Abstract
Proliferative diabetic retinopathy (PDR) is a common cause of blindness in the developed world’s working adult population and affects those with type 1 and type 2 diabetes. We identified Runt-related transcription factor 1 (RUNX1) as a gene upregulated in CD31+ vascular endothelial cells obtained from human PDR fibrovascular membranes (FVMs) via transcriptomic analysis. In vitro studies using human retinal microvascular endothelial cells (HRMECs) showed increased RUNX1 RNA and protein expression in response to high glucose, whereas RUNX1 inhibition reduced HRMEC migration, proliferation, and tube formation. Immunohistochemical staining for RUNX1 showed reactivity in vessels of patient-derived FVMs and angiogenic tufts in the retina of mice with oxygen-induced retinopathy, suggesting that RUNX1 upregulation is a hallmark of aberrant retinal angiogenesis. Inhibition of RUNX1 activity with the Ro5–3335 small molecule resulted in a significant reduction of neovascular tufts in oxygen-induced retinopathy, supporting the feasibility of targeting RUNX1 in aberrant retinal angiogenesis.
Introduction
Neovascularization is a pathological feature of proliferative diabetic retinopathy (PDR), wet age-related macular degeneration, retinopathy of prematurity, cancer, and other conditions (1). Antiangiogenic therapies typically target vascular endothelial growth factor (VEGF) and are effective treatments for a number of neovascular ocular diseases and some solid tumors (2). Despite the success of anti-VEGF therapies, considerable interest remains in identifying new therapeutic targets for aberrant angiogenesis as anti-VEGF therapies may acutely trigger hemorrhages and tractional retinal detachments in patients with PDR, whereas extended therapy may lead to tissue atrophy, ischemia, and reperfusion injury (3–6).
A major challenge to understanding neovascularization in PDR is that animal models of diabetes do not develop the proliferative phase of diabetic retinopathy consistently (7). Fibrovascular membranes (FVMs) in patients are surgically removed to relieve retinal traction with associated retinal detachment and remain largely understudied as they are often discarded after ocular surgery. To establish platforms for discovering the mechanisms underlying aberrant angiogenesis, we developed methods for the isolation and characterization of vascular endothelial cells (ECs) from patient-derived PDR FVMs (8).
Research Design and Methods
The Internal Review Board of Massachusetts Eye and Ear (MEE) approved this study. Research protocols adhered to the ARVO Statement on Human Subjects and the tenets of the Declaration of Helsinki. All participants gave informed consent prior to surgery and inclusion in the study. Surgical samples were collected at MEE. Control retinal samples were obtained from cadaver eyes of subjects without a diagnosis of diabetes through an approved internal review board protocol from Massachusetts General Hospital (Supplementary Table 1).
Whole-Transcriptome Sequencing
CD31+ cells were isolated from FVMs as previously described (8). RNA-sequencing was performed using a HiSEq 2000 (Illumina), aligned to reference genome UCSC hg19/GRCh37 with TopHat, and analyzed using Partek Flow (Partek), CuffLinks, EdgeR, and DESeq2 (9). A mixed-model ANOVA was used with a threshold false discovery rate of <0.05 and fold change ≥ ±2 for significance. Gene ontology was determined using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (10).
Oxygen-Induced Retinopathy Model
Mouse care and experimental procedures were in accordance with MEE Institutional Animal Care and Use Committee regulations. Oxygen-induced retinopathy (OIR) was induced in wild-type C57BL/6J mice as previously described (11). Intravitreal injections with 1 µL of 75 μmol/L Ro5–3335 Runt-related transcription factor 1 (RUNX1) inhibitor or DMSO were performed on left eyes only at postnatal day (P)13 and P15 under ketamine/xylazine anesthesia. Pups were euthanized at P17, and eyes were collected, fixed in 4% paraformaldehyde, and used for retinal flat mounts (nvehicle = 7; nRo5–3335 = 9). Avascular and neovascular areas were quantified using Photoshop (Adobe Systems) as previously described (12).
