Abstract
Although over 40 type 1 diabetes (T1D) risk loci have been mapped in humans, the causative genes and variants for T1D are largely unknown. Here, we investigated a candidate gene in the 21q22.3 risk locus—UBASH3A, which is primarily expressed in T cells where it is thought to play a largely redundant role. Genetic variants in UBASH3A have been shown to be associated with several autoimmune diseases in addition to T1D. However, the molecular mechanism underlying these genetic associations is unresolved. Our study reveals a previously unrecognized role of UBASH3A in human T cells: UBASH3A attenuates the NF-κB signal transduction upon T-cell receptor (TCR) stimulation by specifically suppressing the activation of the IκB kinase complex. We identify novel interactions of UBASH3A with nondegradative polyubiquitin chains, TAK1 and NEMO, suggesting that UBASH3A regulates the NF-κB signaling pathway by an ubiquitin-dependent mechanism. Finally, we show that risk alleles at rs11203203 and rs80054410, two T1D-associated variants in UBASH3A, increase UBASH3A expression in human primary CD4+ T cells upon TCR stimulation, inhibiting NF-κB signaling via its effects on the IκB kinase complex and resulting in reduced IL2 gene expression.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease arising from the destruction of the insulin-producing pancreatic β-cells. T1D is a common, complex disease with multiple genetic and environmental risk factors. Although genome-wide association studies have discovered over 40 chromosomal regions where there is significant statistical evidence of association with T1D (1), the causative genes and variants located in most of these regions have yet to be identified and their mechanisms of action determined. The current study focuses on one such locus, on chromosome 21q22.3, containing two genes, TMPRSS3 and UBASH3A, of which the latter is generally considered the most likely candidate for this T1D risk locus (2–9). Besides T1D, single nucleotide polymorphisms (SNPs) in the 21q22.3 chromosomal region are associated with several other autoimmune diseases, suggesting that this locus plays a broad role in autoimmunity (10–12).
UBASH3A (also known as STS-2, TULA, and CLIP4) is expressed primarily in T cells (13) and encodes a protein called ubiquitin-associated and SH3 domain–containing A (UBASH3A). The UBASH3A protein has three functional domains: the N-terminal UBA (ubiquitin-associated), SH3 (SRC homology 3), and the COOH-terminal histidine phosphatase (also referred to as phosphoglycerate mutase-like [PGM]) domains. It has been shown that the UBA domain binds to monoubiquitin (Ub); the SH3 domain interacts with CBL—an E3 ubiquitin ligase—and dynamin; and the PGM domain mediates self-dimerization (14–18). UBASH3A has four identified ubiquitination sites at lysine residues 15, 202, 309, and 358. Monoubiquitination at Lys 202 causes UBASH3A to adopt a closed conformation, which prevents the binding of the UBA domain to substrates in trans (17).
UBASH3A has a paralogue, UBASH3B (also known as STS-1 and TULA-2), which shares the same domain structure as UBASH3A. UBASH3B differs from UBASH3A in several significant ways. UBASH3B is ubiquitously expressed and has not been associated with any autoimmune or immune-mediated disorder in genome-wide association studies. UBASH3B displays significant protein tyrosine phosphatase activity both in vitro and in vivo and suppresses T-cell receptor (TCR) signaling by dephosphorylating ZAP-70 and Syk (19–23). In contrast, UBASH3A exhibits very weak, possibly acid-dependent, phosphatase activity in vitro; in vivo, knockout of the murine homolog of UBASH3A results in only a modest increase in phosphorylation of ZAP-70 (19,24).
Mice lacking either Ubash3a or Ubash3b alone, or in combination, exhibit no overt defects without immune challenge (13). However, T cells from Ubash3a−/−Ubash3b−/− double-knockout mice are hyperresponsive to TCR stimulation compared with T cells from wild-type (WT) mice, whereas T cells from Ubash3a−/− and Ubash3b−/− single-knockout mice display only a modest increase in proliferation (19). A similar hierarchical response is seen in the trinitrobenzene sulfonic acid–induced colitis model, where knockout of either Ubash3a or Ubash3b increases both inflammation and T-cell responses, but the Ubash3a−/−Ubash3b−/− double-knockout mice display a more severe phenotype than either of the single-knockout mice (25). These findings suggest that Ubash3a, in combination with Ubash3b, acts to inhibit T-cell activation and function, albeit by an as-yet-unresolved mechanism.
In the current study, we define the roles of UBASH3A and its genetic variants in T1D. Our study reveals novel interactions between UBASH3A, TAK1, and NEMO, which regulate TCR-induced NF-κB signaling. T1D risk alleles in UBASH3A are shown to be associated with increased UBASH3A expression and decreased IL2 expression in activated human primary CD4+ T cells.
Research Design and Methods
Sample Information
Frozen viable peripheral blood mononuclear cells (PBMCs) from healthy subjects of European ancestry were obtained from the Type 1 Diabetes Genetics Consortium (T1DGC) and from STEMCELL Technologies. UBASH3A genotyping data used in this study were either obtained from T1DGC or generated by PCR and Sanger sequencing. All biospecimens and data were represented by only nonidentifying codes. This study was approved by the University of Florida Institutional Review Board.
Generation of UBASH3A−/− and UBASH3A-Overexpressing Cell Clones
To knockout UBASH3A in Jurkat cells, a CRISPR construct targeting exon 2 of the UBASH3A gene was generated (26) using the guide sequence 5′-CACGGGGAGGAAGACGGCGG-3′ and the pSpCas9n(BB)-2A-Puro plasmid (Addgene plasmid #48141, a gift from Feng Zhang, Massachusetts Institute of Technology). To overexpress UBASH3A in Jurkat cells, a cDNA of the full-length, predominantly expressed transcript of UBASH3A was cloned into the pEF-DEST51 vector (Thermo Fisher Scientific). The CRISPR and pEF-DEST51 constructs were delivered into Jurkat cells by electroporation. Cell clones were obtained by limiting dilution. UBASH3A−/− clones were screened by PCR and Sanger sequencing, and UBASH3A-overexpressing clones were identified by immunoblotting with anti-V5.
Stimulation and Lysis of Cells
For stimulation longer than 30 min, cells were stimulated with 5 μg/mL plate-bound anti-CD3 (clone OKT3; BioLegend) with or without 5 μg/mL soluble anti-CD28 antibody (clone CD28.2; BioLegend). For stimulation shorter than 30 min, cells were starved of serum for 4 h, and then incubated for 30 min on ice with 10 μg/mL soluble anti-CD3 and 10 μg/mL soluble anti-CD28. Next, 10 μg/mL goat anti-mouse IgG (SouthernBiotech) was added to the cells, followed by incubation at 37°C for the indicated periods of time. Mock stimulation was performed with only cell culture medium.
