Abstract
The genus Culicoides includes vectors of important animal diseases such as bluetongue and Schmallenberg virus (BTV and SBV). This genus includes 1300 species classified in 32 subgenera and 38 unclassified species. However, the phylogenetic relationships between different subgenera of Culicoides have never been studied. Phylogenetic analyses of 42 species belonging to 12 subgenera and 8 ungrouped species of genus Culicoides from Ecuador, France, Gabon, Madagascar and Tunisia were carried out using two molecular markers (28S rDNA D1 and D2 domains and COI mtDNA). Sequences were subjected to non-probabilistic (maximum parsimony) and probabilistic (Bayesian inference (BI)) approaches. The subgenera Monoculicoides, Culicoides, Haematomyidium, Hoffmania, Remmia and Avaritia (including the main vectors of bluetongue disease) were monophyletic, whereas the subgenus Oecacta was paraphyletic. Our study validates the subgenus Remmia (= Schultzei group) as a valid subgenus, outside of the subgenus Oecacta. In Europe, Culicoides obsoletus, Culicoides scoticus and Culicoides chiopterus should be part of the Obsoletus complex whereas Culicoides dewulfi should be excluded from this complex. Our study suggests that the current Culicoides classification needs to be revisited with modern tools.
Keywords: Culicoides spp., Phylogeny, Ecuador, France, Gabon, Madagascar, Tunisia, 28S, COI
Abstract
Le genre Culicoides comprend des vecteurs de maladies animales importantes telles que la fièvre catarrhale et le virus Schmallenberg. Ce genre comprend 1300 espèces classées dans 32 sous-genres et 38 espèces non classées. Cependant, les relations phylogénétiques entre les différents sous-genres de Culicoides n’ont jamais été étudiées. Des analyses phylogénétiques de 42 espèces appartenant à 12 sous-genres et 8 espèces non groupées du genre Culicoides d’Équateur, de France, du Gabon, de Madagascar et de Tunisie ont été réalisées en utilisant deux marqueurs moléculaires (ADNr 28S domaines D1 et D2 et ADNmt COI). Les séquences ont été soumises à des approches non probabilistes (maximum de parcimonie) et probabilistes (inférence bayésienne). Les sous-genres Monoculicoides, Culicoides, Haematomyidium, Hoffmania, Remmia et Avaritia (y compris les principaux vecteurs de la fièvre catarrhale du mouton) étaient monophylétiques alors que le sous-genre Oecacta était paraphylétique. Notre étude valide le sous-genre Remmia (= groupe Schultzei) sous la forme d’un sous-genre valide, en dehors du sous-genre Oecacta. En Europe, Culicoides obsoletus, Culicoides scoticus et Culicoides chiopterus devraient faire partie du complexe Obsoletus alors que Culicoides dewulfi devrait être exclu de ce complexe. Notre étude suggère que la classification actuelle de Culicoides doit être revisitée avec des outils modernes.
Introduction
Biting midges of the genus Culicoides Latreille 1809 (Diptera: Ceratopogonidae) are among the world’s smallest haematophagous flies, measuring from 1 to 3 mm, and are described worldwide, except in Antarctica and New Zealand [45]. They are mainly known as vectors of bluetongue virus (BTV), Schmallenberg virus (SBV) and Oropouche virus (OROV) [12].
Currently, approximately 1300 living and 42 fossil species of Culicoides have been described worldwide. Their classification includes 32 subgenera [9] and 38 groups although 13% of occurring species remain ungrouped [11]. This classification is exclusively typological, based on common morphological similarities (e.g. characteristics of reproductive organs, wings, antennae and palps), without any phylogenetic considerations. As most species feature spotted wings, the accurate identification of adults is largely based on subtle variations in size, shape and position of spots that form wing patterns [61–63].
In Africa, Asia and Europe, Culicoides imicola and the Obsoletus complex (both from the subgenus Avaritia Fox) are considered the most important vectors of BTV, SBV and epizootic haemorrhagic disease virus [20, 35, 45, 60]. Other groups of Culicoides are also involved in the transmission of these viruses, such as the Schultzei group (now in the subgenera Remmia Glukhova and sometimes synonymised with Oecacta Poey) [4, 11], Culicoides pulicaris and C. punctatus (Culicoides Latreille), C. circumscriptus (Beltranmyia Vargas) [45], and C. paraensis (Haematomydium Goeldi) only for OROV in South America [12].
Since the recent European bluetongue epizootic outbreak, there has been growing interest in DNA barcoding of Culicoides based on the mitochondrial DNA (mtDNA) cytochrome oxidase I (COI) gene, ribosomal (rDNA) regions such as internal transcribed spacer 1 (ITS1) and internal transcribed spacer 2 (ITS2), and the nuclear CAD gene [29]. The rise of DNA barcoding and the lack of taxonomic experts thus enabled COI sequencing to become a tool for rapid identification of Culicoides species [1].
Ribosomal DNA markers have been used to investigate phylogenies of closely related species (ITS1 and ITS2: [25, 26, 49]; 28S: [27, 28]), interspecific genetic distances (ITS1 [43, 47]) and population structure (ITS1: [53]) within Culicoides. The sequences obtained with ITS1 are generally of low quality [47]. Polymerase chain reaction (PCR) products with ITS2 seem to include many different sequences, even from one individual sample [38].
The lack of phylogenetic data about Culicoides does not allow hypotheses about the vector competence for diseases caused by different Culicoides-borne viruses. Due to the wide distribution and the great economic importance of veterinary diseases transmitted by biting midges, it seems important to build a modern classification of these insects based on phylogenetic studies to help in epidemiological analyses.
In this study, we carried out a phylogenetic analysis of 42 Culicoides species from Europe, America and Africa (including Madagascar) using specimens available in our laboratory. Our sampling included major proven vectors of diseases (i.e. subgenera Avaritia, Culicoides, Haematomydium, and Schultzei group). In each case, the mtDNA COI and the D1 and D2 regions of the 28S rDNA were analysed. The latter regions were chosen based on the fact that they appear to contain major phylogenetic information at the considered taxonomic level [18, 30, 51] especially for Culicoides [27, 28, 58].
Material and methods
Collection of Culicoides and identification
Midges were collected in Ecuador, France, Gabon, Madagascar and Tunisia between 2009 and 2010 using ultraviolet CDC traps and standard CDC miniature light traps (John W. Hock Company, Gainesville, FL, USA). Insects were stored in ethanol 95°. Specimens were identified to species, species group or subgenera (Table 1; Figs. 1–4) using different morphological keys [13, 14, 17, 21, 22, 24, 31, 36, 62].
Table 1.
List of Culicoides spp. used in the phylogenetic analyses, classification of Borkent, 2014.
