Abstract
Neonatal diabetes is frequently part of a complex syndrome with extrapancreatic features: 18 genes causing syndromic neonatal diabetes have been identified to date. There are still patients with neonatal diabetes who have novel genetic syndromes. We performed exome sequencing in a patient and his unrelated, unaffected parents to identify the genetic etiology of a syndrome characterized by neonatal diabetes, sensorineural deafness, and congenital cataracts. Further testing was performed in 311 patients with diabetes diagnosed before 1 year of age in whom all known genetic causes had been excluded. We identified 5 patients, including the initial case, with three heterozygous missense mutations in WFS1 (4/5 confirmed de novo). They had diabetes diagnosed before 12 months (2 before 6 months) (5/5), sensorineural deafness diagnosed soon after birth (5/5), congenital cataracts (4/5), and hypotonia (4/5). In vitro studies showed that these WFS1 mutations are functionally different from the known recessive Wolfram syndrome–causing mutations, as they tend to aggregate and induce robust endoplasmic reticulum stress. Our results establish specific dominant WFS1 mutations as a cause of a novel syndrome including neonatal/infancy-onset diabetes, congenital cataracts, and sensorineural deafness. This syndrome has a discrete pathophysiology and differs genetically and clinically from recessive Wolfram syndrome.
Introduction
Neonatal diabetes is diagnosed before 6 months of age and reflects a severe reduction in β-cell number or function. A genetic diagnosis is possible in 82% of patients with 23 genetic causes identified to date (1,2). Thirty-nine percent of patients with neonatal diabetes have a genetic etiology that results in the development of at least one extrapancreatic feature in addition to diabetes (1). This subtype is phenotypically and genetically heterogeneous with mutations in 18 genes known to cause syndromic forms of neonatal diabetes (1). Fifteen percent of patients with syndromic neonatal diabetes do not have a mutation in one of the known etiological genes, suggesting undescribed novel genetic syndromes.
The identification of novel etiological genes in patients with syndromic neonatal diabetes has been revolutionized by the introduction of next-generation sequencing. In outbred pedigrees, exome or whole-genome sequencing of an affected case and their unaffected, unrelated parents is a powerful tool to identify de novo heterozygous mutations. We have successfully used this approach to identify two novel etiological genes for syndromic neonatal diabetes in our cohort (2,3).
Neonatal diabetes has not been described in patients with recessive loss-of-function mutations in WFS1, the genetic cause of Wolfram syndrome, even though early-onset diabetes is a cardinal feature of this multisystem disease. In these patients, diabetes onset occurs in the first decade of life (median age at onset 6 years, range 1–32 years [4–6]). Additional extrapancreatic features (optic atrophy, diabetes insipidus, and hearing loss) usually develop sequentially between the first and third decade of life.
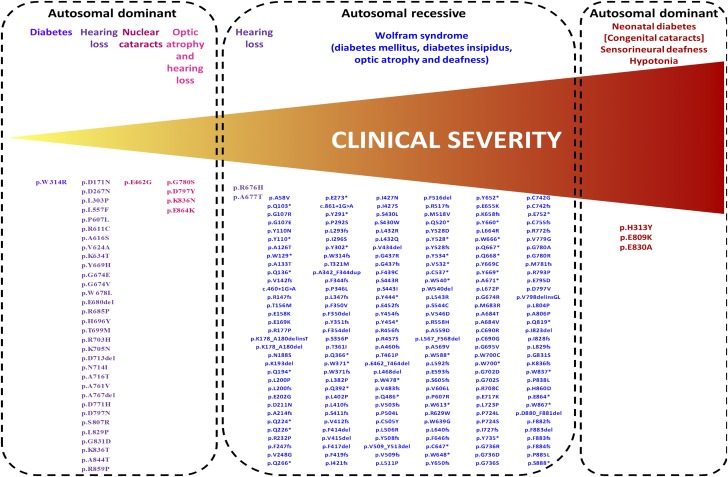
Heterozygous mutations in WFS1 have been found to cause less severe phenotypes than Wolfram syndrome–associated recessive mutations. In particular, dominant WFS1 mutations have been reported as causing isolated adult-onset diabetes (7), isolated low-frequency hearing loss (8,9), optic atrophy and hearing impairment (10–12), and isolated congenital nuclear cataracts (13) (Fig. 1).
Figure 1.
Phenotypic spectrum caused by autosomal dominant and autosomal recessive WFS1 mutations. The figure includes WFS1 mutations reported as disease causing in the Human Gene Mutation Database that have a minor allele frequency <0.00447 in the ExAC database (maximum allele frequency possible for Wolfram syndrome–causing mutations, assuming a disease frequency of 1/100,000, as calculated by the alleleFrequencyApp [http://cardiodb.org/allelefrequencyapp/]).
We used a trio-based exome sequencing strategy to identify dominant WFS1 mutations as the cause of a novel syndrome characterized by permanent neonatal/infancy-onset diabetes, congenital or early-onset sensorineural deafness, and congenital cataracts.
Research Design and Methods
Genetic Analysis
We performed exome sequencing on samples from an affected proband and parents using SureSelect Human All Exon Kit V4 (Agilent). Paired-end sequencing was performed on an Illumina HiSeq2500, using 100 bp reads. The resulting reads were aligned to the hg19 reference genome with BWA. Variants were called with GATK UnifiedGenotyper and annotated using ANNOVAR and SeattleSeq Annotation server as previously described (3).
Replication studies were performed in a cohort of 311 patients diagnosed with diabetes before 12 months of age in whom mutations in the 22 known neonatal diabetes genes had been excluded. All the patients in the cohort were analyzed using our targeted next-generation sequencing assay that includes baits for WFS1 as previously described (14).