Whole-cell lysates were extracted as previously described (27). For some experiments as indicated in the figure legends, whole-cell lysates were extracted with cell lysis buffer containing 25 mmol/L HEPES, pH 7.0, 150 mmol/L NaCl, 0.5% NP-40, 1 mmol/L EDTA, protease and phosphatase inhibitors, and 5 mmol/L N-ethylmaleimide (NEM), a deubiquitinase inhibitor.
Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific), according to the manufacturer’s protocol.
Transfection of HEK293T Cells
A cDNA of the full-length transcript of UBASH3A was cloned into the pcDNA3.1 vector (Thermo Fisher Scientific). Expression constructs encoding WT (Addgene plasmid #17608), lysine-48 (K48)-only (Addgene plasmid #17605), and lysine-63 (K63)-only Ub (Addgene plasmid #17606) tagged with hemagglutinin (HA) were gifts from Ted Dawson (Johns Hopkins University) (28). HEK293T cells were transfected using the X-tremeGENE HP DNA Transfection Reagent (Roche).
Coimmunoprecipitation and Immunoblotting
Coimmunoprecipitation and immunoblotting were performed as previously described (27), and antibodies used for these experiments are provided in Supplementary Table 1.
Quantitative PCR
Frozen PBMCs from healthy subjects were thawed, and primary CD4+ T cells were negatively selected using the Human CD4+ T Cell Isolation kit and LS MACS columns (Miltenyi). Cells were stimulated as described above for 6 h, and then total RNA was extracted using the RNeasy Plus Mini kit (QIAGEN). First-strand cDNA was synthesized using oligo(dT)20 primer and the iScript Select cDNA Synthesis kit (Bio-Rad). PCRs containing SYBR Green I were performed on a LightCycler 480 II real-time PCR instrument (Roche). All samples were tested in duplicate, and CT values were generated by the second derivative maximum method provided by the Roche software. Relative gene expression levels were calculated using the 2ΔCT method, where ΔCT = mean CT (GAPDH) – mean CT (gene of interest). The primers used for the assay are provided in Supplementary Table 2.
IL-2 ELISA
Cells were stimulated as described above for 24 h, and the Human IL-2 ELISA Max Deluxe kit (BioLegend) was used to measure IL-2 production in the culture supernatants.
Glutathione S-Transferase Pull-Down Assays
cDNA sequences of the UBA (residues 20–60), SH3 (residues 241–300), and PGM (residues 316–623) domains of UBASH3A were cloned into the pGEX-6P-1 vector (GE Healthcare). Glutathione S-transferase (GST)-tagged UBA, SH3, and PGM domains of UBASH3A were expressed in Escherichia coli and purified.
Whole-cell lysate from unstimulated Jurkat cells was precleared by incubation with Glutathione Sepharose 4B resin for 2 h at 4°C. One milligram of the precleared lysate was incubated for 2 h at 4°C with 20 μL of packed, fresh Glutathione Sepharose 4B resin, and with one of the following at the same molar concentration: 11.6 μg GST-tagged UBA, 12.6 μg GST-tagged SH3, or 22.9 μg GST-tagged PGM domain of UBASH3A and 10 μg GST. No lysate controls contained all the same ingredients as the experimental reactions except for the precleared lysate. Eluates were extracted from the resin using LDS sample buffer and analyzed by immunoblotting.
Binding of UBASH3A to Ubiquitin Oligomers
Five hundred micrograms of whole-cell lysate from HEK293T cells expressing V5-tagged UBASH3A was incubated with 0.5 μg of anti-V5 antibody for 4 h at 4°C. Next, 20 μL Protein G Dynabeads were added to the lysate, followed by 1-h incubation at 4°C. The beads were washed four times with the cell lysis buffer, and the supernatant was discarded. The beads were resuspended in the cell lysis buffer, and 2.5 μg of a specific type of ubiquitin oligomers was added to the beads, K48-linked Ub oligomers (K482–7), K63-linked Ub oligomers (K632–7), or methionine-1 (M1)-linked tetra-ubiquitin oligomers (M14) (Boston Biochem). After 2-h incubation at 4°C and four washes of the beads, the captured Ub oligomers and proteins were eluted using LDS sample buffer containing 50 mmol/L DTT. The eluates were subjected to immunoblotting with a polyclonal antiubiquitin antibody.
Statistical Analyses
The Prism software v. 6.0 (GraphPad) was used to perform Student t tests and linear regression analysis and to calculate Pearson correlation coefficient.
Results
UBASH3A Downregulates IL-2 Production
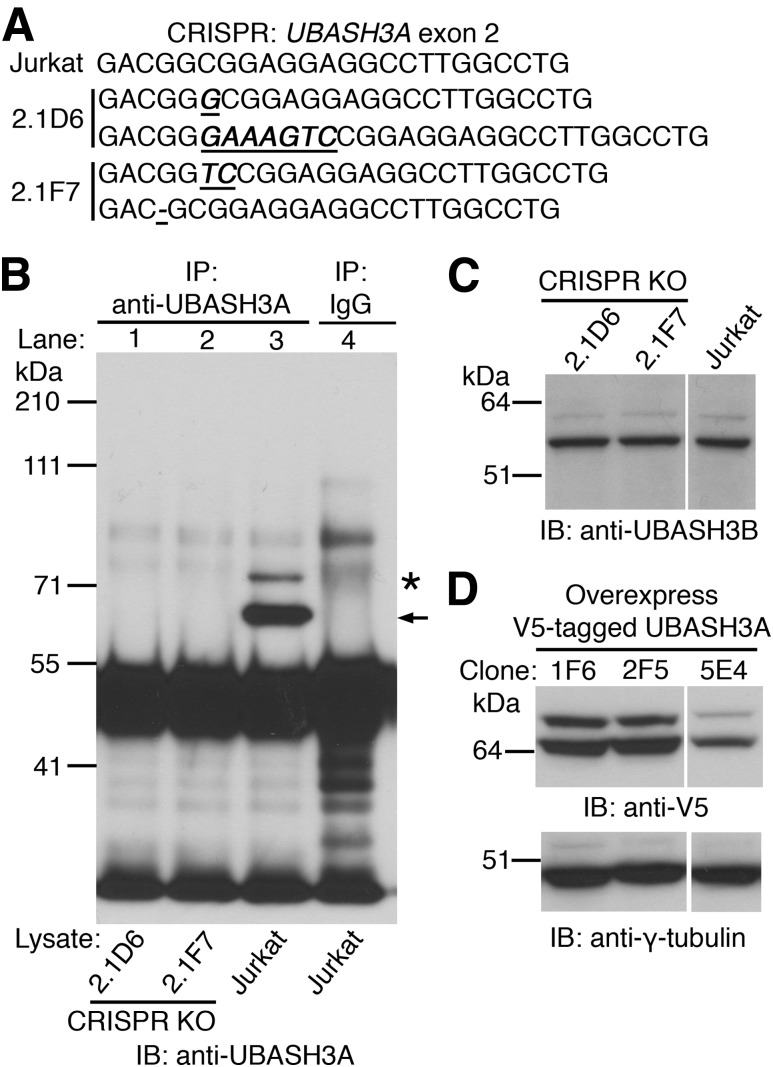
To explore the function of UBASH3A in humans, Jurkat lymphoblastic T cells null for UBASH3A were generated using CRISPR/Cas9. Two clones, 2.1D6 and 2.1F7, with distinct frameshifting mutations in exon 2 of the UBASH3A gene on both alleles, were obtained by limiting dilution (Fig. 1A). Full-length UBASH3A protein could not be detected in the whole-cell lysates from 2.1D6 and 2.1F7 cells (Fig. 1B), whereas normal levels of UBASH3B could (Fig. 1C), confirming the specificity of the knockouts. In parallel, a COOH-terminal V5-tagged version of the 623-amino–acid, major isoform of UBASH3A in humans was expressed in WT Jurkat cells, and three clones were generated by limiting dilution—1F6, 2F5, and 5E4. The expression levels of V5-tagged UBASH3A and its monoubiquitinated form were comparable in 1F6 and 2F5 cells but higher than those in 5E4 cells (Fig. 1D).