Cell Culture
Human retinal microvascular ECs (HRMECs) (Cell Systems) and human umbilical vein ECs (HUVECs) (Lonza) were grown at 37°C with 5% CO2 using endothelial growth media plus antibiotics (Lonza) and 2% FBS (Atlanta Biologicals). For the quantitative RT-PCR (qRT-PCR) gene candidate screen in high glucose, cells were treated for 48 h with endothelial basal media-2 with 2% platelet-poor plasma-derived serum (Alfa Aesar) and d-glucose (Sigma-Aldrich) or osmotic control (mannitol and L-glucose; Sigma-Aldrich).
Small Interfering RNA Gene Knockdown
Small interfering RNA (siRNA) (75 nmol/L) (Integrated DNA Technologies; sequences in Supplementary Table 2) were transfected for 6–8 h using DharmaFECT 1 (GE Life Sciences/Dharmacon) in Opti-MEM (Life Technologies).
Immunostaining
FVMs and cells were incubated with mouse anti-RUNX1 (Santa Cruz Biotechnology) followed by biotinylated secondary antibody (Vector Laboratories), horseradish peroxidase–labeled avidin (PerkinElmer), tyramide signal amplification (PerkinElmer), Vector Red chromogenic substrate (Vector Laboratories), and Gill no. 3 hematoxylin counterstain (Sigma-Aldrich). Immunofluorescent staining for CD31 (mouse anti-CD31; DakoCytomation) and Ki67 (rabbit anti-Ki67; Novus Biologicals) was performed as previously described (8). The number of Ki67-positive nuclei was averaged for three fields of view per sample at ×20 original magnification.
Mouse retinal whole mounts were blocked with 1% BSA, 0.1% Triton X-100, and 3% serum in PBlec buffer, incubated with primary (isolectin IB4 Alexa Fluor 488 Conjugate, Life Technologies; rat anti-CD31, BD Pharmingen; and rabbit anti-RUNX1, LifeSpan BioSciences) and secondary antibodies (donkey anti-rat Alexa Fluor 594 and donkey anti-rabbit Alexa Fluor 647 or goat anti-rabbit Alexa Fluor 594; Life Technologies). Samples were imaged with an Axioskop 2 MOT Plus microscope (Carl Zeiss) or TCS-SP5 confocal microscope (Leica Microsystems).
qRT-PCR Analysis
RNA was extracted using RNeasy Mini Kits (Qiagen). Primers were purchased from Integrated DNA Technologies for 200 candidate genes (Supplementary Table 2). Genes were excluded from further analysis if there was no amplification or amplification was outside the linear range. This resulted in a final list of 101 genes analyzed by qRT-PCR. cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad Laboratories) and probed using FastStart Universal SYBR Green Master (Hoffmann-La Roche). Fluorescent intensities were normalized to spike-in controls (ERCC RNA Spike-In Mix; Life Technologies), hypoxanthine phosphoribosyltransferase 1, and B2-microglobulin.
Western Blot Analysis
Cells were lysed using RIPA buffer (Cell Signaling Technology). Samples were separated on a 4–15% SDS-PAGE, transferred to polyvinylidene difluoride membranes, blocked with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE), and probed with mouse anti-RUNX1 (Santa Cruz Biotechnology Inc.), rabbit anti–β-actin (Cell Signaling Technology), IRDye 680RD donkey anti-rabbit, and IRDye 800CW donkey anti-mouse (LI-COR Biosciences) antibodies. Immunoreactive bands were visualized using the Odyssey Infrared Imaging System, and band intensities normalized to β-actin were quantified using Image Studio version 2.1 (LI-COR Biosciences).
Scratch-Wound Migration Assay
Migration was assessed with the scratch-wound assay (13). One scratch was generated per well and imaged on an EVOS imaging system every 3 h for 12 h. Images were analyzed with the TScratch Matlab module (14).
Tube-Forming Assay
HRMECs were treated overnight with 75 μmol/L Ro5–3335 or DMSO before plating 20,000 cells onto wells precoated with basement membrane extract (Trevigen). Cells were imaged 6 h after plating, and tube formation was quantified using the Angiogenesis Analyzer plugin for ImageJ (National Institutes of Health).
