, and Cideb in adipose and liver from obese pigs were significantly higher than that from the lean breed. These data indicated that higher mRNA abundance for Cidea and Cidec in adipose tissue and skeletal muscle, and Cideb in liver from Lantang breeds, might play an important role in the development of fat deposition.
The molecular basis for CIDE-modulated lipid metabolism has been elucidated. CIDE deletion in mice resulted in significant reductions of the expression of SREBP-1c and its downstream target genes (ACC1, FASN, Elovl6, and SCD1) (Abu-Elheiga et al., 2003; Li et al., 2007), which were required for fatty acid synthesis (Shimomura et al., 1999) and TAG synthesis (Horton et al., 2002). Similarly, we found that CIDE-enriched Lantang pigs exhibited greater mRNA abundance for SREBP-1c and FASN. Therefore, it has been indicated that CIDEs could positively modulate the expression of SREBP-1c and its downstream target genes, and lead to increased TAG secretion and lipogenesis (Gong et al., 2009). PGC-1α and HNF-4α mRNA expression, which were critical to de novo lipogenesis and gluconeogenesis (Herzig et al., 2001) and controlling the expression of Cidea and Cideb (Hallberg et al., 2008; Chen et al., 2010; Yu et al., 2013), were higher in adipose and liver tissue from obese pigs than in those from their lean counterparts. At the same time, skeletal muscle PGC-1α mRNA abundance from Lantang pigs was increased compared to the DLY pigs. In addition, adipose DGAT1 and DGAT2 mRNA expression, which play a central modulation role in animal fat deposition (Nishizuka, 1992), were higher in Lantang pigs that in DLY pigs. Skeletal muscle DGAT2 mRNA abundance was also increased in DLY pigs. The present data uncovered that the mRNA levels of perilipin2 in adipose, liver, and skeletal muscle tissue from CIDE-enriched Lantang pigs were lower than those from DLY pigs. Our result was consistent with the report by Singaravelu et al. (2013) who demonstrated that Cideb over-expression resulted in a significant down-regulation of perilipin2 protein levels. In addition, a study by Li et al. (2012) indicated that Cideb and perilipin2 played opposite roles in modulating TAG accumulation, with Cideb as a positive regulator and perilipin2 as a negative regulator of lipid droplet size in hepatocytes. Collectively, these results revealed that the CIDE gene family modulated fat deposition through controlling the expression of lipid metabolism-related genes.
CIDE-sufficient Lantang pigs also exhibited higher plasma NEFA, TAG, and glucose levels than DLY pigs. Our results agreed with the reports that CIDE deficiency resulted in a reduction of plasma TAG and fatty acid levels (Zhou et al., 2003; Li et al., 2007; Toh et al., 2008). We also found that Lantang pigs had greater BFT and AFM than DLY pigs (Lu et al., 2008). Therefore, there may be a close association between CIDE mRNA expression and fat deposition. The further correlation analysis results uncovered an interesting finding that Cidea mRNA expression in adipose was positively correlated with BFT, AFM, TAG, and glucose, and in skeletal muscle was positively correlated with BFT, AFM, and NEFA in the two breeds of pigs. Adipose and liver Cideb mRNA abundance and BFT, AFM, TAG, NEFA, and glucose had a positively significant correlation in the two breeds. Cidec mRNA in adipose but not in liver or skeletal muscle was positively correlated with BFT, AFM, TAG, NEFA, and glucose in Lantang and DLY pigs. We made a correlation analysis between porcine CIDE mRNA and fat deposition-related factors. These results further suggested that CIDE genes contributed to fat deposition of the fatty Lantang pigs.
5. Conclusions
In conclusion, our data suggest that the porcine CIDEs possibly modulated lipid metabolism and contributed to the development of fat deposition and obesity.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31402086), the China Agriculture Research System (CARS-36), the National Basic Research Program (973) of China (Nos. 2013CB127301 and 2013CB127304), the Guangdong International Science and Technology Cooperation Program (No. 2014A050503049), and the Science and Technology Program of Guangdong Province (No. 2016A020210041), China
Compliance with ethics guidelines: Yue-qin QIU, Xue-fen YANG, Xian-yong MA, Yun-xia XIONG, Zhi-mei TIAN, Qiu-li FAN, Li WANG, and Zong-yong JIANG declare that they have no conflict of interest.