Figure 1.
Generation and characterization of Jurkat-derived clones deficient in or overexpressing UBASH3A. A: Two Jurkat-derived UBASH3A−/− clones, 2.1D6 and 2.1F7, were generated by CRISPR targeting exon 2 of the UBASH3A gene. Partial exon 2 sequences of parental Jurkat cells and the two UBASH3A−/− clones are shown. Insertions and deletions are underscored and italicized. B: Whole-cell lysates from 2.1D6, 2.1F7, and Jurkat cells were immunoprecipitated with either anti-UBASH3A (lanes 1–3) or IgG (lane 4). The immunoprecipitates were subjected to immunoblotting with a different anti-UBASH3A antibody. UBASH3A and its monoubiquitinated form are indicated by arrow and asterisk, respectively. C: Immunoblot analysis of whole-cell lysates from 2.1D6, 2.1F7, and Jurkat cells using anti-UBASH3B. D: Whole-cell lysates from three Jurkat-derived clones overexpressing UBASH3A with a COOH-terminal V5 tag were subjected to immunoblotting with anti-V5 and subsequently with anti–γ-tubulin after stripping. The blots in B–D are representative of two independent experiments. KO, knockout.
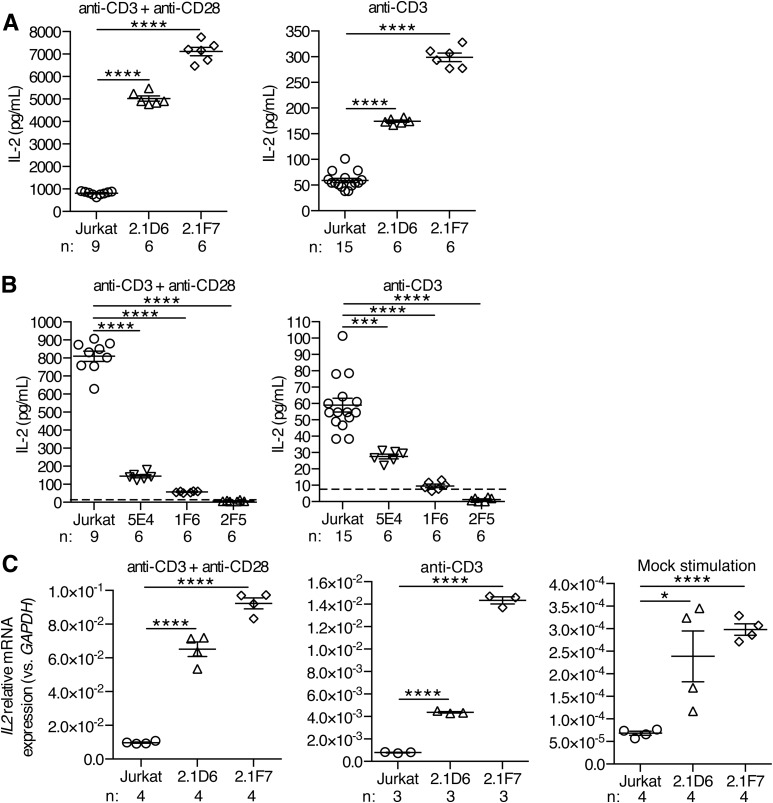
To examine the effect of modulating UBASH3A expression levels on T-cell activation and function, the UBASH3A−/− clones and the UBASH3A-overexpressing clones were stimulated for 24 h with either anti-CD3 alone or anti-CD3 plus anti-CD28. IL-2 production, with either stimulation regimen, was two- to eightfold higher in UBASH3A−/− 2.1D6 and 2.1F7 cells compared with parental Jurkat cells (P < 0.0001) (Fig. 2A). Overexpression of UBASH3A resulted in a comparably significant attenuation of IL-2 production upon stimulation (Fig. 2B). A dose effect was apparent among the three UBASH3A-overexpressing clones: the 5E4 clone had the lowest level of exogenous UBASH3A expression and produced the largest quantity of IL-2 (Figs. 1D and 2B).
Figure 2.
IL-2 production in Jurkat cells and in Jurkat-derived clones deficient in or overexpressing UBASH3A. A and B: ELISA measurements of IL-2 in the culture supernatants of Jurkat cells and of UBASH3A−/− (A) and UBASH3A-overexpressing (B) clones after 24-h stimulation with either anti-CD3 alone or anti-CD3 plus anti-CD28. The dashed lines indicate the ELISA detection limit (i.e., 7.8 pg/mL), and data points below the dashed lines represent values extrapolated from the IL-2 ELISA standard curve. The data are pooled from four ELISA experiments. C: Relative mRNA levels of IL2 in Jurkat cells and in UBASH3A−/− 2.1D6 and 2.1F7 cells. The cells were stimulated for 6 h as described in research design and methods. The data are pooled from two quantitative PCR experiments. Each data point in A–C represents an individual measurement of one sample. Numbers of samples (n) are shown. The mean and SEM values are indicated by solid lines and error bars, respectively. Unpaired two-tailed Student t tests were performed to assess statistical significance: *P = 0.02, ***0.0001 ≤ P < 0.0002, ****P < 0.0001.
Quantification of IL2 transcripts in the UBASH3A−/− clones revealed the same inverse correlation with UBASH3A expression as observed for IL-2 secretion (Fig. 2C). Thus, the effect of modulating UBASH3A expression on IL-2 production in Jurkat cells is largely attributable to the effect on IL2 transcription.
UBASH3A Suppresses TCR-Induced NF-κB Signaling
The NF-κB family of transcription factors plays a central role in innate and adaptive immune responses by regulating the proinflammatory and antiapoptotic gene transcription programs. In unstimulated T cells, NF-κB dimers are retained in an inactive state in the cytoplasm bound to IκB proteins. Upon TCR engagement, the IκB kinase (IKK) complex, consisting of IKKα and IKKβ (two kinases) and the regulatory component NEMO, is phosphorylated and activated and, in turn, phosphorylates IκB proteins leading to their degradation in proteasomes. Once released from the binding of IκB proteins, cytosolic NF-κB dimers translocate to the nucleus, bind specific DNA sequences, and promote transcription of target genes, including those encoding critical cytokines and chemokines, such as IL2, in T cells (29,30).