The bioinformatics tools SIFT, PolyPhen-2, and Align GVGD were accessed through the Alamut software (Interactive Biosoftware, Rouen, France) to predict the effect of novel variants on the WFS1 protein. If the variant was deemed likely pathogenic or of unknown significance, mutation testing was performed in the patient (to confirm the mutation) and the parents (when available) to assess whether the mutation had arisen de novo. Microsatellite analysis of parent/proband trios using the PowerPlex kit (Promega, Southampton, U.K.) was used to confirm family relationships for the putative de novo mutations. In silico modeling of the WFS1 p.Glu809Lys and p.Glu830Ala mutations was carried out using the SWISS-MODEL server (15). Additionally, as templates of only limited sequence similarity could be identified, the PredictProtein server was used to predict secondary structure and functional motifs from primary sequence alone (16).
Immunostaining
COS7 cells were transiently transfected with dominant and recessive WFS1 mutations for 48 h. The cells were fixed in 2% paraformaldehyde for 30 min at room temperature and then permeabilized with 0.1% Triton X-100 for 2 min. The fixed cells were washed with PBS/Tween 0.1%, blocked with Image-iT FX signal enhancer (Invitrogen) for 30 min, and incubated in primary antibody overnight at 4°C. The cells were washed three times in PBS/Tween 0.1% and incubated with secondary antibody for 1 h at room temperature. Images were obtained with an FSA 100 microscope (Olympus). Anti-FLAG antibody and anti-calnexin antibody were obtained from Cell Signaling Technology (Danvers, MA) and Proteintech Group (Chicago, IL), respectively.
INS-1 cells were plated in eight-chamber slides (Thermo Fisher Scientific) at a density of 50,000 cells per chamber and transiently transfected with dominant and recessive GFP-tagged WFS1 mutant constructs for 48 h. The cells were fixed in 4% paraformaldehyde/PBS for 30 min at room temperature, washed three times with PBS, and then permeabilized with 0.1% Triton X-100/PBS for 10 min at room temperature. Cells were blocked overnight in a 2% BSA solution of 0.1% Triton X-100/PBS and washed three times the next day with PBS. Slides were prepared with antifade mounting medium containing DAPI (Vector Laboratories), and images were obtained with an FSA 100 microscope.
Immunoblotting
HeLa cells were transfected with full-length human WFS1 cDNA tagged with a FLAG epitope, as well as p.Pro724Leu, p.Glu809Lys, p.Glu830Ala, and p.His313Tyr mutant WFS1 cDNA. After transfection for 48 h, cells were lysed for 15 min in ice-cold buffer (10 mmol/L Tris-HCl, 50 mmol/L Hepes pH 7.4, 1% v/v Triton X-100, 1 mmol/L EDTA) containing protease inhibitors. Insoluble material was recovered by centrifugation at 13,000 rpm for 15 min and solubilized in 10 mmol/L Tris-HCl and 1% w/v sodium dodecyl sulfate for 10 min at room temperature. After the addition of 4 volumes of lysis buffer (20 mmol/L Hepes pH 7.4, 1% v/v Triton X-100, 150 mmol/L NaCl, 10% v/v glycerol, 1 mmol/L EDTA), samples were sonicated for 30 s. Lysates were separated using 4–20% linear gradient SDS-PAGE (Bio-Rad) and then electroblotted. Primary antibodies used in this study were anti-FLAG (Cell Signaling Technology) and anti–α-tubulin (Cell Signaling Technology).
Luciferase Reporter Assay
For reporter assays, HeLa cells were cotransfected with the endoplasmic reticulum stress response element (ERSE) (rat promoter GRP78)–luciferase construct (17) and various constructs as indicated. Prior to lysis at 24 h after transfection, cells were treated with or without 100 nmol/L of thapsigargin (TG) for 8 h. Firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and normalized to Renilla luciferase values of the cotransfected pRL-TK vector (Promega) to control for differences in transfection efficiency. Statistical analysis of the data was performed by one-way ANOVA followed by Dunnett test using SPSS 22 (IBM).
Results
Genetic Analysis
We performed exome sequencing analysis on a male patient (patient 1) (Table 1) born to unrelated parents who did not have diabetes. He was diagnosed with neonatal diabetes at the age of 13 weeks. He also had congenital cataracts, congenital sensorineural deafness, and bilateral club feet. This patient died at the age of 2 years from sepsis.
Table 1.