Statistical Analysis
Results are presented as mean ± SEM. The Student t test was performed for comparisons between two groups, and one-way ANOVA (Kruskal-Wallis test) was used for comparisons between multiple groups. A P value <0.05 was considered significant.
Results
RNA Sequencing of CD31+ Cells From FVMs
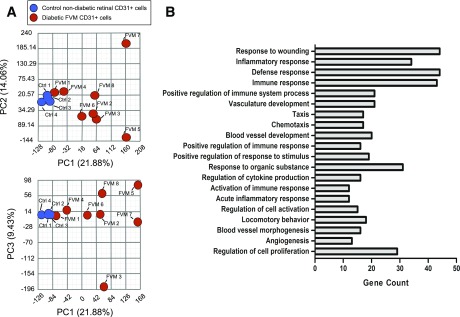
Whole transcriptomic profiles were constructed for CD31+ cells from FVMs and compared with transcriptomes of CD31+ cells from postmortem retinas isolated from individuals without diabetes (Supplementary Fig. 1 and Supplementary Table 1). Postmortem retinas were used to define expression baseline because there is no normal correlate of FVMs. CD31+ cells were identified as vascular ECs based on their expression of five vascular endothelial markers: CD93, CD31, KLF4, ESAM, and VEGFR1 (Supplementary Table 3). However, because a single marker, namely CD31, was used for cell isolation, the presence of other cell types cannot be ruled out. Principal component analysis of the transcriptomes demonstrated congruent expression profiles for control samples, whereas profiles of cells from the FVM samples had more variable gene expression patterns (Fig. 1A). The data sets are accessible through the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE94019) (15).
Figure 1.
Principle component analysis of transcriptomes and gene ontology of candidate genes. A: Two-dimensional representations of the three most significant principle components (PCs). PC1, PC2, and PC3 account for 45.37% of the observed sample variance. The wide distribution of transcriptomes from FVM-derived CD31+ cells contrasts the congruity of transcriptomes of cells from normal retinal samples, indicating significant variability in FVM gene expression levels. B: The top 20 pathways are presented in order of descending significance with the number of genes from the candidate gene list enriching the pathway.
Two hundred genes were identified as differentially expressed in CD31+ cells from FVMs involving gene ontology categories related to wound response, vessel development, angiogenesis, and other categories (Fig. 1B and Supplementary Table 4).
RUNX1 Is Upregulated in Cultured ECs Exposed to High Glucose
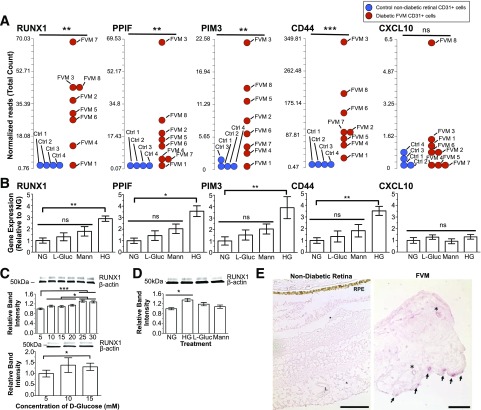
Hyperglycemia is a major risk factor for progression to PDR in patients with diabetes (16). High glucose-dependent genes from the candidate gene list were identified in primary HRMEC culture using a qRT-PCR screen. Four out of 101 genes were glucose responsive consistent with transcriptomic data (fold change ± SEM): RUNX1 (2.9 ± 0.2), peptidyl-prolyl cis-trans isomerase F (PPIF) (3.6 ± 0.5), serine/threonine-protein kinase PIM3 (PIM3) (3.9 ± 1.0), and CD44 (3.5 ± 0.4) (Fig. 2A and B). We focused on RUNX1 for further studies because of its documented roles in EC biology (17–19). Exposure of HRMEC and HUVEC cultures to high glucose induced a 30% increase in RUNX1 protein, whereas osmotic controls had no significant effect in HRMEC and HUVECs (Fig. 2C and D). Lastly, we confirmed increased RUNX1 protein expression in vessels from FVMs compared with retina from control subjects without diabetes using immunohistochemical staining (Fig. 2E).