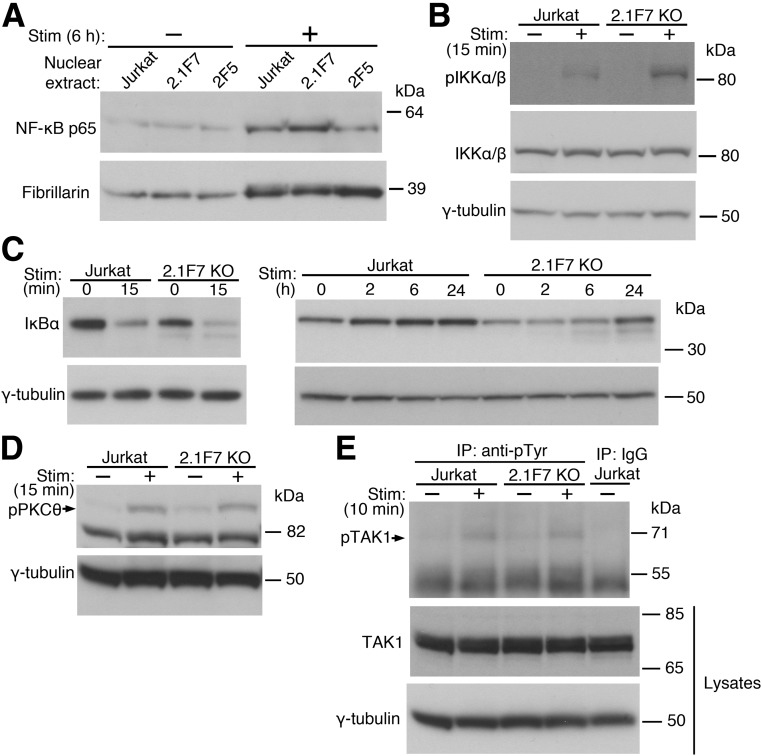
To determine whether the observed effect of UBASH3A on IL2 transcription is mediated through NF-κB signaling, nuclear extracts were prepared and immunoblotted for NF-κB p65 (also known as RelA). The baseline levels of nuclear NF-κB p65 were similar in UBASH3A−/− 2.1F7 cells, UBASH3A-overexpressing 2F5 cells, and Jurkat cells (Fig. 3A). However, upon 6-h anti-CD3 plus anti-CD28 stimulation, UBASH3A−/− 2.1F7 cells had increased level of NF-κB p65 in the nucleus relative to Jurkat cells, whereas 2F5 cells appeared to have less (Fig. 3A and Supplementary Table 3). These findings suggest that the effect of UBASH3A on IL-2 production is mediated by NF-κB and that UBASH3A acts upstream of the nuclear translocation of NF-κB.
Figure 3.
UBASH3A inhibits TCR-induced NF-κB signaling. A: Nuclear extracts were prepared from Jurkat, UBASH3A−/− 2.1F7, and UBASH3A-overexpressing 2F5 cells after mock stimulation or stimulation (Stim) with anti-CD3 plus anti-CD28 for 6 h and then subjected to immunoblotting with anti-NF-κB p65 followed by anti-fibrillarin (as loading control) after stripping. B–E: Jurkat and UBASH3A−/− 2.1F7 cells were mock stimulated or stimulated with anti-CD3 plus anti-CD28 for the indicated periods of time, and whole-cell lysates from the cells were analyzed. B: Immunoblotting with an antibody recognizing both phospho-IKKα (Ser176/180) and phospho-IKKβ (Ser177/181) and then, after stripping, with anti-IKKα/β and anti–γ-tubulin sequentially. C and D: Immunoblotting with anti-IκBα (C) or anti–phospho-PKCθ (Thr538) (D) and subsequently with anti–γ-tubulin after stripping. E: The lysates, which contained 5 mmol/L NEM, were immunoprecipitated with anti-phosphotyrosine or IgG, and the immunoprecipitates were subjected to immunoblotting with anti-TAK1. The lysates were also directly analyzed by immunoblotting with anti-TAK1 and subsequently with anti–γ-tubulin after stripping. The blots in A–E are representative of three independent experiments. KO, knockout.
To assess the effect of manipulating UBASH3A levels on the activity of the IKK complex, the amount of active, phosphorylated IKKα and IKKβ was measured after TCR/CD28 costimulation. Upon activation, UBASH3A−/− 2.1F7 cells had increased levels of phosphorylated IKKα/β compared with Jurkat cells (Fig. 3B and Supplementary Table 3). Consistent with this observation, the degradation of IκBα was accelerated in UBASH3A−/− 2.1F7 cells compared with Jurkat cells, either in the absence of stimulation or after anti-CD3 plus anti-CD28 stimulation for various periods of time ranging from 15 min to 6 h (Fig. 3C and Supplementary Table 3).
Knocking out UBASH3A did not markedly alter the activation of PKCθ and TAK1 upon TCR/CD28 costimulation, as similar amounts of phosphorylated PKCθ and phosphorylated TAK1 were observed in UBASH3A−/− 2.1F7 cells compared with parental Jurkat cells (Fig. 3D and E and Supplementary Table 3). PKCθ is selectively required for the TCR-induced NF-κB signaling (31) and is activated via PLCγ1 and diacylglycerol as a result of TCR ligation, as well as via costimulatory signals from CD28 and PI3K activation. PKCθ phosphorylates the membrane-associated protein CARMA1, which in turn recruits BCL10 and MALT1, forming the CBM signaling complex. The active CBM complex leads to the phosphorylation and activation of TAK1, a kinase that phosphorylates IKKα and IKKβ and hence activates the IKK complex (29,32). The increased activity of the IKK complex in TCR-stimulated UBASH3A−/− cells, compared with Jurkat cells, without comparable increases in the activation of the upstream signaling molecules PKCθ and TAK1 suggests that UBASH3A specifically suppresses the activation of the IKK complex by a previously unrecognized mechanism.
UBASH3A Both Binds to and Is Modified by K63-Mediated Ubiquitin Chains
Polyubiquitination plays a key role in propagating TCR-induced NF-κB signaling, acting both as a signal for protein turnover and as a scaffold for protein complex assembly. Upon TCR stimulation, IκB proteins are conjugated with K48-linked polyubiquitin (pUb) chains and then degraded via the ubiquitin-proteasome pathway. MALT1 of the CBM complex binds to TRAF2 and TRAF6, leading to TRAF2/6 oligomerization and activation of the E3 ubiquitin ligase activity of TRAF2/6. TRAF2/6 then catalyzes K63-linked polyubiquitination, which facilitates the activation of the TAK1 complex—consisting of TAK1, TAB1, and TAB2/3—and the IKK complex. In addition, MALT1, BCL10, TRAF6, and NEMO are all polyubiquitinated upon TCR stimulation, which promotes the NF-κB signaling (32,33).