Clinical characteristics of patients
| Patient ID |
|||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Birth weight (g)/gestation (weeks) |
1,570/31 (SDS −0.23, centile 40.58) |
2,500/40 (SDS −2.07, centile 1.9) |
3,010/38 (SDS −0.28, centile 39.09) |
2,400/41 (SDS −2.69, centile 0.35) |
2,670/40 (SDS −1.89, centile 2.96) |
| Sex |
Male |
Female |
Male |
Female |
Male |
| WFS1 mutations |
p.Glu809Lys (c.2425G>A) |
p.Glu809Lys (c.2425G>A) |
p.Glu809Lys (c.2425G>A) |
p.Glu830Ala (c.2489A>C) |
p.His313Tyr (c.937C>T) |
| De novo |
Yes |
Yes |
Maternal sample N/A |
Yes |
Yes |
| Age at last assessment |
Deceased (2 years) |
1.5 years |
8 years |
8 years |
15 months |
| Diabetes |
Yes |
Yes |
Yes |
Yes |
Yes |
| Age at diabetes diagnosis (weeks) |
13 |
24 |
50 |
35 |
36 |
| Insulin dose (units/kg/day) |
0.5–0.6 |
2 |
0.75–0.94 |
0.5–0.6 |
0.18 |
| HbA1c (%) |
6.8 |
7.2 |
7–10.2 |
7 |
10.1 |
| Cataracts |
Yes, congenital |
Yes, congenital |
Yes (age 2.5 years) |
Yes, congenital |
No |
| Glaucoma |
No |
Yes, congenital |
No |
Yes, congenital |
No |
| Other eye abnormalities |
Micropthalmia, bilateral microcornea, and iris coloboma |
Optic atrophy secondary to congenital glaucoma |
Optic atrophy at age 3.1 years |
— |
No |
| Deafness |
Yes, sensorineural (diagnosed at age 1 year) |
Yes, sensorineural (diagnosed at age 18 months) |
Yes (congenital) |
Yes, sensorineural (congenital) |
Yes, sensorineural (congenital) |
| Dysmorphic features |
Yes |
Yes |
— |
Yes (possibly due to facial hypotonia) |
No |
| Hypotonia |
Yes (generalized) |
Yes (limbs) |
— |
Yes |
Yes |
| Additional features | Bilateral club feet, left thumb contracture, asymmetrical ventricles on CT scan L>R | Microcephaly, widely spaced lateral ventricles on CT scan, clinodactyly | Hypothyroidism (12 months), motor and intellectual developmental delay | Subclinical hypothyroidism, growth hormone deficiency | No |
SDS, standard deviation score.
Exome sequencing identified two de novo coding variants in patient 1: WFS1 c.2425G>A p.Glu809Lys and ZNF513 c.1516G>A p.Ala506Thr. The variant in ZNF513 was excluded from further analysis as it was listed in the ExAC (Exome Aggregation Consortium) database (minor allele frequency 0.000008268) (Cambridge, MA; October 2016). The WFS1 c.2425G>A p.Glu809Lys variant was not listed in 59,030 exomes in ExAC and affects a highly conserved residue located in the endoplasmic reticulum (ER) domain of the WFS1 protein.
As WFS1 recessive loss-of-function mutations usually cause childhood-onset diabetes and parental carriers are unaffected, we hypothesized that the heterozygous p.Glu809Lys mutation was causing neonatal diabetes through a dominant-negative mechanism rather than haploinsufficiency. Therefore we investigated the presence of heterozygous missense (but not nonsense or frameshift) WFS1 mutations in a cohort of 311 individuals with diabetes diagnosed in the first year of life in whom mutations in the known neonatal diabetes genes had been excluded using our targeted next-generation sequencing assay (14).
We identified four additional patients harboring likely pathogenic variants in WFS1. Patients 2 and 3 were heterozygous for the same p.Glu809Lys variant identified in patient 1. Patient 4 had a heterozygous c.2489A>C p.Glu830Ala variant and patient 5 carried the c.937C>T p.His313Tyr mutation. Both the p.Glu830Ala and p.His313Tyr variants affect highly conserved residues and are predicted to be pathogenic by in silico analysis. The p.Glu830Ala mutation affects a residue located in the ER domain, whereas the p.His313Tyr involves an amino acid located in the first trans-membrane region of the WFS1 protein. These mutations were found to have arisen de novo in patients 1, 2, 4, and 5. Paternal inheritance was excluded in patient 3, but the maternal sample was not available for testing.
Clinical Features
The clinical features of the five patients are summarized in Table 1. All five individuals had congenital sensorineural deafness and early-onset diabetes diagnosed before 12 months of age (range 13–51 weeks) that was treated with a full replacement dose of insulin. In all five patients, the birth weight was low (median standard deviation score −1.89 [interquartile range −2.07, 0.28], median centile 2.96), consistent with reduced insulin secretion in utero, a feature not described in patients with “classic” Wolfram syndrome.
Congenital cataracts and additional eye abnormalities were present in four of five patients (patients 1, 2, 3, and 4). Two of them also had congenital glaucoma (patients 2 and 4), which had progressed to optic atrophy in patient 2. Patient 1 did not have congenital glaucoma but was diagnosed with micropthalmia, bilateral microcornea, and iris coloboma. Patient 3 was diagnosed with optic atrophy at the age of 3.1 years.
Additional common clinical features included hypotonia (present in patients 1, 2, 4, and 5), hypothyroidism (diagnosed in patients 3 and 4), and structural neurological defects identified by computed tomography (CT) scanning (in patients 1 and 2).