| Subgenus | Taxa present |
Country | No. (codification) | GenBank accession number |
||
|---|---|---|---|---|---|---|
| Group | Species | COI | D1D2 | |||
| Anilomyia | C. metagonatus | EC | EC-meta-1-D458 | KY707782 | KF286339 | |
| C. chiopterus | FR | FR-chio1-P6C61 | KY707805 | KF286340 | ||
| C. dewulfi | FR | FR-dew1-P5C46 | HM022877 | KF286341 | ||
| FR-dew2- P3C17* | HM022878 | |||||
| C. dubitatus | MA | MA-dub1-D358 | KY707796 | KF286342 | ||
| GA | GA-dub2- D558 | KY707795 | KF286343 | |||
| MA | MA-dub3 D379 | KY707797 | KF286344 | |||
| Avaritia | C. imicola | TU | TU-im1-S4Cf3 | KJ729975 | KF286345 | |
| TU-im2-S6Cf111* | KJ729976 | |||||
| C. kibatiensis | MA | MA-kib1-D364* | KY707781 | KF286348 | ||
| MA-kib2-D401 | ||||||
| C. miombo | MA | MA-mio1-D394* | KY707800 | KF286349 | ||
| MA-mio2-D412 | ||||||
| C. sp. | GA | GA-img1-D439 | KY707791 | KF286346 | ||
| GA-img2-D550 | KY707790 | KF286347 | ||||
| C. obsoletus | FR | FR-obs-1-P2C12* | ||||
| FR-obs-2-D223 | HM022852 | KF286350 | ||||
| C. scoticus | FR | FR-sco1-P7C5 | HM022875 | KF286351 | ||
| FR-sco2 P6C25 | HM022857 | KF286352 | ||||
| Beltranmyia | C. circumscriptus | TU | TU-cir2-B1Cf31* | KJ729971 | KF286353 | |
| C. impunctatus | FR | FR-del2-D91 | KY707808 | KF286355 | ||
| FR-del3-D94* | ||||||
| Culicoides | C. lupicaris | FR | FR-lup1-P5C34 | KY707776 | KF286354 | |
| C. newsteadi | TU | TU-new1-S3CM42 | KKJ729989 | KF286356 | ||
| TU-new2-S6Cf51 | KJ729990 | KF286357 | ||||
| C. punctatus | FR | FR-pun1-D327 | KY707806 | KF286358 | ||
| FR-pun2-D242* | ||||||
| FR-pun3-D250* | ||||||
| Haematomyidium | C. limonensis | EC | EC-para2-D460 | KY707809 | KF286360 | |
| C. paraensis | EC | EC-para1-D423 | KF286359 | |||
| Hoffmania | Guttatus group | C. diabolicus | EC | EC-bat1-D451 | KY707783 | KF286361 |
| EC-bat2-D453 | KY707787 | KF286362 | ||||
| C. guttatus | EC | EC-gu1-D481 | KY707785 | KF286363 | ||
| Hylas group | C. hylas | EC | EC-hyl1-D449 | KF286364 | ||
| C. pseudoheliconiae | EC | EC-hyl2-D505 | KY707784 | KF286365 | ||
| Meijerehelea | C. distinctipennis | GA | GA-leu1 D555 | KY707792 | KF286366 | |
| Monoculicoides | C. nubeculosus | FR | FR-nub-D179 | KF178273 | KF286367 | |
| C. parroti | FR | FR-par-D27 | KF178276 | KF286368 | ||
| C. puncticollis | TU | TU-pco1-B7CM49 | KJ730002 | KF286369 | ||
| TU-pco2-B7Cf60* | KJ29998 | KJ730024 | ||||
| Oecacta | C. cataneii/ | TU | TU-cag1-B2CM132 | KJ729968 | KF286388 | |
| C. gejgelensis | TU-cag2-B2Cf34 | KJ729967 | KF286389 | |||
| C. festivipennis | FR | FR-fes2-D66 | KY707777 | KF286377 | ||
| FR-fes3-D103* | ||||||
| C. jumineri | TU | TU-jum1-S3Cf124 | KJ729979 | KF286383 | ||
| TU-jum2-S3CM1* | KJ729980 | KF286384 | ||||
| TU-jum3-S3CM71 | KJ729982 | KF286384 | ||||
| C. sahariensis | TU | TU-sah2-B28Cf6* | KJ30004 | KF286387 | ||
| Remmia | C. enderleini | MA | MA-scg2-D378* | KF186429 | KF286379 | |
| MA-scg3-D363 | ||||||
| C. kingi | TU | TU-kin1-S3Cf167 | KJ729985 | KF286338 | ||
| C. nevilli | MA | MA-scg1-D357 | KF186428 | KF286378 | ||
| Trithecoides | C. fulvithorax/C. ochrothorax | GA | GA-fuo1-D437 | KY707793 | KF286371 | |
| Wirthomyia | C. segnis | FR | FR-seg1-D113* | KY707778 | KF286372 | |
| FR-seg2-D108 | ||||||
| Unplaced 1 | Carpenteri group | C. belemensis | EC | EC-bel1-D477 | KY707786 | KF286373 |
| Unplaced 2 | Fluvialis group | C. castillae | EC | EC-cast1-D474 | KF286374 | |
| EC-cast2-D472* | ||||||
| C. tetrathyris like | EC | EC-fug3-D470 | KY707788 | KF286375 | ||
| C. tetrathyris | EC | EC-tetra1-D517 | KF286370 | |||
| Unplaced 3 | Leoni group | C. glabellus | EC | EC-leg1-D528 | KY707789 | KF286376 |
| Unplaced 4 | Milnei group | C. moreli | MA | MA-mor1-D420 | KY707804 | KF286382 |
| C. zuluensis | MA | MA-mig1-D388 | KY707802 | KF286380 | ||
| MA-mig2-D365 | KY707803 | KF286381 | ||||
| Unplaced 5 | C. paolae | TU | TU-pao2-S5CM1 | KJ729992 | KF286385 | |
| TU-pao3-S3Cf66 | KJ729991 | KF286386 | ||||
EC: Ecuador; FR: France; GA: Gabon; MA: Madagascar; TU: Tunisia. D1D2 rDNA sequences (*specimens having identical sequences) and COI sequences).
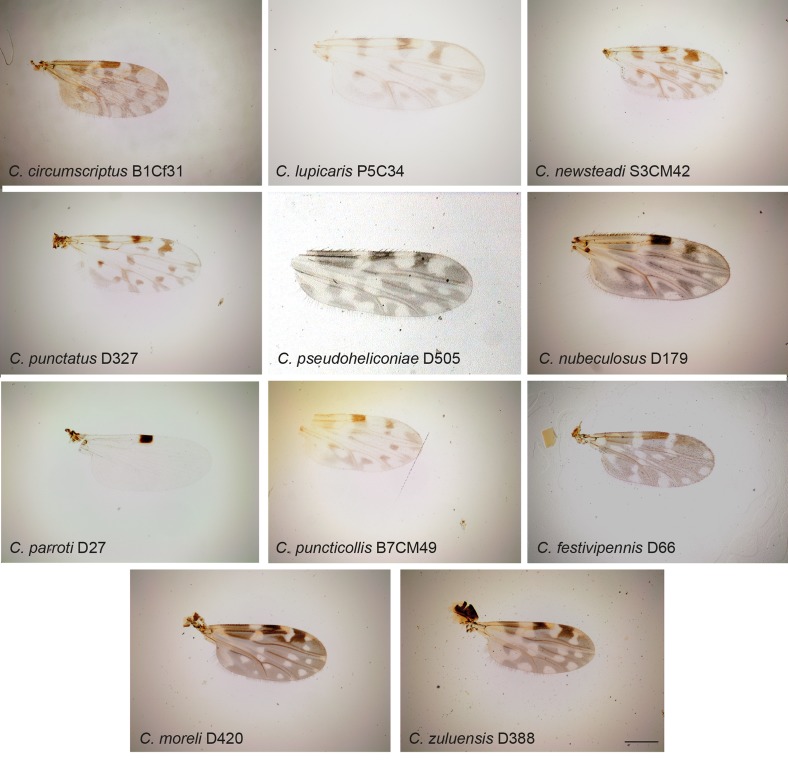
Figure 1.
Culicoides wing pattern details of species included in our study. The specimen codes are linked with the table. The wings were photographed using a ×4 lens. Bars = 200 μm.
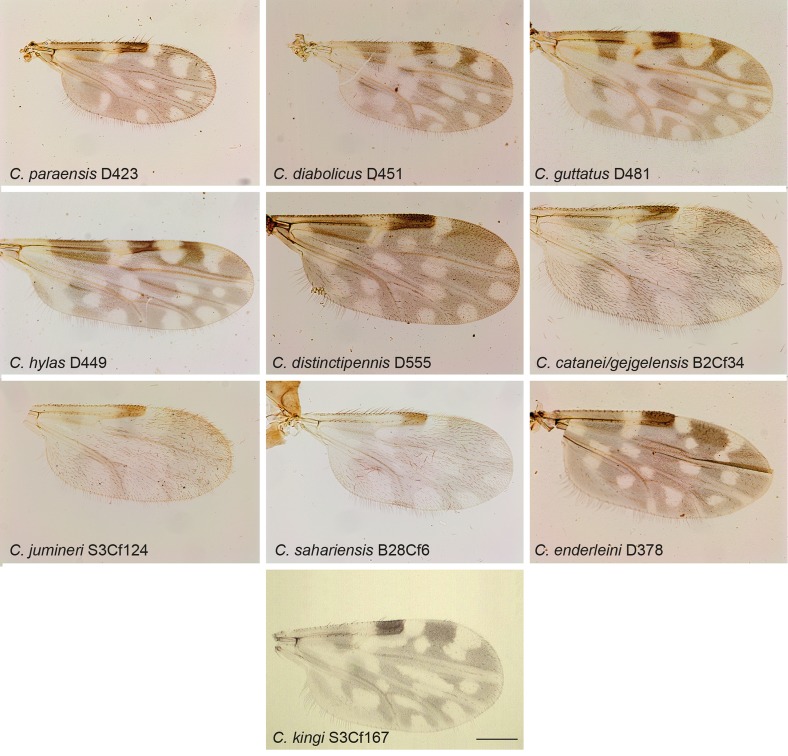
Figure 4.