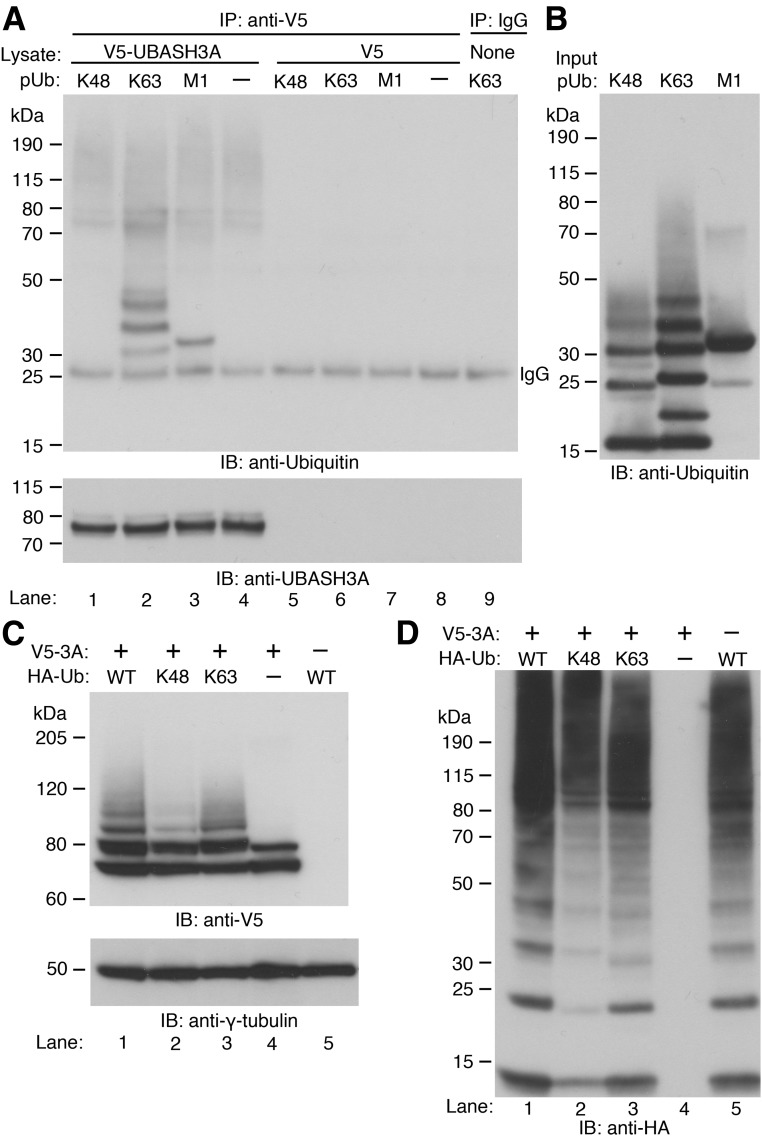
The importance of polyubiquitination in transducing TCR-induced NF-κB signaling and the presence of the functional ubiquitin-binding UBA domain in UBASH3A raise the possibility that UBASH3A might inhibit the NF-κB signaling pathway by either regulating or responding to polyubiquitination events (15–17). To determine the specificity of the UBASH3A UBA domain for ubiquitin linkages, V5-tagged UBASH3A was expressed in HEK293T cells and then immunoprecipitated using Protein G Dynabeads coupled with anti-V5. The binding of V5-tagged UBASH3A to synthetic Ub oligomers, including K48-pUb (2–7 Ub), K63-pUb (2–7 Ub), and M1-linked tetra-ubiquitin (4 Ub), was assessed by immunoblotting with an anti-ubiquitin antibody. V5-tagged UBASH3A preferentially bound to the K63-linked pUb chains (K63-linked chains with 2 and 3 ubiquitin moieties were detected with longer exposure of the blot shown in Fig. 4A) and the M1-linked tetra-Ub chain (Fig. 4A and B). However, no binding to K48-linked pUb chains was observed even with extended exposure of the blot shown in Fig. 4A (data not shown).
Figure 4.
Binding of UBASH3A to ubiquitin oligomers and ubiquitination of UBASH3A. A: Whole-cell lysates from HEK293T cells expressing V5-tagged UBASH3A (lanes 1–4) or V5 (lanes 5–8) were incubated with Protein G Dynabeads coupled with anti-V5. In lane 9, no lysate was incubated with Protein G Dynabeads coupled with IgG. After washing, the Dynabeads were incubated with equal amounts of K48-pUb (2–7 Ub), K63-pUb (2–7 Ub), or M1-linked tetra-ubiquitin (4 Ub) chains. The ubiquitin oligomers captured by the Dynabeads were detected by immunoblotting with anti-ubiquitin and subsequently with anti-UBASH3A after stripping. B: Two micrograms of each of the input ubiquitin oligomers used in A was subjected to immunoblotting with the same anti-ubiquitin antibody used in A. C and D: In lanes 1–3, HEK293T cells were cotransfected with constructs encoding V5-tagged UBASH3A and the indicated HA-tagged ubiquitin—WT, K48-only, and K63-only ubiquitin. In lanes 4 and 5, HEK293T cells were transfected with the construct encoding V5-tagged UBASH3A or the construct encoding HA-tagged WT ubiquitin, and whole-cell lysates containing 5 mmol/L NEM were subjected to immunoblotting with anti-V5 and subsequently with anti–γ-tubulin after stripping in C and with anti-HA in D. All the blots in A–D are representative of two independent experiments.
To determine what type of pUb chains can be conjugated to UBASH3A, HEK293T cells were cotransfected with constructs encoding V5-tagged UBASH3A and one of the following HA-tagged Ub—WT Ub, K48-only Ub, or K63-only Ub. The latter two types of Ub had all lysine residues mutated except for the indicated lysine residue, thus forcing polyubiquitin linkage via the indicated lysine residue. Immunoblotting with anti-V5 showed that all three types of Ub chains could be conjugated to UBASH3A (Fig. 4C). Less K48-polyubiquitinated UBASH3A than K63-polyubiquitinated UBASH3A was detected in the whole-cell lysates from the transfected HEK293T cells (Fig. 4C), but this difference might result from the slightly lower expression level of the exogenous K48-only Ub compared with that of the K63-only Ub (Fig. 4D).