In Silico Protein Analysis
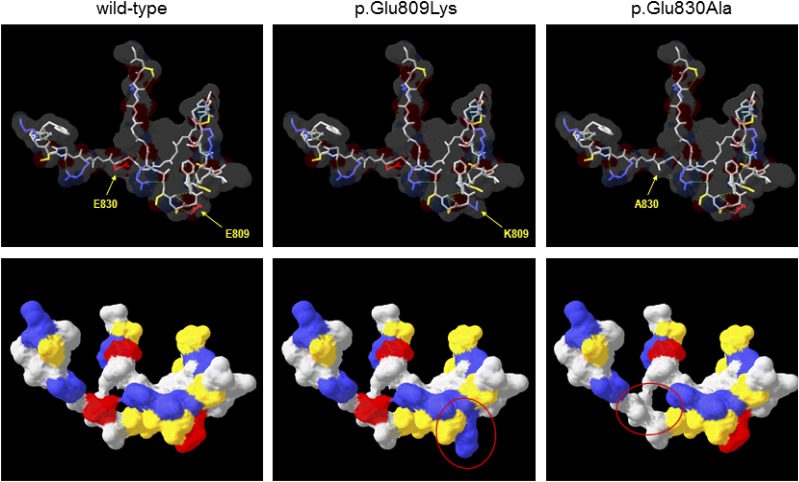
As the WFS1 p.His313Tyr mutation had been reported to have arisen de novo in two patients before (18), we focused our in silico investigations on the p.Glu809Lys and p.Glu830Ala variants. There are currently no 3-D molecular structures for any part of the WFS1 protein; therefore, to assess the potential effect of the p.Glu809Lys and p.Glu830Ala substitutions on protein structure, the sequence of the ER lumenal domain containing these residues (amino acids 653–869) was submitted to the SWISS-MODEL server to identify templates for comparative modeling, with models constructed on the best-scoring template. Predicted molecular structures suggested that both variants have significant effects on the surface properties of the molecule in terms of charge and hydrophobicity (Fig. 2). Given the lack of underlying secondary structure in these models, such changes might be expected to have significant effects on folding of this region in vivo, which are not apparent from in silico modeling. However, as the sequence similarity between WFS1 and the template was low (27%), these models may be of limited reliability, prompting further sequence analysis using the PredictProtein server. This predicted the presence of a number of short secondary structural motifs (mostly β-strand) as well as regions of intrinsic disorder spanning ∼30% of the lumenal domain (data not shown). As with 3-D comparative modeling, the p.Glu809Lys and p.Glu830Ala substitutions were predicted to have local effects on the extent of these regions as well as on solvent accessibility. Again, such changes might affect protein folding, and the presence of intrinsically disordered regions is particularly interesting as misfolding of such regions has been implicated in other diseases such as synucleinopathies (19).
Figure 2.
Comparative protein modeling of WFS1. The COOH-terminal lumenal domain of WFS1 (residues 653–869) was submitted to SWISS-MODEL for template identification; models were then constructed for residues 798–838 using the top-scoring template (PDB identifier 1ltl; MCM protein from Methanothermobacter thermautotrophicus; 27% sequence similarity over region modeled). Upper panels show the protein backbone with selected side chains as indicated in stick format for wild-type and variant WFS1, the predicted molecular surface is shown as a transparent layer, and broken green lines indicate predicted hydrogen bonds. Lower panels show the same structures but rotated ∼45° away from the viewer around a horizontal axis and with the predicted molecular surface shown as a solid layer. In all panels, amino acids are colored by type (red, acidic; blue, basic; yellow, uncharged polar; gray, nonpolar/hydrophobic); regions of significantly altered surface properties are indicated by red ovals in the lower panels. A830, p.Ala830; E809, p.Glu809; E830, p.Glu830; K809, p.Lys809.
Functional Studies
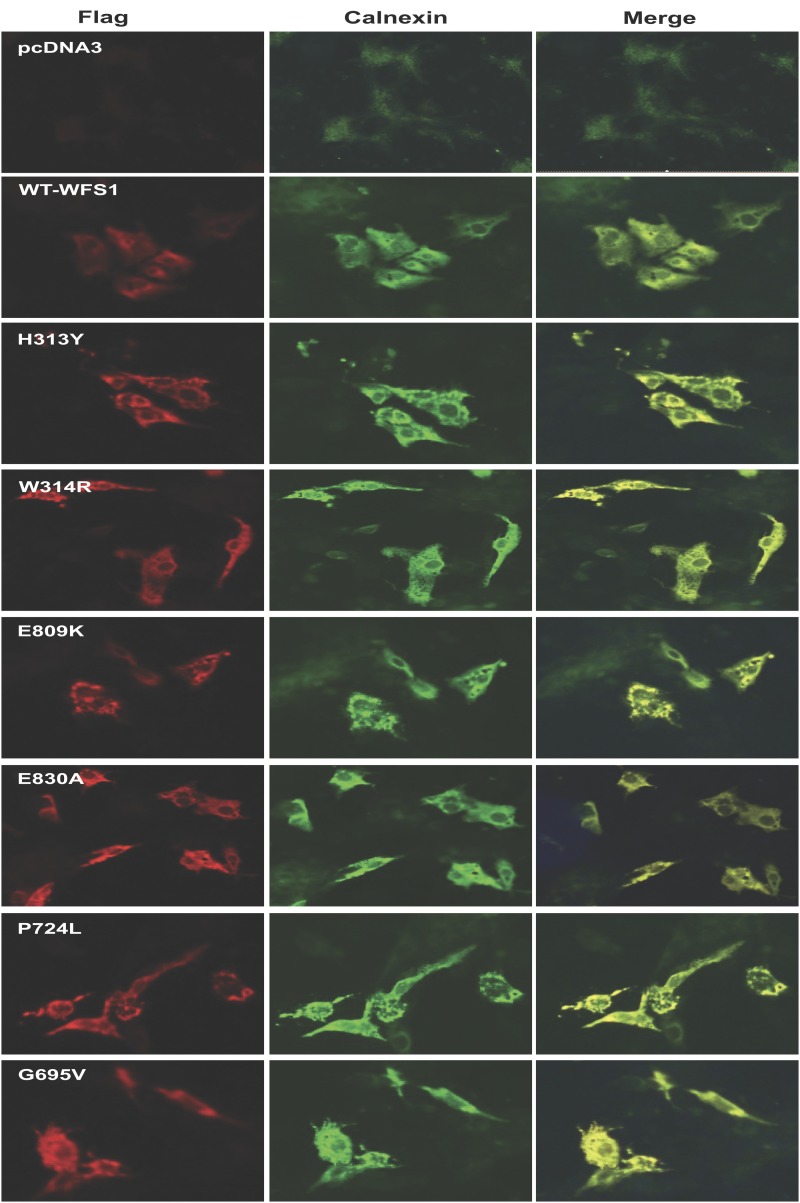