Culicoides wing pattern details of species included in our study. The specimen codes are linked with the table. The wings were photographed using a ×10 lens. Bars = 200 μm.
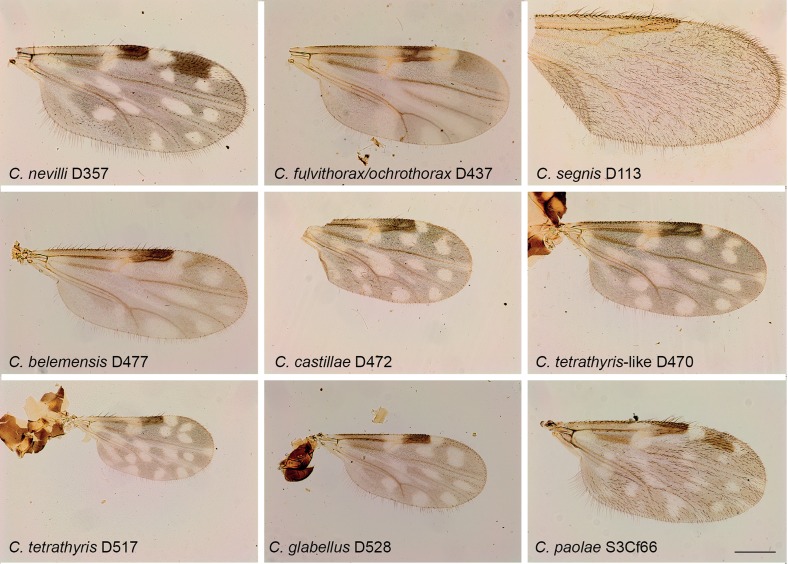
Figure 2.
Culicoides wing pattern details of species included in our study. The specimen codes are linked with the table. The wings were photographed using a ×10 lens. Bars = 200 μm.
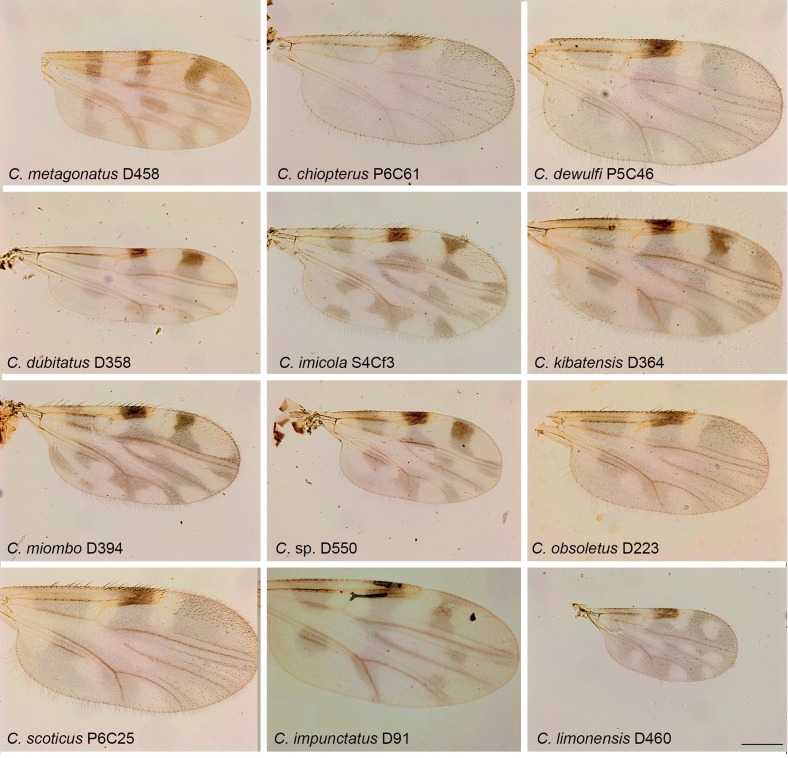
Figure 3.
Culicoides wing pattern details of species included in our study. The specimen codes are linked with the table. The wings were photographed using a ×10 lens. Bars = 200 μm.
Specimen identification was performed after mounting the head, wings and spermathecae on microscope slides, leaving the thorax and legs for subsequent DNA extraction [2]. Consequently, we were unable to identify C. fulvithorax and C. ochrothorax without their thorax that includes their discriminant character. Moreover, the accurate identification of females of some closely related specimens, such as C. cataneii and C. gejgelensis, was not possible [36]. Two specimens from Gabon, belonging to subgenus Avaritia, present new morphological characters compared with currently known species; hereafter we will refer to these specimens as Culicoides sp. At least two specimens of each species were sequenced, except for 13 species from which only one specimen was available (Table 1; Figs. 1–4).
A total of 68 specimens belonging to 42 species were analysed: 34 species belonging to the subgenera Anilomyia, Avaritia, Beltranmyia, Culicoides, Haematomyidium, Hoffmania, Meijerehelea, Monoculicoides, Oecacta, Remmia, Trithecoides and Wirthomyia, and 8 species belonging to unclassified groups [11]. Species distribution included: (i) Ecuadorian specimens (12 species) assigned to subgenera Anilomyia, Haematomyidium and Hoffmania and the unclassified groups Carpenteri group, Fluvialis group and Leoni group; (ii) French specimens (11 species) assigned to subgenera Avaritia, Culicoides, Monoculicoides, Oecacta and Wirthomyia; (iii) four Gabonese specimens assigned to subgenera: Avaritia, Meijerehelea and Trithecoides; (iv) Malagasy specimens (7 species) assigned to subgenera Avaritia, Remmia and to the Milnei group and (v) Tunisian specimens (8 species) assigned to six subgenera (Avaritia, Beltranmyia, Culicoides, Monoculicoides, Oecacta, Remmia) and 1 species was C. paolae (incertae sedis).
DNA extraction and PCR amplification
DNA was extracted from individual Culicoides using the QIAmp DNA Mini Kit (Qiagen GmbH, Hilden, Germany), following the manufacturer’s instructions. Polymerase chain reactions (PCRs) for D1-D2 and cytochrome oxidase genes were performed in a 50 μL volume using 5 μL of DNA solution and 50 pmol of primers C′1 (5′-ACCCGCTGAATTTAAGCAT-3′) and D2 (5′-TCCGTGTTTCAAGACGGG-3′) for D1-D2 [18] and C1J1718 (5′-GGAGGATTTGGAAATTGATTAGT-3′), C1N2191 (5′-CAGGTAAAATTAAAATATAAACTTCTGG-3′) or LepF (5′-ATTCAACCAATCATA AAGATA TTGG-3′) and LepR (5′-TAAACTTCTGGATGTCCAAAAAATCA-3′) for COI [2, 57].
Amplification conditions for D1-D2 were: initial denaturation step at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and extension at 68 °C for 60 s followed by a final extension at 68 °C for 10 min. For COI amplification, conditions included: (1) initial denaturation step at 95 °C for 15 min, then 5 cycles at 95 °C for 40 s, 45 °C for 40 s, 72 °C for 1 min, were followed by 45 cycles at 95 °C for 40 s, 50 °C for 40 s, 72 °C for 1 min and a final extension step at 72 °C for 20 min for C1J1718/C1N2191, and (2) initial denaturation step at 94 °C for 3 min, 5 cycles of denaturation at 94 °C for 30 s, annealing at 45 °C for 90 s and extension at 68 °C for 60 s were followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 51 °C for 90 s, and extension at 68 °C for 60 s and a final extension at 68 °C for 10 min for LepF1/LepR. Amplicons were analysed by electrophoresis in 1.5% agarose gel stained with 0.1% ethidium bromide. All sequences obtained are available in GenBank (Table 1).
Phylogenetic analyses
Most sequences of COI and D1-D2 genes were analysed separately and concatenated, except for four specimens (Culicoides castillae, C. hylas, C. paraensis and C. tetrathyris). The phylogenetic tree was constructed using both non-probabilistic (maximum parsimony, MP) and probabilistic approaches (Bayesian inference, BI), using Atrichopogon sp. and Forcipomyia sp. as outgroups [7].