UBASH3A Interacts With TAK1 and NEMO via Its SH3 Domain
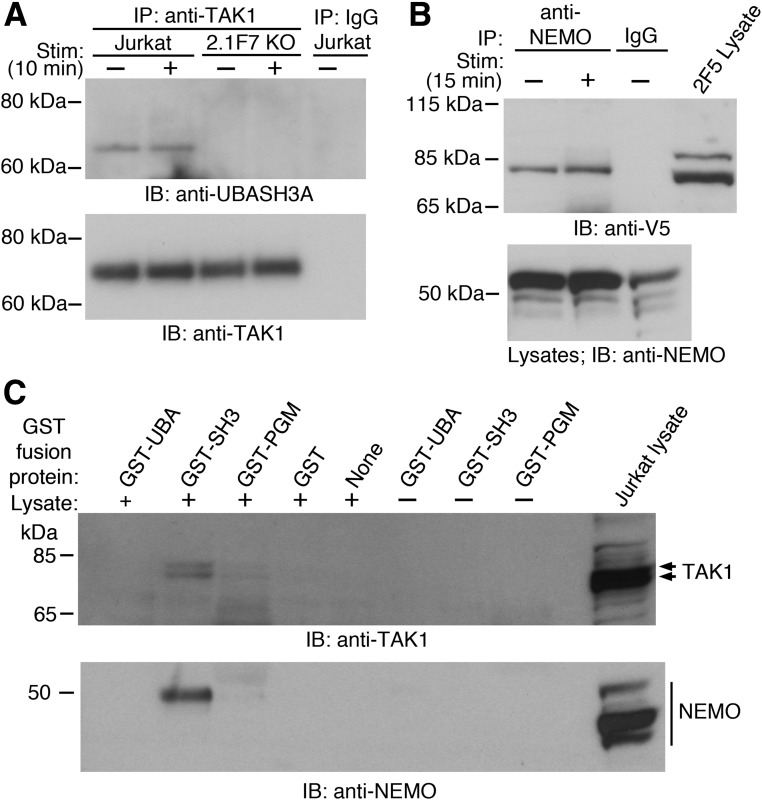
UBASH3A did not noticeably affect the polyubiquitination of NEMO and TRAF6 upon TCR/CD28 costimulation, as similar amounts of polyubiquitinated NEMO and polyubiquitinated TRAF6 were observed in Jurkat cells, UBASH3A−/− 2.1F7 cells, and UBASH3A-overexpressing 2F5 cells (Supplementary Fig. 1). UBASH3A could suppress the activation of the IKK complex upon TCR stimulation by interacting with subunits of the IKK and/or the TAK1 complex, either directly or by binding to the Ub chains that are associated with those molecules. UBASH3A coimmunoprecipitated with TAK1 in Jurkat cells with or without anti-CD3 plus anti-CD28 stimulation (Fig. 5A). Interaction between V5-tagged UBASH3A and NEMO was detected by coimmunoprecipitation in 2F5 cells, with or without anti-CD3 plus anti-CD28 stimulation (Fig. 5B).
Figure 5.
UBASH3A interacts with TAK1 and NEMO. A: Jurkat and UBASH3A−/− 2.1F7 cells were mock stimulated or stimulated (Stim) with anti-CD3 plus anti-CD28 for 10 min, and whole-cell lysates from the cells, which contained 5 mmol/L NEM, were immunoprecipitated with either anti-TAK1 or IgG. The immunoprecipitates were subjected to immunoblotting with anti-UBASH3A and subsequently with anti-TAK1 after stripping. B: UBASH3A-overexpressing 2F5 cells were mock stimulated or stimulated with anti-CD3 plus anti-CD28 for 15 min, and whole-cell lysates from the cells, which contained 5 mmol/L NEM, were immunoprecipitated with either anti-NEMO or IgG. The immunoprecipitates and the lysates were subjected to immunoblotting with anti-V5 and anti-NEMO, respectively. C: GST pull-down assays using GST-tagged UBA, SH3, and PGM domains of UBASH3A with and without whole-cell lysate from unstimulated Jurkat cells. The pull-down products and the Jurkat lysate were subjected to immunoblotting with anti-TAK1 and anti-NEMO. Arrows indicate isoforms of TAK1. All the blots in A–C are representative of two to three independent experiments. KO, knockout.
To map the interaction of UBASH3A with TAK1 and NEMO, the UBA, SH3, and PGM domains of UBASH3A were fused with GST and expressed and purified from E. coli (Supplementary Fig. 2). GST pull-down assays were carried out using whole-cell lysates from unstimulated Jurkat cells and each of the purified GST-tagged domains of UBASH3A. The UBASH3A SH3 domain was able to pull down both TAK1 and NEMO (Fig. 5C).
Two T1D-Associated SNPs in UBASH3A Regulate UBASH3A and IL2 Expression in Human Primary CD4+ T Cells
Our studies indicate that modulating UBASH3A expression levels in Jurkat cells results in considerable changes in both TCR-induced NF-κB signaling and IL-2 production. To extend these results to primary human T cells, the effects of T1D-associated SNPs on UBASH3A and IL2 expression were explored. A fine-mapping study using ImmunoChip identified rs11203203 (c.554–365G>A) and rs80054410 (c.554–541T>C)—two highly correlated (r2 = 1) SNPs in intron 4 of UBASH3A—as credible causative variants associated with T1D with high statistical significance (9). The minor allele of each of these SNPs confers risk for T1D and occurs at a frequency of approximately 0.3 in the HapMap CEU population. Previous studies showed that rs11203203 and rs80054410 are located in a putative enhancer or superenhancer region in human primary CD4+ T cells (9,34).