To study the subcellular localization of the dominant disease-causing variants, we performed immunostaining of COS7 cells transiently expressing 1) the dominant mutations, p.Glu809Lys, p.Glu830Ala, and p.His313Tyr; 2) a dominant mutation causing isolated adult-onset diabetes, p.Trp314Arg (7); 3) two recessive Wolfram syndrome–causing mutations, p.Pro724Leu and p.Gly695Val (20); or 4) wild-type WFS1 tagged at its C-terminus with a FLAG epitope. Immunostaining of cells expressing wild-type WFS1 showed a diffuse reticular pattern that colocalized with the ER marker calnexin (Fig. 3). In contrast, both dominant and recessive WFS1 mutants showed a punctate staining pattern in the ER, suggesting a tendency for these WFS1 mutants to misfold and aggregate. We also carried out similar experiments in insulin-secreting rat INS-1 cells using GFP-tagged WFS1 constructs. These studies were suboptimal due to low transfection efficiency (<10%) and expression levels. Interestingly, in these circumstances, the distribution of wild-type WFS1 and the dominant variants p.Glu809Lys, p.Glu830Ala, and p.His313Tyr was similar to that observed in COS7 cells, but the recessive mutation p.Pro724Leu showed a diffuse reticular staining pattern similar to wild-type WFS1 rather than a punctate appearance as seen for the dominant mutations (Supplementary Fig. 1). This suggests that, although all these mutants may have a tendency to misfold, there might be a spectrum of severity for different mutations. Such differences are likely to only become apparent at relatively low levels of expression at which variants carrying less severe mutations can still be folded efficiently. Because WFS1 is highly expressed in β-cells, it is also possible that INS-1 cells have a greater capacity to efficiently fold WFS1 proteins carrying less severe (i.e., recessive) mutations compared with that of COS7 cells.
Figure 3.
Double immunofluorescence staining of COS7 cells transiently transfected with dominant and recessive variants of WFS1. COS7 cells were transiently transfected with 1) control (pcDNA3); 2) wild-type WFS1 tagged at its C-terminus with a FLAG epitope (WT); 3) the dominant mutations p.His313Tyr (H313Y), p.Glu809Lys (E809K), and p.Glu830Ala (E830A); 4) a dominant variant causing isolated adult-onset diabetes, p.Trp314Arg (W314R) (7); and 5) two recessive Wolfram syndrome–causing mutations, p.Pro724Leu (P724L) and p.Gly695Val (G695V) (20). The cells were stained with anti-FLAG (red fluorescence, left) and anti-calnexin (green fluorescence, center). Right panels: Merged images are shown. n = 4; magnification is 10×.
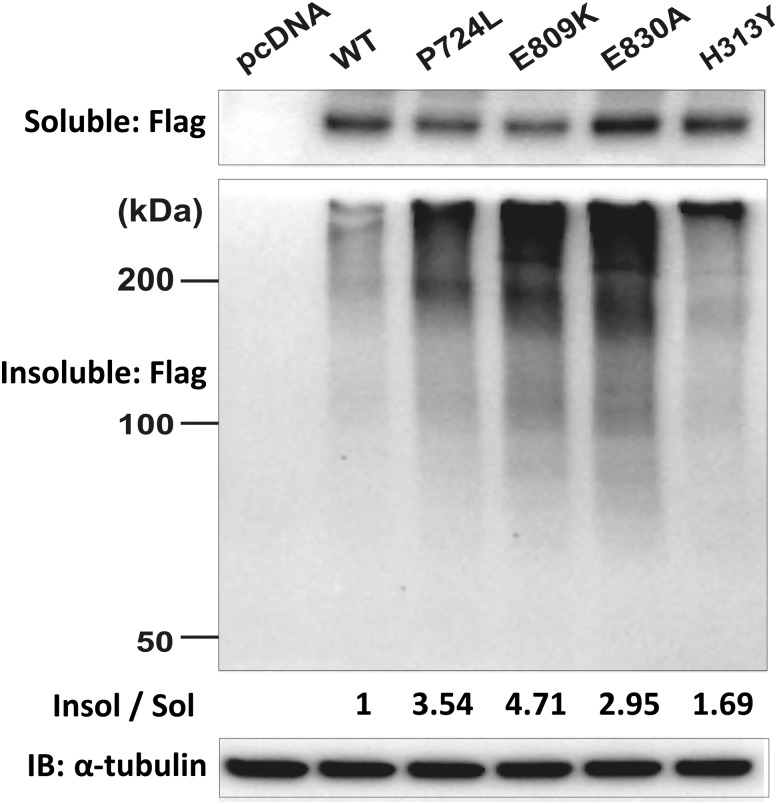
To further assess misfolding and subsequent aggregation of the dominant mutations p.Glu809Lys, p.Glu830Ala, and p.His313Tyr, as well as the recessive Wolfram syndrome–causing mutation p.Pro724Leu, we performed immunoblot analysis of detergent-soluble and detergent-insoluble lysates from HeLa cells transiently expressing these disease-causing mutations or wild-type WFS1. We found that the formation of insoluble and high–molecular weight complexes was much more prominent in cells expressing disease-causing variants than in cells expressing wild-type WFS1 (Fig. 4). These results suggest that folding defects could be a common feature of WFS1 missense mutations. We did not observe any clear differences between the recessive missense mutation and the dominant missense mutations in these two experiments; this may be a consequence of the high levels of protein expression in Cos7 and HeLa cells, potentially overwhelming the cell’s ability to efficiently fold even proteins with WFS1 variants resulting in minor folding defects.