Maximum parsimony analysis was carried out with PAUP* 4.0b10 [59] by selecting the heuristic search option with tree bisection reconnection branch swapping (TBR) and 1000 random sequence addition (RSA). All sites were equally weighed but a step matrix (ponderation TS/TV = 2) was applied. Sequences were edited and aligned manually using Se-Al [52]. The insertion of several interlocked gap zones was therefore necessary to align sequences. Sequence alignment was performed respecting the criteria defined by [6]: (1) to minimise the number of inferred mutations (number of steps); (2) to prefer substitution to insertion-deletion, and (3) to prefer transitions to transversions because they have a higher probability of occurrence.
A total of 423 bp and 613 bp were analysed for COI and D1-D2, respectively.
For the model-based approach, the best-fit model of nucleotide substitutions was computed with jModelTest v2.1.4 [16] using the Akaike Information Criterion (AIC). The Hasegawa, Kishino and Yano (HKY) +I+Γ model was indicated as the best-fit model for the mitochondrial COI gene. The general time reversible (GTR) +I+Γ model was indicated as the best-fit model for both D1-D2 and concatenated markers (rDNA marker D1D2 and COI). Bayesian analyses were carried out using MrBayes 3.1.2 [54] with 4,000,000 generations, 10,000 of the saved trees were discarded, and the 30,000 remaining were used to construct the resulting BI tree. The robustness of tree nodes was assessed by clade posterior probability values (CPP).
A first maximum parsimony analysis on COI sequences showed trees of 2786 steps with a consistency index (CI of 0.225) and a retention index (RI of 0.443). The codon position 3 in the COI gene was found to have saturated transition information as compared to position 1+2 (data not shown). Therefore, we decided to remove the codon position 3. A new analysis was performed with COI codon position 1+2 including 10,000,000 generations. As many as 25,000 of the saved trees were discarded. Both COI and D1-D2 sequences were analysed independently using the BI and MP approaches and a concatenated fragment using the BI approach.
Results
A first run of amplification was carried out using the C1J1718/C1N2191 primers. Pseudogenes were amplified for six specimens from Ecuador (C. castillae, C. hylas, C. pseudoheliconiae, C. guttatus, C. tetrathyris and C. diabolicus) and one specimen from Gabon (C. distinctipennis). Consequently, the COI of these specimens was tentatively amplified and sequenced using the LepF1/LepR primers. Finally, the COI of all these specimens was obtained, except for Culicoides castillae and C. tetrathyris.
For the parsimony analysis, the sequences for COI with 127 variable characters, of which 97 were parsimony-informative were analysed. The most parsimonious trees obtained were 557 steps long. With D1D2 sequences, 203 variable characters were found, out of which 169 were parsimony-informative. The most parsimonious trees obtained were 639 steps long. The ribosomal gene had a much higher proportion of parsimony-informative sites than the mitochondrial gene (Table 2).
Table 2.
Information about DNA sequences used in this study, in relation to one of the most parsimonious trees resulting from the combined analysis and mapped in a combined matrix.
| DNA region | COI | D1-D2 |
|---|---|---|
| Aligned matrix (bp)/number of characters included | 423 | 661 |
| Number of constant sites | 352 | 456 |
| Number of variable sites | 127 | 203 |
| Number of parsimoniously informative characters | 97 | 169 |
| Number of variable parsimony uninformative characters | 30 | 34 |
| Tree length (L) | 557 | 639 |
| CI | 0.227 | 0.436 |
| RI | 0.361 | 0.689 |
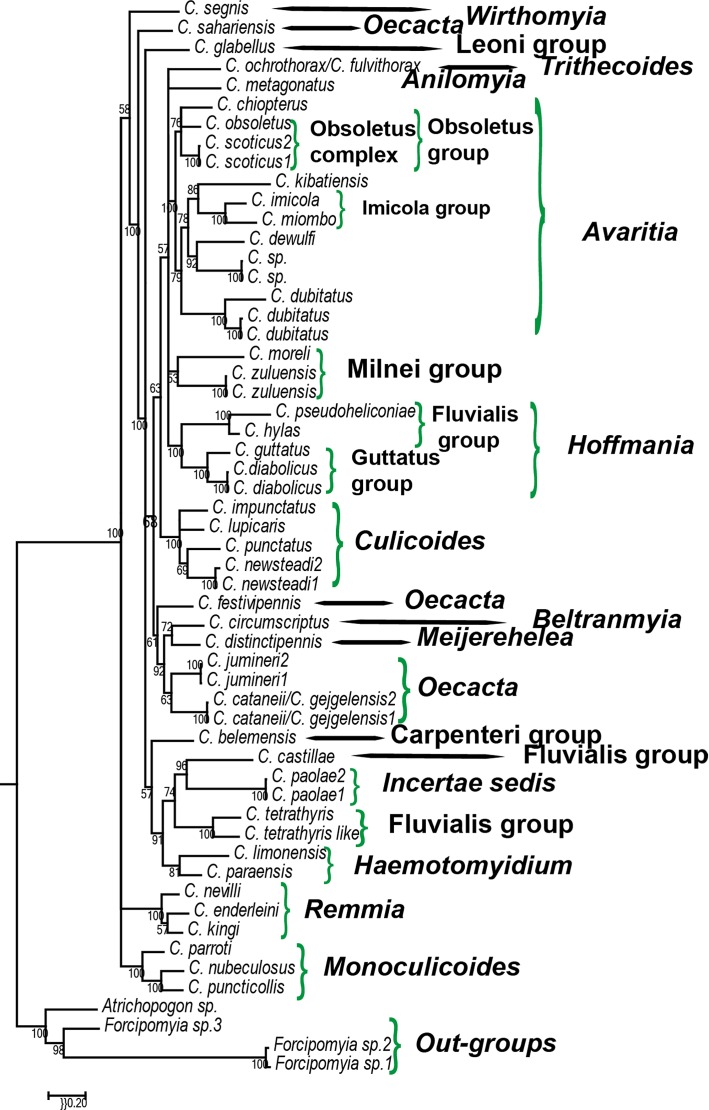
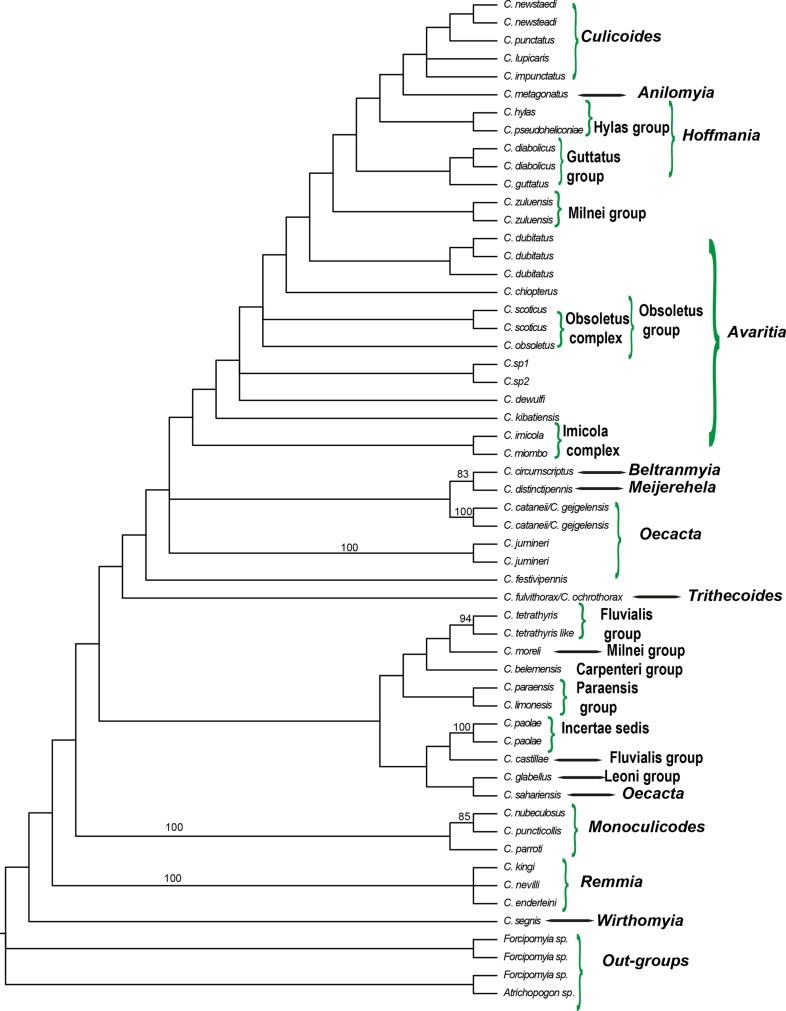
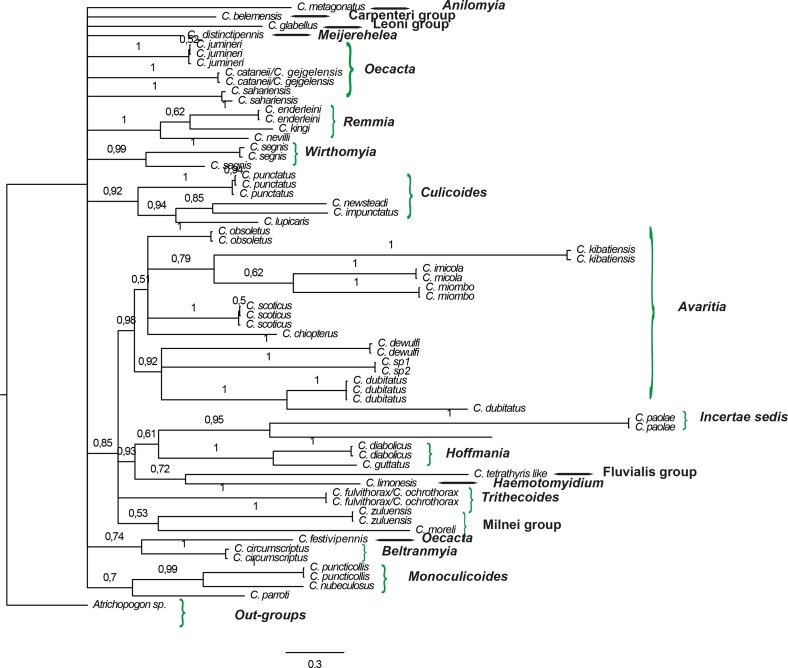
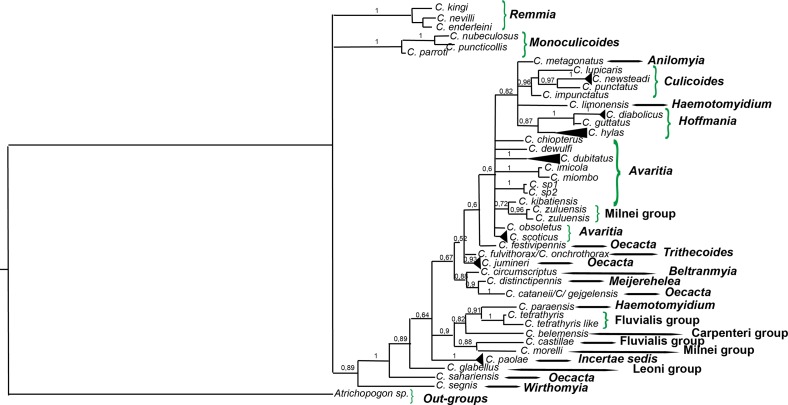
Topologies of the trees obtained by MP and BI are presented in the Appendices. Our main findings were that: (i) the MP COI tree (data not shown) is unusable; (ii) the MP D1D2 tree (Appendix 1) shows that the subgenera Monoculicoides, Culicoides, Haematomyidium and Remmia were monophyletic, whereas the subgenera Hoffmania and Avaritia were paraphyletic; (iii) the BI COI tree (Appendix 2) shows that the subgenera Avaritia, Culicoides, Hoffmania, Monoculicoides and Remmia were monophyletic, whereas the subgenus Oecacta was paraphyletic, and (iv) the BI D1D2 tree (Appendix 3) shows that the subgenera Culicoides, Hoffmania, Monoculicoides and Remmia were monophyletic, whereas the subgenus Oecacta was paraphyletic.
Results of the concatenated Bayesian inference analysis of Culicoides relationships are shown in Figure 5 and presented below.
Figure 5.
Bayesian tree resulting from the phylogenetic analysis of the concatenated dataset according to the best-fit partitioning strategy. Robustness of nodes is indicated by the posterior probability values (%).