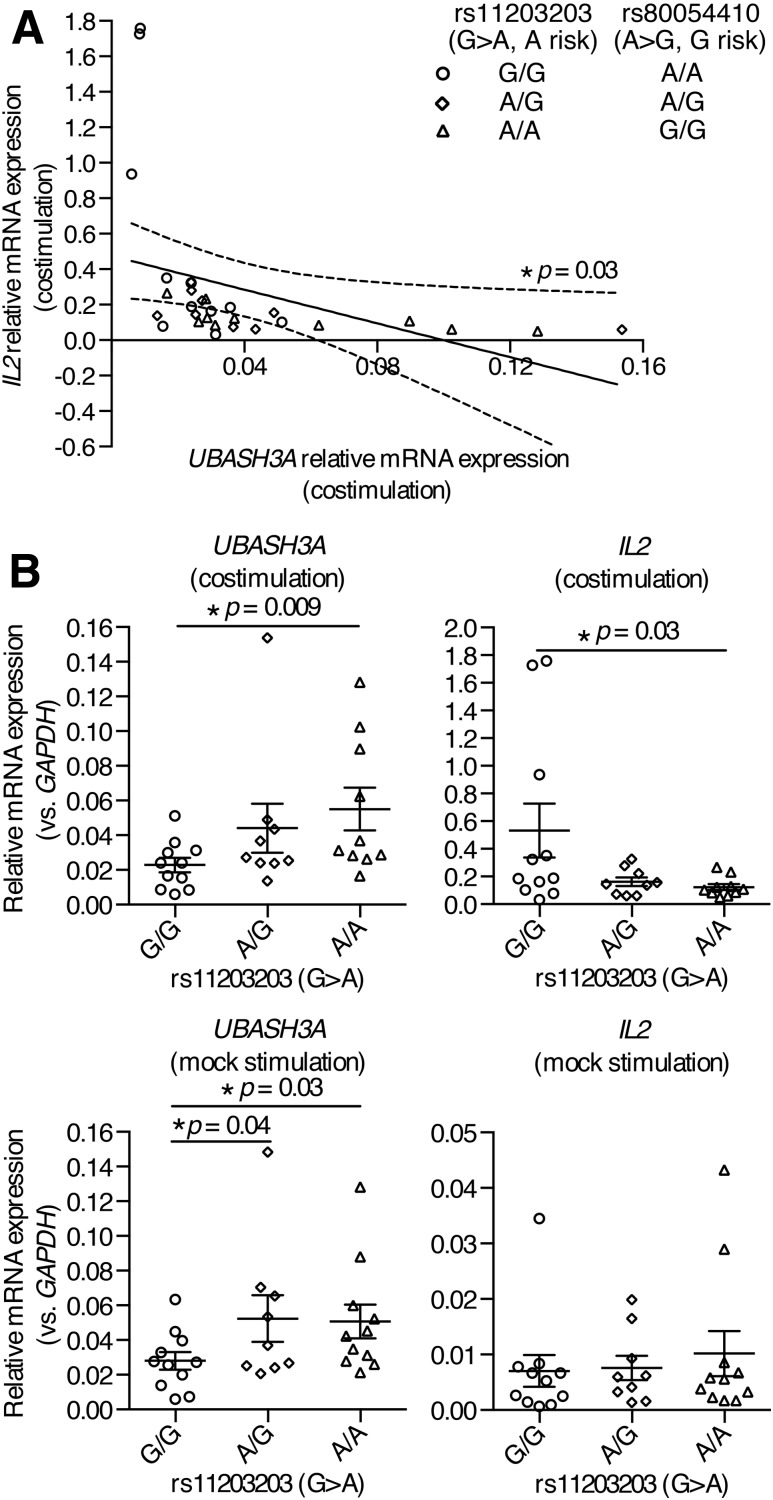
To delineate the functions of rs11203203 and rs80054410, CD4+ T cells were purified by negative selection from PBMCs from healthy subjects of European ancestry with different genotypes at these two SNPs (Fig. 6A, genotypes at rs80054410 are completely correlated with those at rs11203203). Quantitative PCR was conducted to measure the relative amounts of UBASH3A and IL2 transcripts in CD4+ T cells that were stimulated with anti-CD3 plus anti-CD28, or mock stimulated, for 6 h. In stimulated CD4+ T cells, the relative mRNA level of IL2 was inversely correlated with that of UBASH3A (Pearson r = −0.39, two-tailed P = 0.03) (Fig. 6A), consistent with the finding in Jurkat cells that UBASH3A downregulated IL-2 production upon TCR stimulation. In mock-stimulated CD4+ T cells, the relative mRNA levels of UBASH3A in subjects heterozygous at rs11203203 and in subjects homozygous for the minor risk allele (A) of rs11203203 were significantly higher than that in subjects homozygous for the major allele (G) (Fig. 6B). To isolate the effects of rs11203203 from other loci, allele-specific expression was assayed using mRNA-seq data on CD4+ T cells from 38 T1D subjects of European ancestry. The risk allele of the noncoding SNP rs11203203 led to imbalanced expression of the alleles of rs2277798, a coding SNP in exon 1 of UBASH3A (Supplementary Fig. 3). In stimulated CD4+ T cells, subjects homozygous for the minor, risk allele of rs11203203 had significantly more UBASH3A mRNA and less IL2 mRNA, compared with subjects homozygous for the major allele of rs11203203 (Fig. 6B).
Figure 6.
rs11203203 and rs80054410 regulate the expression of UBASH3A and IL2 in human primary CD4+ T cells. A and B: Human primary CD4+ T cells were negatively selected from PBMCs from healthy subjects and then stimulated with anti-CD3 plus anti-CD28 or with culture medium for 6 h. The relative mRNA levels of UBASH3A and IL2 are shown. Each data point represents one subject, and the data are pooled from six quantitative PCR experiments. In A, the best-fit line generated by linear regression is represented by a solid line, and the 95% CI bands of the best-fit line are indicated by dashed curves. Genotypes at rs11203203 and rs80054410 are shown. In B, the mean and SEM values are indicated by solid lines and error bars, respectively. Unpaired one-tailed Student t tests were performed to assess statistical significance.
Discussion
We previously identified a statistically significant association between SNPs in the chromosome 21q22.3 region and T1D in a linkage study in T1D multiplex families (2). Fine-mapping further localized the association, pointing toward UBASH3A as a candidate for the causal gene in the region (9). Most studies of UBASH3A have focused on its potential regulatory role as a phosphatase; however, UBASH3A has only very weak phosphatase activity relative to UBASH3B, and there is no clear difference in the specificity of the phosphatase domains of the two proteins that would explain the exclusivity of the observed genetic associations of multiple autoimmune diseases with UBASH3A. This led us to explore the function and specificity of the UBA and SH3 domains of UBASH3A in T cells.
We found that UBASH3A preferentially binds to the nondegradative K63- and M1-linked Ub chains but not detectably to the degradative K48-linked Ub chains and that UBASH3A downregulates TCR-induced NF-κB signaling by specifically suppressing the activation of the IKK complex. These results suggest that UBASH3A acts on the NF-κB signaling pathway through a mechanism involving ubiquitin-mediated protein complex formation rather than protein turnover. Our identification of two novel binding partners for UBASH3A, TAK1 and NEMO, lends further support to this idea as both proteins play key roles in the NF-κB signaling pathway. NEMO binds to M1- and K63-linked Ub chains (35,36) and to K63/M1-linked hybrid Ub chains (37). These interactions of NEMO with nondegradative Ub chains play critical roles in the NF-κB signaling pathway as unanchored K63-linked pUb chains can directly activate TAK1 and the IKK complex in vitro, possibly by inducing complex formation and trans-autophosphorylation (38), and residues of NEMO that are necessary for binding M1-linked linear ubiquitin chains are required for NF-κB activation by TNF-α and other agonists (36). We propose that besides its limited role as a phosphatase, UBASH3A specifically inhibits the activation of the IKK complex by competing with NEMO for the binding of K63-linked and/or M1-linked Ub chains and/or by altering the conformation of the IKK complex.
The 21q22.3 locus is associated with T1D (P = 1.2 × 10−15), although with a modest odds ratio of 1.16 (9). There are multiple SNPs, including rs11203203 and rs80054410, proximal to, or within, the transcription unit of UBASH3A that display statistically significant associations with T1D, and these SNPs may have independent, or even opposing, effects on the disease risk via different mechanisms (9). Indeed, a large number of cis-eQTL (expression quantitative trait loci) SNPs affecting UBASH3A expression in human primary CD4+ T cells have been identified (39). Controlling for the effects of these multiple variants considerably reduces the number of subjects available for any one genotype. Thus, in our study, we focused only on genotypes at rs11203203 and rs80054410, without controlling for the genotypes at other T1D-associated SNPs in UBASH3A. We controlled generally for genetic background by using only healthy subjects of European ancestry; it is possible that in T1D subjects, risk alleles at other loci, and possibly the disease itself, might amplify the effects of rs11203203 and rs80054410 on NF-κB signaling. Nevertheless, we were able to identify a modest, but statistically significant, effect of genotype at rs11203203 on UBASH3A transcript levels and a corresponding change in IL2 transcript levels in activated human primary CD4+ T cells. These observations in primary cells were completely consistent with the results we obtained when we artificially manipulated UBASH3A levels in Jurkat cells by knockout or overexpression.