Figure 4.
High–molecular weight complexes of WFS1 mutants in detergent-insoluble fractions. HeLa cells were transfected with control (pcDNA), FLAG-tagged wild-type WFS1 (WT), mutant WFS1 p.Pro724Leu (p.P724L), p.Glu809Lys (p.E809K), p.Glu830Ala (p.E830A), or mutant WFS1 p.His313Tyr (p.H313Y) expression plasmid and separated into detergent-soluble (Sol) and detergent-insoluble (Insol) fractions with anti-FLAG antibody. IB, immunoblot.
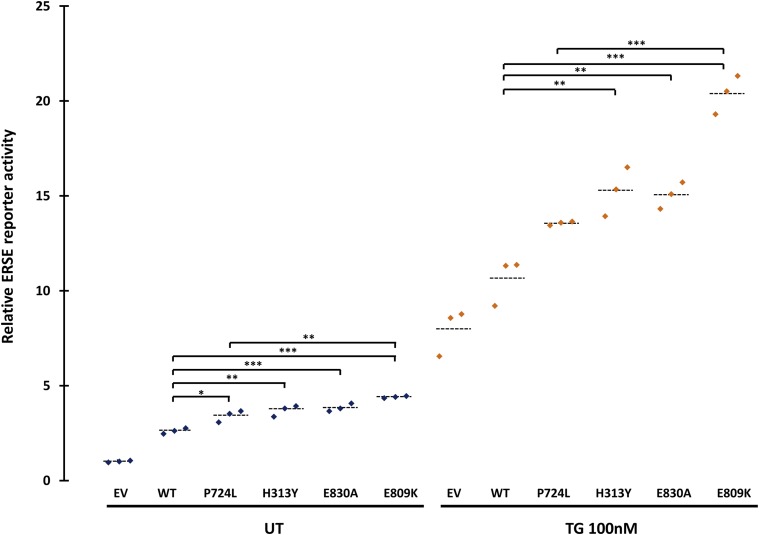
WFS1 is known to negatively regulate the cellular response to ER stress, thereby raising the possibility that dominant WFS1 mutations could alter WFS1 function by causing or enhancing a dysregulated ER stress response (17,21). To test this hypothesis, we investigated the effects of the p.Glu809Lys, p.Glu830Ala, and p.His313Tyr variants, as well as the recessive p.Pro724Leu mutation, on the ER stress response. We cotransfected HeLa cells with a luciferase reporter construct containing an ERSE and 1) control (pcDNA); 2) wild-type WFS1; 3) the dominant mutations p.Glu809Lys, p.Glu830Ala, and p.His313Tyr; or 4) the recessive mutation Pro724Leu expression vector. The ERSE reporter reflects activation levels of the ER stress response. In the absence of the ER stress inducer TG, all of the dominant disease-causing variants activated the ERSE reporter significantly more than wild-type WFS1 (Fig. 5). The dominant p.Glu809Lys mutation results in significantly more ERSE reporter activation than the recessive p.Pro724Leu mutation (4.40 vs. 3.41, P = 0.0003), but the other mutations were not significantly different (mutation p.His313Tyr 3.70 vs. p.Pro724Leu 3.41, P = 0.35; mutation p.Glu830Ala 3.84 vs. p.Pro724Leu 3.41, P = 0.09). After inducing ER stress with TG, all the disease-causing dominant mutations exhibited significantly higher ERSE activity in TG-treated conditions than wild-type WFS1 (Fig. 5). Again the dominant p.Glu809Lys mutation showed the highest ERSE activity, which was significantly different from the recessive p.Pro724Leu mutation (21.32 vs. 13.44, P < 0.0001) (Fig. 5). Collectively, these results suggest that the dominant WFS1 mutations p.Glu809Lys, p.Glu830Ala, and p.His313Tyr strongly induce ER stress, whereas recessive variants, including the p.Pro724Leu variant, are less potent in inducing ER stress.
Figure 5.
Luciferase reporter assays in HeLa cells transfected with the ERSE reporter together with control (pcDNA) (EV), wild-type WFS1 (WT), mutant c.2171C>T p.Pro724Leu (P724L), mutant c.937C>T p.His313Tyr (p.H313Y), mutant c.2489A>C p.Glu830Ala (p.E830A), or mutant c.2425G>A p.Glu809Lys (p.E809K) expression plasmid. Cells were untreated (UT) or treated with TG (100 nmol/L) for 8 h. Relative intensity of luciferase (Promega Dual-Luciferase Reporter Assay System) was then measured (n = 3; dashed lines represent mean). Transfections were normalized with the pRL-TK vector (Promega) as an internal control. Asterisks indicate a significant difference analyzed by one-way ANOVA followed by Dunnett test: *P < 0.01, **P < 0.001, ***P < 0.0001.
Discussion
We report five patients with a novel congenital syndrome characterized by neonatal/infancy-onset diabetes, congenital cataracts, and congenital sensorineural deafness caused by heterozygous de novo WFS1 missense mutations. Functional analysis suggests that these dominant mutations tend to aggregate in the ER and induce robust ER stress. It is likely that strong ER stress induced by these dominant WFS1 mutations causes early cell death (probably starting in utero), resulting in the severe syndrome identified in our patients.