According to the combined data analysis, the genus Culicoides was clearly monophyletic.
The subgenus Culicoides was also monophyletic: Tunisian specimens of C. newsteadi were grouped in one clade (CPP = 100) with French specimens of C. punctatus, C. lupicaris and C. impuctatus.
The subgenus Hoffmania (species from Ecuador) displayed two clusters with C. batesi and C. guttatus as the sister group of the Hylas group. The tree shows that the subgenus Hoffmania was monophyletic (CPP = 100), and in the position of sister species of the Milnei group and the subgenera Trithecoides, Anilomyia and Avaritia.
The subgenus Avaritia was also monophyletic (CPP = 100). C. chiopterus, C. obsoletus and C. scoticus were grouped together as a clade, the sister group of all other members of the subgenus Avaritia (CPP = 78). C. dewulfi was shown to be closely related to the new Culicoides species from Gabon (CPP = 92). The analysis also revealed that the Imicola group was monophyletic. C. imicola s. st. is the sister species of C. miombo (CPP = 100) and C. kibatiensis is the sister species of them (CPP = 86).
The Schultzei group, which includes C. enderleini, C. nevilli and C. kingi, was monophyletic (CPP = 100).
The subgenus Monoculicoides, including C. nubeculosus, C. parroti and C. puncticollis, was monophyletic (CPP = 100).
Culicoides distinctipennis (subgenus Meijerehela) from Madagascar and C. festivipennis (subgenus Oecacta) from France exhibited similar wing patterns (white spots on the wing apex) but they were not grouped in our cladogram (CPP = 61).
The Milnei group, which includes C. zuluensis and C. moreli, was monophyletic (CPP = 53).
Bayesian analysis showed that the subgenus Oecacta was paraphyletic: C. circumscriptus – subgenus Beltranmyia and Culicoides distinctipennis – subgenus Meijerehela are included within the members of this subgenus, and C. sahariensis is separated from the other members of subgenus Oecata.
The monophyly of the Fluvialis group (CPP = 74) is discussed due to the inclusion in this clade of the incertae sedis C. paolae. The subgenus Haematomyodium is the sister group of the Fluvialis group (BI, CPP = 91).
Discussion
The present study is, to our knowledge, the first systematic molecular analysis carried out on the genus Culicoides at a large taxonomic level, not focusing on closely related species (42 species belonging to 12 subgenera and unclassified groups collected in Afrotropical, Neotropical and Palaearctic areas). The data, based on COI and D1-D2 sequences, were subjected to a range of MP and BI analyses in order to explore the phylogenetic signal. To date, other molecular phylogenies, based mainly on COI sequences, were commonly restricted to a single subgenus or group located in Afrotropical or Palaearctic areas [29], except for one containing 37 Palaearctic species representing 10 subgenera [1]. Phylogenies studies based on ITS1 and ITS2 have also been reported on 25 French species [49] and 9 Italian species [26], respectively.
Pseudogenes are homologous sequences arising from currently or evolutionarily active genes that have lost their ability to function as a result of disrupted transcription or translation. They may contain stop codons, repetitive elements, have frame shifts and/or lack of transcription. However, they might retain gene-like features [65]. Pseudogenes have been identified in the mitochondrial genome of insects, also widely used in phylogenetic studies, with the risk of obtaining erroneous results during phylogenetic reconstruction [34]. To our knowledge, this is the first report of pseudogenes in Culicoides.
The subgenera Anilomyia, Beltranmyia, Meijerehelea, Trithecoides, Wirthomyia and some groups are only represented by a single species. Consequently, further studies are required to discuss their monophylies.
We demonstrate here the monophyly of the subgenus Monoculicoides that is in agreement with previous studies (ITS1: [49]; COI: [1, 55]).
Similarly, our results suggest that the subgenus Culicoides is also monophyletic, as previously reported by another study based on COI sequences [1], whereas studies based on ITS1 [32, 49], ITS2 [32] and COI [55] sequences suggested the paraphyly of this subgenus. Within this group, our findings suggest (according to molecular [33, 43, 48] and morphological studies [17, 38, 57]) the validity of C. lupicaris, whereas it is sometimes synonymised with C. delta [11].
Monophyly of the subgenus Avaritia is clearly supported by the present study, which corroborates previous results obtained from both morphological [46] and molecular data based on COI [15] and ITS1 [32, 49]. However, the paraphyly of the subgenus Avaritia has also been reported by analysis of COI [1, 50, 55] and ITS2 [32] sequences, taking into account the phylogenetic position of C. dewulfi outside this subgenus.