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- 1.Abu-Elheiga L, Oh W, Kordari P, et al. Acetyl-CoA carboxylase 2 mutant mice are protected against obesity and diabetes induced by high-fat/high-carbohydrate diets. Proc Natl Acad Sci USA. 2003;100(18):10207–10212. doi: 10.1073/pnas.1733877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahima RS, Flier JS. Adipose tissue as an endocrine organ. Trends Endocrinol Metab. 2000;11(8):327–332. doi: 10.1016/S1043-2760(00)00301-5. [DOI] [PubMed] [Google Scholar]
- 3.Bell M, Wang H, Chen H, et al. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57(8):2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernlohr DA, Jenkins AE, Bennaars AA. Adipose tissue and lipid metabolism. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes, 4th Ed. Elsevier, Amsterdam; 2002. pp. 263–289. [DOI] [Google Scholar]
- 5.Chen ZJ, Norris JY, Finck BN. Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) stimulates VLDL assembly through activation of cell death-inducing DFFA-like effector B (CideB) J Biol Chem. 2010;285(34):25996–26004. doi: 10.1074/jbc.M110.141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZM, Qi XH, Zhang H, et al. Changes of leptin and leptin receptor gene expression in subcutaneous fat and hypothalamus of Lantang and Landrace pigs. J Huazhong Agric Univ. 2010;29(1):67–70. (in Chinese) [Google Scholar]
- 7.Danesch U, Hoeck W, Ringold GM. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J Biol Chem. 1992;267(10):7185–7193. [PubMed] [Google Scholar]
- 8.Girousse A, Langin D. Adipocyte lipases and lipid droplet-associated proteins: insight from transgenic mouse models. Int J Obes (Lond) 2012;36(4):581–594. doi: 10.1038/ijo.2011.113. [DOI] [PubMed] [Google Scholar]
- 9.Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol. 2009;20(2):121–126. doi: 10.1097/MOL.0b013e328328d0bb. [DOI] [PubMed] [Google Scholar]
- 10.Hallberg M, Morganstein DL, Kiskinis E, et al. A functional interaction between RIP140 and PGC-1α regulates the expression of the lipid droplet protein CIDEA. Mol Cell Biol. 2008;28(22):6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 12.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulver MW, Berggren JR, Cortright RN, et al. Skeletal muscle lipid metabolism with obesity. Am J Physiol Endocrinol Metab. 2003;284(4):E741–E747. doi: 10.1152/ajpendo.00514.2002. [DOI] [PubMed] [Google Scholar]
- 14.Inohara N, Koseki T, Chen S, et al. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17(9):2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang JP, Zhou J, Chen J, et al. Effect of chicken egg yolk antibody against adipose tissue plasma membranes on carcass composition and lipogenic hormones and enzymes in pigs. Livestock Sci. 2007;107(2-3):235–243. doi: 10.1016/j.livsci.2006.09.020. [DOI] [Google Scholar]
- 16.Keller P, Petrie JT, de Rose P, et al. Fat-specific protein 27 regulates storage of triacylglycerol. J Biol Chem. 2008;283(21):14355–14365. doi: 10.1074/jbc.M708323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan LT, Huang LS, Ma JW, et al. Experiment for comparing the performance of Erhualian pig double cross combinations and that of Duroc×(Landrace×Large Yorkshire) three-way cross combination. J Southwest Univ Natl. 2004;30(6):741–744. [Google Scholar]
- 18.Leonhardt M, Langhans W. Fatty acid oxidation and control of food intake. Physiol Behav. 2004;83(4):645–651. doi: 10.1016/j.physbeh.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 19.Li JZ, Ye J, Xue B, et al. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 2007;56(10):2523–2532. doi: 10.2337/db07-0040. [DOI] [PubMed] [Google Scholar]