The clinical features identified in our patients are distinct from those described in patients with classic Wolfram syndrome, with the presentation of symptoms at such an early age being the major difference. All our patients were diagnosed with diabetes before 12 months of age, with two patients being diagnosed in the neonatal period. In contrast, patients with genetically confirmed recessive Wolfram syndrome develop insulin-dependent diabetes at a median age at diagnosis of 6 years (range 1–32 years) (22). Diabetes is often the presenting feature of Wolfram syndrome, with optic atrophy and deafness appearing months to years later (median age 11 and 14 years, respectively). In contrast, our patients were all diagnosed with sensorineural deafness soon after birth. Four out of five patients in our cohort also had eye manifestations, but only two of them had optic atrophy. In fact, congenital cataracts were the most common eye feature in our patients (diagnosed in four of five). Although cataracts have been reported in some patients with Wolfram syndrome, they are not a consistent feature of the disease and are not congenital. Interestingly, isolated congenital nuclear cataracts have been previously reported in one family with a heterozygous WFS1 mutation (13) (Fig. 1), highlighting the importance of WFS1 in function and/or development of the eye lens. The fourth cardinal feature of Wolfram syndrome, diabetes insipidus, is also the least consistent, being diagnosed in ∼40% of Wolfram syndrome patients, at a median age of 13 years (22). Currently none of our patients have diabetes insipidus, but they are all younger than the median age that this is diagnosed. We therefore cannot exclude the possibility that they might develop this feature in the future. Clinical follow-up in these patients will be needed to establish the presence of any developmental and/or growth delay. Another striking difference between our patients and typical Wolfram syndrome patients is the birth weight. Patients with classic Wolfram syndrome generally have a normal weight at birth (Exeter monogenic diabetes database, unpublished data), whereas all our patients have birth weight below the median for their gestational age and sex, which is consistent with impaired insulin secretion in utero.
The novel syndrome we are reporting is genetically different from classic Wolfram syndrome as it results from spontaneous heterozygous mutations rather than being recessively inherited. Additional dominantly acting WFS1 mutations have been previously reported to cause autosomal dominant forms of both optic atrophy and hearing loss (10–12), low-frequency hearing loss (8,9), congenital cataracts (13), and isolated adult-onset diabetes (7). In these patients, the dominant phenotype is less severe than typically seen in classic recessive Wolfram syndrome (Fig. 1).
The phenotype observed in our patients is more severe than that resulting from homozygous complete loss-of-function mutations, suggesting that it is not just loss of WFS1 activity that results in this phenotype. In vitro functional studies showed that the dominant mutations we report may act through a different mechanism than recessive Wolfram syndrome mutations, as they strongly induce ER stress both basally and in the presence of TG (Fig. 5). Protein aggregation is unlikely to play a major role in the pathogenesis of recessive Wolfram syndrome as 1) this condition is known to often result from complete absence of WFS1 protein caused by recessive null mutations and 2) heterozygous carriers of Wolfram syndrome–causing missense mutations are clinically unaffected. On the contrary, functional data on the dominant mutations identified in our patients support a role for protein aggregation. Taken together, our results suggest that the dominant mutations actively induce ER stress. Additional functional studies on β-cell lines and primary islets are needed to determine whether the combined toxic effects of protein aggregation and ER stress have a direct effect on insulin secretion, possibly as a consequence of β-cell death. Recessive missense and null WFS1 mutations result in Wolfram syndrome due to the inability to regulate the unfolded protein response rather than directly inducing ER stress. Functional studies on the mutation causing autosomal dominant adult-onset diabetes without other features (p.Trp314Arg) suggest that this mutation is also functionally different from the mutations we studied, as it impairs the protein’s ability to suppress the ER stress response after its activation (7). The mechanisms underlying the broad phenotypic spectrum caused by specific dominantly acting mutations in WFS1 are currently unknown. Further studies investigating the effect of specific dominant and recessive WFS1 mutations in other cell lines and animal models are needed to further our understanding of the pathophysiology underlying this broad spectrum of disease.
The results of our functional studies suggest a direct genotype–phenotype correlation within the dominant mutations we report, which could be explained by the differences in functional impact seen in the in vitro experiments. The three patients with the p.Glu809Lys mutation included the two patients with the most severe clinical phenotype (diabetes diagnosed before 6 months and additional severe neurological features). Cells transfected with the WFS1 construct harboring the p.Glu809Lys mutation showed the highest level of ERSE reporter activity both at baseline and after induction of ER stress in the luciferase assay experiments (Fig. 5). The heterozygous p.Glu809Lys mutation has been identified previously in two patients with features that included early-onset diabetes (diagnosed at 6 months and 2 years of age, respectively), deafness, and eye abnormalities (cataracts and optic atrophy) (4,6). In both patients, parental samples were not available for testing and the authors hypothesized the presence of a second, undetected WFS1 mutation. We suggest that these two patients have the same autosomal dominant syndrome we report. Patient 5, who is heterozygous for the previously reported p.His313Tyr mutation, does not have the full triad of symptoms identified in the other patients: he has early-onset diabetes, congenital deafness, and severe hypotonia but no cataracts or other eye features. In keeping with this being a less severe dominant phenotype, three previously reported patients had diabetes occurring outside the first year of life and no reports of eye abnormalities (5,18). The delayed manifestation of disease in these patients is consistent with the results of our in vitro experiments where the p.His313Tyr mutation resulted in a lower level of ER stress compared with the p.Glu809Lys mutation (Fig. 5). Further studies looking at the evolution of disease in patients with this rare syndrome will be needed to precisely define a genotype–phenotype relationship.
In conclusion, we describe a novel congenital syndrome characterized by neonatal/early-onset diabetes, sensorineural deafness, and congenital cataracts caused by dominant WFS1 mutations. Our findings highlight a new disease mechanism linked to WFS1 mutations and establish this gene as a cause of syndromic neonatal/early-onset diabetes. Elucidation of the complex roles played by WFS1 in regulation of insulin secretion and ER stress will be fundamentally important for current studies looking into therapeutic options for patients with Wolfram syndrome (23).