Most authors erroneously included C. dewulfi in the obsoletus complex or the obsoletus group [25, 41]. Our results clearly suggest that C. dewulfi does not belong to this group (Fig. 1) as previously emphasised by different molecular markers [25, 28, 43, 56]. Based on morphological, morphometrical and molecular data [2, 23, 28, 46, 47], C. obsoletus and C. scoticus are two closely related species belonging to the Obsoletus complex [5, 28, 60]. According to [42], the Imicola group in the Afrotropical region includes C. imicola and C. miombo. C. kibatiensis could present similar characters to C. trifasciellus, and subgroup trifasciellus is distinct from Culicoides imicola and the Imicola group [24]. C. trifasciellus belongs to the Orientalis group of the Afrotropical region [41] and its taxonomic position with C. kibatiensis is not resolved. Our results show that C. imicola is a cryptic species of C. miombo, C. kibatiensis being the sister species of the clade formed by these two species. Future studies are needed to carry out investigations at a subgeneric scale in order to determine the species status of the most important vector subgenera. Interestingly, the specimens collected in Gabon belong to the subgenus Avaritia and show unique morphological characters and nucleotide substitutions (COI). Therefore, a morphological description of this new species of Culicoides is in progress.
Our study validates the subgenus Remmia (= Schultzei group) as a valid subgenus, outside of the subgenus Oecacta [3, 11], and includes C. kingi [4, 14]. The status of the Schultzei group and its subgeneric affiliation has been disputed. It is sometimes included in the subgenus Remmia Glukhova [11, 19, 43], the latter considered by some authors as a junior synonym of the subgenus Oecacta (Poey) [14, 64] or sometimes unclassified [62]. Unlike many authors, we believe the subgenus Oecacta includes only the Furens group and/or perhaps the Schultzei group, if the first group is excluded from the latter as previously suggested [3, 14]. Future integrative taxonomy studies [27–29] should define this group with more precision, especially its relationships with the subgenus Oecacta.
C. paolae from Tunisia is included in the Fluvialis group constituted by New World species in our sampling (C. castillae, C. fluvialis, C. tetrathyris and C. tetrathyris like). The hypothesis that Culicoides paolae could be a synonym of the Central American species C. jamaicensis seems fair. Indeed, C. paolae and C. jamaicensis present huge morphological similarities [8, 44], thus raising the possibility that it was introduced into the Mediterranean Region at the time of Columbus, and was only discovered 500 years later and named C. paolae [44].
The subgenus Hoffmania is monophyletic with two clades (Hylas and Guttatus groups). To our knowledge, no recent data exist in the literature. This subgenus includes 82 extant species from South America and Asia. The wide geographic distribution warrants further studies of the subgenus Hoffmania using phylogenetic and integrative approaches at several scales.
The Milnei group is monophyletic. The species constituting this group have been described from the Afrotropical region [11] and transmit several pathogenic organisms, e.g. Akabane virus, BTV, Letsitele virus, unidentified virus isolates (Cul. 5/69), Dipetalonema perstans and Dipetalonema streptocerca [10]. To our knowledge, this group has been understudied and future integrative taxonomy studies [27–29] should more precisely define this group [24].
Palaearctic, Neotropical and Afrotropical Culicoides are mixed in our cladogram (Fig. 5) and there is no geographic clustering, indicating that the palaeobiogeography of the genus Culicoides does not follow the generalised tracks [37]. Consequently, Culicoides settled in different areas by wind [45], animal carriage [39] or by human activities [44]. For example, C. imicola specimens collected in Laos, Thailand, Vietnam and Reunion Island are of African origin [40]. In a Culicoides catalogue [11], 42 fossils were recorded from ambers from the Dominican Republic, the USA, Canada, Germany, Poland, the Baltic area and Russia, suggesting a Laurasian origin of the genus.
In conclusion, this study showed that the subgenera Monoculicoides, Culicoides, Haematomyidium, Hoffmania and Avaritia (including the main vectors of bluetongue disease) are monophyletic, whereas the subgenus Oecacta is paraphyletic. As proposed by Harrup et al. [29], a cladistic reinterpretation of the subgeneric classification of Culicoides, and species delimitations, should represent Culicoides taxonomy. Integrative taxonomy (including morphological, mitochondrial and other markers) and modern morphometric analysis (based on wing characteristics including type specimens) can help taxonomists as suggested by Hadj-Henni et al.
Acknowledgments
The authors thank Christophe Paupy (Centre International de Recherches Médicales de Franceville – CIRMF) and COMILOG/ERAMET (Parc de La Lékédi Bakoumba) for providing biting midges from Gabon. The authors thank Stavana Strutz for proofreading this manuscript.
Appendices
Appendix 1.
Parsimonious tree based on D1-D2 28S rDNA nucleotide sequences. Majority-rule 50% consensus: values between 50% and 100% are indicated on the branches.
Appendix 2.
Bayesian tree resulting from the phylogenetic analysis of the COI mtDNA according to the best-fit partitioning strategy. Robustness of nodes was indicated by the posterior probability values.
Appendix 3.
Bayesian tree resulting from the phylogenetic analysis of the D1D2 28S rDNA according to the best-fit partitioning strategy. Robustness of nodes was indicated by the posterior probability values.
Cite this article as: Augot D, Mathieu B, Hadj-Henni L, Barriel V, Zapata Mena S, Smolis S, Slama D, Randrianambinintsoa FJ, Trueba G, Kaltenbach M, Rahola N & Depaquit J: Molecular phylogeny of 42 species of Culicoides (Diptera, Ceratopogonidae) from three continents. Parasite, 2017, 24, 23.
References
- 1. Ander M, Troell K, Chirico J. 2013. Barcoding of biting midges in the genus Culicoides: a tool for species determination. Medical and Veterinary Entomology, 27(3), 323–331. [DOI] [PubMed] [Google Scholar]
- 2. Augot D, Sauvage F, Jouet D, Simphal E, Veuille M, Couloux A, Kaltenbach ML, Depaquit J. 2010. Discrimination of Culicoides obsoletus and Culicoides scoticus, potential bluetongue vectors, by morphometrical and mitochondrial cytochrome oxidase subunit I analysis. Infection Genetics & Evolution, 10, 629–637. [DOI] [PubMed] [Google Scholar]
- 3. Augot D, Randrianambinintsoa FJ, Gasser A, Depaquit J. 2013. Record of two species of Culicoides (Diptera: Ceratopogonidae) new for Madagascar and molecular study showing the paraphylies of the subgenus Oecacta and the Schultzei group. Bulletin de la Société de Pathologie Exotique, 106, 201–205. [DOI] [PubMed] [Google Scholar]
- 4. Bakhoum MT, Fall M, Fall AG, Bellis GA, Gottlieb Y, Labuschagne K, Venter GJ, Diop M, Mall I, Seck MT, Allène X, Diarra M, Gardès L, Bouyer J, Delécolle JC, Balenghien T, Garros C. 2013. First record of Culicoides oxystoma Kieffer and diversity of species within the Schultzei group of Culicoides Latreille (Diptera: Ceratopogonidae) biting midges in Senegal. PLoS One, 8(12), e84316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balenghien T, Pagès N, Goffredo M, Carpenter S, Augot D, Jacquier E, Talavera S, Monaco F, Depaquit J, Grillet C, Pujols J, Satta G, Kasbari M, Setier-Rio ML, Izzo F, Alkan C, Delécolle JC, Quaglia M, Charrel R, Polci A, Bréard E, Federici V, Cêtre-Sossah C, Garros C. 2014. The emergence of Schmallenberg virus across Culicoides communities and ecosystems in Europe. Preventive Veterinary Medicine, 116(4), 360–369. [DOI] [PubMed] [Google Scholar]