In summary, our study reveals a previously unrecognized role for UBASH3A in human T cells: UBASH3A inhibits the TCR-induced NF-κB signaling pathway and hence IL2 expression, specifically by suppressing the activation of the IKK complex through interactions with TAK1 and NEMO and specific nondegradative ubiquitin chains. These functions of UBASH3A are enhanced by the risk alleles of two previously identified T1D-associated SNPs in UBASH3A, indicating that a reduced capacity of CD4+ T cells to express IL2, and possibly other critical genes regulated by NF-κB, in response to TCR stimulation contributes to T1D risk in a UBASH3A-dependent manner.
Supplementary Material
Article Information
Acknowledgments. This research uses resources from the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, and JDRF. The authors thank Matthew Mika (University of Virginia) for reagents, Jocyndra A. Wright (University of Florida) for technical assistance, and David A. Ostrov (University of Florida) for help with purification of GST-fusion proteins.
Funding. This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants DK106718, DK046635, and DK085678 (to P.C.) and by JDRF Advanced Postdoctoral Fellowship award 3-APF-2016-177-A-N (to Y.G.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.G. and P.C. conceived the study, interpreted the data, and wrote the manuscript. Y.G. and T.K.P. performed the experiments. Y.G. analyzed the data. J.R.B.N. and L.M.M. aligned and analyzed the mRNA-seq data. P.C. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1023/-/DC1.
References
- 1.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med 2009;360:1646–1654 [DOI] [PubMed] [Google Scholar]
- 2.Concannon P, Onengut-Gumuscu S, Todd JA, et al.; Type 1 Diabetes Genetics Consortium . A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes 2008;57:2858–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008;359:2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant SFA, Qu H-Q, Bradfield JP, et al.; DCCT/EDIC Research Group . Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes 2009;58:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plagnol V, Howson JMM, Smyth DJ, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 2011;7:e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson K, Wong R, Barriga KJ, et al. rs11203203 is associated with type 1 diabetes risk in population pre-screened for high-risk HLA-DR,DQ genotypes. Pediatr Diabetes 2012;13:611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frederiksen BN, Steck AK, Kroehl M, et al. Evidence of stage- and age-related heterogeneity of non-HLA SNPs and risk of islet autoimmunity and type 1 diabetes: the diabetes autoimmunity study in the young. Clin Dev Immunol 2013;2013:417657 [DOI] [PMC free article] [PubMed]
- 9.Onengut-Gumuscu S, Chen W-M, Burren O, et al.; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 2010;362:1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhernakova A, Stahl EA, Trynka G, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet 2011;7:e1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Gallo L-M, Sánchez E, Ortego-Centeno N, et al. Evidence of new risk genetic factor to systemic lupus erythematosus: the UBASH3A gene. PLoS One 2013;8:e60646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpino N, Turner S, Mekala D, et al. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity 2004;20:37–46 [DOI] [PubMed] [Google Scholar]
- 14.Wattenhofer M, Shibuya K, Kudoh J, et al. Isolation and characterization of the UBASH3A gene on 21q22.3 encoding a potential nuclear protein with a novel combination of domains. Hum Genet 2001;108:140–147 [DOI] [PubMed] [Google Scholar]
- 15.Feshchenko EA, Smirnova EV, Swaminathan G, et al. TULA: an SH3- and UBA-containing protein that binds to c-Cbl and ubiquitin. Oncogene 2004;23:4690–4706 [DOI] [PubMed] [Google Scholar]
- 16.Kowanetz K, Crosetto N, Haglund K, Schmidt MHH, Heldin C-H, Dikic I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J Biol Chem 2004;279:32786–32795 [DOI] [PubMed] [Google Scholar]
- 17.Hoeller D, Crosetto N, Blagoev B, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol 2006;8:163–169 [DOI] [PubMed] [Google Scholar]
- 18.Bertelsen V, Breen K, Sandvig K, Stang E, Madshus IH. The Cbl-interacting protein TULA inhibits dynamin-dependent endocytosis. Exp Cell Res 2007;313:1696–1709 [DOI] [PubMed] [Google Scholar]
- 19.San Luis B, Sondgeroth B, Nassar N, Carpino N. Sts-2 is a phosphatase that negatively regulates zeta-associated protein (ZAP)-70 and T cell receptor signaling pathways. J Biol Chem 2011;286:15943–15954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell 2007;27:486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal R, Carpino N, Tsygankov A. TULA proteins regulate activity of the protein tyrosine kinase Syk. J Cell Biochem 2008;104:953–964 [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Ren L, Kim S, et al. Determination of the substrate specificity of protein-tyrosine phosphatase TULA-2 and identification of Syk as a TULA-2 substrate. J Biol Chem 2010;285:31268–31276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas DH, Getz TM, Newman TN, et al. A novel histidine tyrosine phosphatase, TULA-2, associates with Syk and negatively regulates GPVI signaling in platelets. Blood 2010;116:2570–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Jakoncic J, Carpino N, Nassar N. Structural and functional characterization of the 2H-phosphatase domain of Sts-2 reveals an acid-dependent phosphatase activity. Biochemistry 2009;48:1681–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman TN, Liverani E, Ivanova E, et al. Members of the novel UBASH3/STS/TULA family of cellular regulators suppress T-cell-driven inflammatory responses in vivo. Immunol Cell Biol 2014;92:837–850 [DOI] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge Y, Onengut-Gumuscu S, Quinlan AR, et al. Targeted deep sequencing in multiple-affected sibships of European ancestry identifies rare deleterious variants in PTPN22 that confer risk for type 1 diabetes. Diabetes 2016;65:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim KL, Chew KCM, Tan JMM, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 2005;25:2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 2012;26:203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 1999;18:6853–6866 [DOI] [PubMed] [Google Scholar]
- 31.Sun Z, Arendt CW, Ellmeier W, et al. PKC-θ is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature 2000;404:402–407 [DOI] [PubMed] [Google Scholar]
- 32.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev 2012;246:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wertz IE, Dixit VM. Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2010;2:a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo Y-C, Lin S-C, Rospigliosi CC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell 2009;33:602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahighi S, Ikeda F, Kawasaki M, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009;136:1098–1109 [DOI] [PubMed] [Google Scholar]
- 37.Emmerich CH, Ordureau A, Strickson S, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci U S A 2013;110:15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z-P, Sun L, Chen X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 2009;461:114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raj T, Rothamel K, Mostafavi S, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 2014;344:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.