Supplementary Material
Article Information
Acknowledgments. The authors thank the families for participating in this study. The authors are grateful to Hana Lango-Allen and Anna-Marie Bussell, of the Department of Molecular Genetics, Royal Devon & Exeter NHS Foundation Trust, for technical assistance.
Funding. A.T.H. and S.E. are the recipients of a Wellcome Trust Senior Investigator award (grant WT098395/Z/12/Z). A.T.H. is employed as a core member of staff within the National Institute for Health Research–funded Exeter Clinical Research Facility and is a National Institute for Health Research Senior Investigator. E.D.F. is a Naomi Berrie Fellow in Diabetes Research. S.E.F. has a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant 105636/Z/14/Z). This work was partly supported by grants from National Institutes of Health (DK020579 and UL1 TR000448) to F.U.
Author Contributions. E.D.F., S.E.F., M.B.J., and S.E. performed the genetic analysis. E.D.F., S.E.F., and A.T.H. wrote the first draft of the manuscript. S.E.F., T.Y., D.A., J.M., R.C., S.E., and F.U. participated in manuscript improvement. T.Y., D.A., and J.M. participated in data acquisition. D.A., J.M., and F.U. participated in data analysis and interpretation. J.M., S.E., F.U., and A.T.H. participated in study conception and design. G.J. and R.C. performed bioinformatic analysis. F.A., M.M., N.L., V.O., and O.P. collected patient samples and clinical data. A.T.H. analyzed the clinical data. All authors reviewed the manuscript. A.T.H. is the guarantor of the clinical and genetic work and F.U. is the guarantor of the functional work, and both had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1296/-/DC1.
References
- 1.De Franco E, Flanagan SE, Houghton JA, et al. The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet 2015;386:957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flanagan SE, Haapaniemi E, Russell MA, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet 2014;46:812–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lango Allen H, Flanagan SE, Shaw-Smith C, et al.; International Pancreatic Agenesis Consortium . GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet 2011;44:20–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsunaga K, Tanabe K, Inoue H, et al. Wolfram syndrome in the Japanese population; molecular analysis of WFS1 gene and characterization of clinical features. PLoS One 2014;9:e106906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohayem J, Ehlers C, Wiedemann B, et al.; Wolfram Syndrome Diabetes Writing Group . Diabetes and neurodegeneration in Wolfram syndrome: a multicenter study of phenotype and genotype. Diabetes Care 2011;34:1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussenot A, Rouzier C, Quere M, et al. Mutation update and uncommon phenotypes in a French cohort of 96 patients with WFS1-related disorders. Clin Genet 2015;87:430–439 [DOI] [PubMed] [Google Scholar]
- 7.Bonnycastle LL, Chines PS, Hara T, et al. Autosomal dominant diabetes arising from a Wolfram syndrome 1 mutation. Diabetes 2013;62:3943–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bespalova IN, Van Camp G, Bom SJ, et al. Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum Mol Genet 2001;10:2501–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesperance MM, Hall JW 3rd, San Agustin TB, Leal SM. Mutations in the Wolfram syndrome type 1 gene (WFS1) define a clinical entity of dominant low-frequency sensorineural hearing loss. Arch Otolaryngol Head Neck Surg 2003;129:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiberg H, Hansen L, Kjer B, et al. Autosomal dominant optic atrophy associated with hearing impairment and impaired glucose regulation caused by a missense mutation in the WFS1 gene. J Med Genet 2006;43:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogewind BF, Pennings RJ, Hol FA, et al. Autosomal dominant optic neuropathy and sensorineual hearing loss associated with a novel mutation of WFS1. Mol Vis 2010;16:26–35 [PMC free article] [PubMed] [Google Scholar]
- 12.Rendtorff ND, Lodahl M, Boulahbel H, et al. Identification of p.A684V missense mutation in the WFS1 gene as a frequent cause of autosomal dominant optic atrophy and hearing impairment. Am J Med Genet A 2011;155A:1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry V, Gregory-Evans C, Emmett W, et al. Wolfram gene (WFS1) mutation causes autosomal dominant congenital nuclear cataract in humans. Eur J Hum Genet 2013;21:1356–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellard S, Lango Allen H, De Franco E, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 2013;56:1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014;42:W252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachdav G, Kloppmann E, Kajan L, et al. PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 2014;42:W337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonseca SG, Ishigaki S, Oslowski CM, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest 2010;120:744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen L, Eiberg H, Barrett T, et al. Mutation analysis of the WFS1 gene in seven Danish Wolfram syndrome families; four new mutations identified. Eur J Hum Genet 2005;13:1275–1284 [DOI] [PubMed] [Google Scholar]
- 19.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008;37:215–246 [DOI] [PubMed] [Google Scholar]
- 20.Inoue H, Tanizawa Y, Wasson J, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet 1998;20:143–148 [DOI] [PubMed] [Google Scholar]
- 21.Fonseca SG, Fukuma M, Lipson KL, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem 2005;280:39609–39615 [DOI] [PubMed] [Google Scholar]
- 22.de Heredia ML, Cleries R, Nunes V: Genotypic classification of patients with Wolfram syndrome: insights into the natural history of the disease and correlation with phenotype. Genet Med 2013;15:497–506 [DOI] [PubMed]
- 23.Lu S, Kanekura K, Hara T, et al. A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proc Natl Acad Sci U S A 2014;111:E5292–E5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.