- 6. Barriel V. 1994. Molecular phylogenies and how to code insertion-deletion events. Comptes Rendus de l’Académie des Sciences III, 317(7), 693–701. [PubMed] [Google Scholar]
- 7. Beckenbach AT, Borkent A. 2003. Molecular analysis of the biting midges (Diptera: Ceratopogonidae), based on mitochondrial cytochrome oxidase subunit 2. Molecular Phylogenetics and Evolution, 27(1), 21–35. [DOI] [PubMed] [Google Scholar]
- 8. Boorman J, Mellor PS, Scaramozzino P. 1996. A new species of Culicoides (Diptera, Ceratopogonidae) from southern Italy. Parassitologia, 38(3), 501–503. [PubMed] [Google Scholar]
- 9. Borkent A, Wirth WW. 1997. World species of biting midges (Diptera: Ceratopogonidae). Bulletin of the American Museum of Natural History, 233, 1–257. [Google Scholar]
- 10. Borkent A. 2005. The biting midges, the Ceratopogonidae (Diptera), in Biology of disease vectors. Marquardt WC, Editor Elsevier: p. 113–126. [Google Scholar]
- 11. Borkent A. 2016. The Subgeneric classification of species of Culicoides-thoughts and a warning. Elsevier Academic Press, Burlington, MA, USA: http://www.inhs.uiuc.edu/research/FLYTREE/CulicoidesSubgenera.pdf [Google Scholar]
- 12. Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. 2013. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Research, 100(1), 102–113. [DOI] [PubMed] [Google Scholar]
- 13. Cornet M, Nevill EM, Walker AR. 1974. Note sur les Culicoides (Diptera: Ceratopogonidae) du groupe de C. milnei Austen, 1909, en Afrique orientale et australe. Cahier ORSTOM série Entomologie Médicale Parasitaire, 12(4), 231–243. [Google Scholar]
- 14. Cornet M, Bruhnes J. 1994. Révision des espèces de Culicoides apparentées à C. schultzei (Enderlein, 1908) dans la région afrotropicale (Diptera, Ceratopogonidae). Bulletin de la Société Entomologique de France, 99(2), 149–164. [Google Scholar]
- 15. Dallas JF, Cruickshank RH, Linton YM, Nolan DV, Patakakis M, Braverman Y, Capela R, Capela M, Pena I, Meiswinkel R, Ortega MD, Baylis M, Mellor PS, Mordue Luntz AJ. 2003. Phylogenetic status and matrilineal structure of the biting midges, Culicoides imicola, in Portugal, Rhodes and Israel. Medical and Veterinary Entomology, 17(4), 379–387. [DOI] [PubMed] [Google Scholar]
- 16. Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9(8), 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delécolle JC. 1985. Nouvelle contribution à l’étude systématique et iconographique des espèces du genre Culicoides (Diptera: Ceratopogonidae) du Nord-Est de la France. Thèse d’Université. Université Louis Pasteur de Strasbourg, UER Sciences Vie et Terre, Strasbourg, France. [Google Scholar]
- 18. Depaquit J, Perrotey S, Lecointre G, Tillier A, Tillier S, Ferté H, Kaltenbach M, Léger N. 1998. Systématique moléculaire des Phlebotominae: étude pilote. Paraphylie du genre Phlebotomus. Comptes Rendus de l’Académie des Sciences, 32(10), 849–855. [DOI] [PubMed] [Google Scholar]
- 19. Dyce AL, Bellis GA, Muller MJ. 2007. Pictorial atlas of Australasian Culicoides wings (Diptera, Ceratopogonidae). Australian Biological Resources Study: Canberra, 88. [Google Scholar]
- 20. Elbers ARW, Meiswinkel R, van Weezep E, Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. 2011. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands. Emerging Infectious Diseases, 19(1), 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Felippe-Bauer ML, Caceres AG, Silva CS, Valderrama-Bazan W, Gonzales-Perez A, Costa JM. 2008. Description of Culicoides pseudoheliconiae sp. n. from Peruvian Amazon and revalidation of Culicoides contubernalis Ortiz & Leon (Diptera: Ceratopogonidae). Memorias do Instituto Oswaldo Cruz, 103(3), 259–262. [DOI] [PubMed] [Google Scholar]
- 22. Forattini OP. 1957. Culicoides da Regiao Neotropical (Diptera, Ceratopogonidae). Arquivos Faculdade Higiene Saude Publica Universidae Sao Paulo, 11, 159–526. [DOI] [PubMed] [Google Scholar]
- 23. Garros C, Mathieu B, Balenghien T, Cêtre-Sossah C, Delécolle JC. 2010. Suggesting synonymies? Comments on Kiehl et al. (2009) The European vectors of bluetongue virus: are there species complexes, single species or races in Culicoides obsoletus and C. pulicaris detectable by sequencing ITS-1, ITS-2 and 18SrDNA. Parasitology Research, 107(3), 731–734. [DOI] [PubMed] [Google Scholar]
- 24. Glick J. 1990. Culicoides biting midges (Diptera: Ceratopogonidae) of Kenya. Journal of Medical Entomology, 27(2), 85–195. [DOI] [PubMed] [Google Scholar]
- 25. Gomulski LM, Meiswinkel R, Delécolle JC, Goffredo M, Gasperi G. 2005. Phylogenetic relationships of the subgenus Avaritia Fox, 1995 including Culicoides obsoletus (Diptera, Ceratopogonidae) in Italy based on internal transcribed spacer 2 ribosomal DNA sequences. Systematic Entomology, 10, 1–13. [Google Scholar]
- 26. Gomulski LM, Meiswinkel R, Delécolle JC, Goffredo M, Gasperi G. 2006. Phylogeny of the subgenus Culicoides and related species in Italy, inferred from internal transcribed spacer 2 ribosomal DNA sequences. Medical and Veterinary Entomology, 20(2), 229–238. [DOI] [PubMed] [Google Scholar]
- 27. Hadj-Henni L, De Meulemeester T, Mathieu B, Depaquit J, Augot D. 2014. Taxonomic assessment of Culicoides brunnicans, C. santonicus and C. vexans (Diptera: Ceratopogonidae) in France: implications in systematic. Infection Genetics and Evolution, 33, 324–331. [DOI] [PubMed] [Google Scholar]
- 28. Hadj-Henni L, Sauvage F, Ninio C, Depaquit J, Augot D. 2014. Wing geometry as a tool for discrimination of Obsoletus group (Diptera: Ceratopogonidae: Culicoides) in France. Infection Genetics and Evolution, 21, 110–117. [DOI] [PubMed] [Google Scholar]
- 29. Harrup LE, Bellis GA, Balenghien T, Garros C. 2015. Culicoides Latreille (Diptera: Ceratopogonidae) taxonomy: current challenges and future directions. Infection Genetics and Evolution, 30, 249–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hwang UW, Kim W, Tautz D, Friedrich M. 1998. Molecular phylogenetics at the Felsenstein zone: approaching the Strepsiptera problem using 5.8S and 28S rDNA sequences. Molecular Phylogenetics. Evolution, 9(3), 470–480. [DOI] [PubMed] [Google Scholar]
- 31. Khamala CPM, Kettle DS. 1971. The Culicoides Latreille (Diptera: Ceratopogonidae) of East Africa. Transactions of the Royal Entomological Society of London, 123, 1–95. [Google Scholar]
- 32. Kiehl E, Walldorf V, Klimpel S, Quraishy SA, Mehlhorn H. 2009. The European vectors of Bluetongue virus: are there species complexes, single species or races in Culicoides obsoletus and C. pulicaris detectable by sequencing ITS-1, ITS-2 and 18S-rDNA? Parasitology Research, 105(2), 331–336. [DOI] [PubMed] [Google Scholar]
- 33. Lassen SB, Nielsen SA, Skovgaard H, Kristensen M. 2012. Molecular differentiation of Culicoides biting midges (Diptera: Ceratopogonidae) from the subgenus Culicoides Latreille in Denmark. Parasitology Research, 110(5), 1765–1771. [DOI] [PubMed] [Google Scholar]
- 34. Leite LAR. 2012. Mitochondrial pseudogenes in insect DNA barcoding: differing points of view on the same issue. Biota Neotropica, 12, 301–308. [Google Scholar]
- 35. Mardulyn P, Goffredo M, Conte A, Hendrickx G, Meiswinkel R, Balenghien T, Sghaier S, Lohr Y, Gilbert M. 2013. Climate change and the spread of vector-borne diseases: using approximate Bayesian computation to compare invasion scenarios for the bluetongue virus vector Culicoides imicola in Italy. Molecular Ecology, 22(9), 2456–2466. [DOI] [PubMed] [Google Scholar]