- 20.Li JZ, Lei Y, Wang Y, et al. Control of cholesterol biosynthesis, uptake and storage in hepatocytes by Cideb. Biochim Biophys Acta. 2010;1801(5):577–586. doi: 10.1016/j.bbalip.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Li XH, Ye J, Zhou LK, et al. Opposing roles of cell death-inducing DFF45-like effector B and perilipin 2 in controlling hepatic VLDL lipidation. J Lipid Res. 2012;53(9):1877–1889. doi: 10.1194/jlr.M026591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YH, Lei T, Chen XD, et al. Molecular cloning, chromosomal location and expression pattern of porcine CIDEa and CIDEc . Mol Biol Rep. 2009;36(3):575–582. doi: 10.1007/s11033-008-9216-5. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Millar JS, Cromley DA, et al. Knockdown of Acyl-CoA: diacylglycerol acyltransferase 2 with antisense oligonucleotide reduces VLDL TG and ApoB secretion in mice. Biochim Biophys Acta. 2008;1781(3):97–104. doi: 10.1016/j.bbalip.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Lu P, Li DF, Yin JD, et al. Flavour differences of cooked longissimus muscle from Chinese indigenous pig breeds and hybrid pig breed (Duroc×Landrace×Large White) Food Chem. 2008;107(4):1529–1537. doi: 10.1016/j.foodchem.2007.10.010. [DOI] [Google Scholar]
- 25.Malaguarnera M, Di Rosa M, Nicoletti F, et al. Molecular mechanisms involved in NAFLD progression. J Mol Med (Berl) 2009;87(7):679–695. doi: 10.1007/s00109-009-0464-1. [DOI] [PubMed] [Google Scholar]
- 26.Nishino N, Tamori Y, Tateya S, et al. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest. 2008;118(8):2808–2821. doi: 10.1172/JCI34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase. Science. 1992;258(5082):607–614. doi: 10.1126/Science.1411571. [DOI] [PubMed] [Google Scholar]
- 28.Nordström EA, Rydén M, Backlund EC, et al. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-α)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes. 2005;54(6):1726–1734. doi: 10.2337/diabetes.54.6.1726. [DOI] [PubMed] [Google Scholar]
- 29.O'Hea EK, Leveille GA. Significance of adipose tissue and liver as sites of fatty acid synthesis in the pig and the efficiency of utilization of various substrates for lipogenesis. J Nutr. 1969;99(3):338–344. doi: 10.1093/jn/99.3.338. [DOI] [PubMed] [Google Scholar]
- 30.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274(4):30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 31.Singaravelu R, Lyn RK, Srinivasan P, et al. Human serum activates CIDEB-mediated lipid droplet enlargement in hepatoma cells. Biochem Biophys Res Commun. 2013;441(2):447–452. doi: 10.1016/j.bbrc.2013.10.080. [DOI] [PubMed] [Google Scholar]
- 32.Tian ZM, Ma XY, Yang XF, et al. Influence of low protein diets on gene expression of digestive enzymes and hormone secretion in the gastrointestinal tract of young weaned piglets. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2016;17(10):742–751. doi: 10.1631/jzus.B1600229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toh SY, Gong J, Du G, et al. Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of Fsp27 deficient mice. PLoS ONE. 2008;3(8):e2890. doi: 10.1371/journal.pone.0002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034–1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walstra P, Merkus GSM. Procedure for assessment of the lean meat percentage as a consequence of the new EU reference dissection method in pig carcass classification. Report ID-DLO 96.014, Zeist, the Netherlands; 1995. [Google Scholar]
- 36.Yonezawa T, Kurata R, Kimura M, et al. Which CIDE are you on? Apoptosis and energy metabolism. Mol Biosyst. 2011;7(1):91–100. doi: 10.1039/c0mb00099j. [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Wang H, Zhao J, et al. Expression of CIDE proteins in clear cell renal cell carcinoma and their prognostic significance. Mol Cell Biochem. 2013;378(1):145–151. doi: 10.1007/s11010-013-1605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Yon Toh S, Chen Z, et al. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet. 2003;35(1):49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]