- 36. Mathieu B, Cêtre-Sossah C, Garros C, Chavernac D, Balenghien T, Carpenter S, Setier-Rio ML, Vignes-Lebbe R, Ung V, Candolfi E, Delécolle JC. 2012. Development and validation of IIKC: an interactive identification key for Culicoides (Diptera: Ceratopogonidae) females from the Western Palaearctic region. Parasites & Vectors, 5, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matile L. 1990. Recherches sur la systématique et l’évolution des Keroplatidae (Diptera, Mycetophiloidea). Mémoires du Muséum National Histoire Naturelle, 148, 1–682. [Google Scholar]
- 38. Matsumoto Y, Yanase T, Tsuda T, Noda H. 2009. Species-specific mitochondrial gene rearrangements biting midges and vector species identification. Medical and Veterinary Entomology, 23, 47–55. [DOI] [PubMed] [Google Scholar]
- 39. Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, Eschweiler J, Hoffmann B, Beer M. 2007. First occurrence of Culicoides obsoletus-transmitted bluetongue virus epidemic in Central Europe. Parasitology Research, 101(1), 219–228. [DOI] [PubMed] [Google Scholar]
- 40. Meiswinkel R. 1991. Afrotropical Culicoides: C. (Avaritia) miombo sp. nov., a widerspread species closely allied to C. (A.) imicola Kieffer, 1913 (Diptera: Ceratopogonidae). The Onderstepoort Journal of Veterinary Research, 58(3), 155–170. [PubMed] [Google Scholar]
- 41. Meiswinkel R, Van Rijn P, Leijs P, Goffredo M. 2007. Potential new Culicoides vector of bluetongue virus in Northern Europe. The Veterinary Record, 161, 564–565. [DOI] [PubMed] [Google Scholar]
- 42. Meiswinkel R. 2004. Adult characters defining and separating the Imicola and Orientalis species complexes of the subgenus Avaritia Fox, 1955 (Culicoides, Diptera: Ceratopogonidae). Veterinaria Italiana, 40(3), 345–351. [PubMed] [Google Scholar]
- 43. Meiswinkel R, Gomulski LM, Delécolle JC, Goffredo M, Gasperi G. 2004. The taxonomy of Culicoides vector complexes – unfinished business. Veterinaria Italiana, 40(3), 151–159. [PubMed] [Google Scholar]
- 44. Meiswinkel R, Labuschagne K, Goffredo M. 2004. Christopher Columbus and Culicoides: was C. jamaicensis Edwards, 1922 introduced into the Mediterranean 500 years ago and later re-named C. paolae Boorman 1996? Veterinaria Italiana, 40(3), 340–344. [PubMed] [Google Scholar]
- 45. Mellor PS, Boorman J, Baylis M. 2000. Culicoides biting midges: their role as arbovirus vectors. Annual Review of Entomology, 45, 307–340. [DOI] [PubMed] [Google Scholar]
- 46. Muñoz-Muñoz F, Talavera S, Carpenter S, Nielsen SA, Werner D, Pagès N. 2014. Phenotypic differentiation and phylogenetic signal of wing shape in western European biting midges, Culicoides spp., of the subgenus Avaritia. Medical and Veterinary Entomology, 28(3), 319–329. [DOI] [PubMed] [Google Scholar]
- 47. Nielsen SA, Kristensen M. 2011. Morphological and molecular identification of species of the obsoletus group (Diptera: Ceratopogonidae) in Scandinavia. Parasitology Research, 109(4), 1133–1141. [DOI] [PubMed] [Google Scholar]
- 48. Pagès N, Muñoz-Muñoz F, Talavera S, Sarto V, Lorca C, Núñez JI. 2009. Identification of cryptic species of Culicoides (Diptera: Ceratopogonidae) in the subgenus Culicoides and development of species-specific PCR assays based on barcode regions. Veterinary Parasitology, 165(3–4), 298–310. [DOI] [PubMed] [Google Scholar]
- 49. Perrin A, Cetre-Sossah C, Mathieu B, Baldet T, Delecolle JC, Albina E. 2006. Phylogenetic analysis of Culicoides species from France based on nuclear ITS1-rDNA sequences. Medical and Veterinary Entomology, 20(2), 219–228. [DOI] [PubMed] [Google Scholar]
- 50. Pettersson E, Bensch S, Ander M, Chirico J, Sigvald R, Ignell R. 2013. Molecular identification of bloodmeals and species composition in Culicoides biting midges. Medical and Veterinary Entomology, 27(1), 104–112. [DOI] [PubMed] [Google Scholar]
- 51. Raghavendra K, Cornel AJ, Reddy BP, Collins FH, Nanda N, Chandra D, Verma V, Dash AP, Subbarao SK. 2009. Multiplex PCR assay and phylogenetic analysis of sequences derived from D2 domain of 28S rDNA distinguished members of the Anopheles culicifacies complex into two groups, A/D and B/C/E. Infection Genetics and Evolution, 9(2), 271–277. [DOI] [PubMed] [Google Scholar]
- 52. Rambaut A. 2002. Se-Al: sequence alignment editor, v.2.0 alpha 11 carbon, University of Oxford: Oxford, United Kingdom, http://tree.bio.ed.ac.uk/software/seal/. [Google Scholar]
- 53. Ritchie A, Blackwell A, Malloch G, Fenton B. 2004. Heterogeneity of ITS1 sequences in the biting midge Culicoides impunctatus (Goetghebuer) suggests a population in Argyll Scotland may be genetically distinct. Genome, 47, 546–558. [DOI] [PubMed] [Google Scholar]
- 54. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. [DOI] [PubMed] [Google Scholar]
- 55. Sarvašová A, Kočišová A, Halán M, Delécolle JC, Mathieu B. 2014. Morphological and molecular analysis of the genus Culicoides (Diptera: Ceratopogonidae) in Slovakia with five new records. Zootaxa, 3872(5), 541–560. [DOI] [PubMed] [Google Scholar]
- 56. Schwenkenbecher JM, Mordue (Luntz) AJ, Piertney SB. 2009. Phylogenetic analysis indicates that Culicoides dewulfi should not be considered part of the Culicoides obsoletus complex. Bulletin of Entomological Research, 99(4), 371–375. [DOI] [PubMed] [Google Scholar]
- 57. Simon C, Frati F, Beckenbach A, Crespi B, Lui H, Flook P. 1994. Evolution, weighing and phylogenetic unity of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annuals of the Entomological Society of America, 8, 651–701. [Google Scholar]
- 58. Slama D, Chaker E, Mathieu B, Babba H, Depaquit J, Augot D. 2014. Biting midges monitoring (Diptera: Ceratopogonidae: Culicoides Latreille) in the governate of Monastir (Tunisia): species composition and molecular investigations. Parasitology Research, 113(7), 2435–2443. [DOI] [PubMed] [Google Scholar]
- 59. Swofford DL. 2002. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0 Beta, Sinauer Associates: Sunderland, MA. [Google Scholar]
- 60. Venail R, Balenghien T, Guis H, Tran A, Baldet T, Setier-Rio ML, Delécolle JC, Mathieu B, Martinez D, Languille J, Baldet T, Garros C. 2012, Assessing diversity and abundance of vector populations at a national scale: example of Culicoides surveillance in France after bluetongue virus emergence, in Parasitology Research Monograph. Mehlhorn H, Editor, vol. 3, Springer-Verlag: Berlin, Heidelberg, p. 77–102 [Mehlhorn H (Series Editor): Arthropods as Vectors of Emerging Diseases]. [Google Scholar]
- 61. Wirth WW, Dyce AL, Peterson BV. 1985. An atlas of wing photographs, with a summary of the numerical characters of the neartic species of Culicoides (Diptera: Ceratopogonidae). Contributions of the American Entomological Institute, 22, 1–46. [Google Scholar]
- 62. Wirth WW, Dyce AL, Spinelli GR. 1988. An atlas of wing photographs, with a summary of the numerical characters of the neotropical species of Culicoides (Diptera: Ceratopogonidae). Contributions of the American Entomological Institute, 25, 2–72. [Google Scholar]
- 63. Wirth WW, Hubert AA. 1989. The Culicoides of Southeast Asia (Diptera: Ceratopogonidae). Memoirs of the American Entomological Institute, 44, 1–508. [Google Scholar]
- 64. Yu YX, Liu JH, Liu GP, Liu ZJ, Hao BS, Yan G, Zhao TS. 2005. Ceratopogonidae of China, Insecta, Diptera, Vol. 1-2 Military Medical Science Press, Beijing, p. 1699 [in Chinese]. [Google Scholar]
- 65. Zuriaga MA, Mas-Coma S, Bargues MD. 2015. A nuclear ribosomal DNA pseudogene in triatomines opens a new research field of fundamental and applied implications in Chagas disease. Memorias Instituto Oswaldo Cruz, 10(3), 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]