Figure 2.
Effect of elevated glucose on HRMEC gene expression. A: Results of FVM RNA-sequencing (reads per kilobase of transcript per million mapped reads) show increased expression of RUNX1, PPIF, PIM3, and CD44 in ECs from patients with PDR compared with patients without diabetes. CXCL10 did not exhibit increased expression in FVMs (ncontrols = 4; nPDR-FVM = 8). B: Corresponding gene expression of RUNX1, PPIF, PIM3, CD44, and CXCL10 measured by qRT-PCR (HG, high d-glucose [30 mmol/L]; NG, normal d-glucose [5 mmol/L]). Each candidate gene had a marked increase in response to d-glucose but no statistically significant changes in response to osmotic controls (L-glucose or mannitol). CXCL10 is not glucose responsive, consistent with RNA-sequencing (n = 3; the experiment was performed in triplicate). C: Increasing d-glucose led to a dose-dependent increase of RUNX1 protein expression in HRMECs (top) and HUVECs (bottom) as determined by Western blot (n = 3; experiment performed in triplicate). D: The increase in RUNX1 protein in HRMECs induced by 30 mmol/L d-glucose was independent of osmotic forces (n = 3; experiment performed in triplicate). E: Normal retinal vessels showed no staining of RUNX1 (L, vessel lumen; RPE, retinal pigment epithelium) (left panel). Scale bars = 100 μm. A subset of vessels in FVM stained positively for RUNX1 (arrows). Asterisks denote nonstaining vessels (left). Scale bar = 50 μm. *P < 0.05; **P < 0.01; ***P < 0.001.
RUNX1 Regulates Migration and Proliferation of Endothelial Cells In Vitro
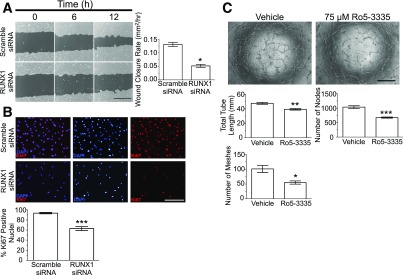
siRNA knockdown of RUNX1 resulted in a 60% decrease in wound closure rate, indicating a significant role of RUNX1 in endothelial cell migration (Fig. 3A). The coefficient of determination (r2 value) for the simple linear regression was calculated comparing wound closure (dependent variable) to time (independent variable) (control r2 = 0.99; scramble siRNA r2 = 0.97; and RUNX1 siRNA r2 = 0.97). This experiment was validated with a second siRNA targeting RUNX1 to rule out possible off-target effects (Supplementary Fig. 2B). ECs transfected with RUNX1 siRNA had 40% fewer Ki67-positive cells compared with controls 24 h after transfection, indicating RUNX1 also contributes to retinal EC proliferation (Fig. 3B).
Figure 3.
Role of RUNX1 in EC function. A: Scratch-wound assay using HRMECs at 0 (left column), 6 (middle column), and 12 h (right column) with scramble siRNA (top) and RUNX1 siRNA (bottom) treatment. Dark gray regions denote wound areas. Scale bar = 400 μm. Quantification of wound closure rates shows that knockdown of RUNX1 effectively inhibits wound closure (n = 10; experiment performed in duplicate). B: Ki67 staining 48 h posttransfection demonstrates significant reduction in cell number and proliferative capacity of RUNX1 siRNA-treated cells compared with cells treated with scramble siRNA. Scale bar = 200 μm. Quantification of percentage of DAPI-positive nuclei colocalized with Ki67 stain (n = 6; experiment performed in duplicate). C: HRMECs treated with Ro5–3335 RUNX1 inhibitor overnight exhibited reduced tube-forming capacity compared with vehicle-treated cells at 6 h after plating. There was statistically significant reduction in tube length, meshes, and nodes. Scale bar = 500 μm. n = 4; experiment performed in duplicate. *P < 0.05; **P < 0.01; ***P < 0.001.
Ro5–3335 RUNX1-Core Binding Factor β Inhibitor Blocks EC Tube Formation In Vitro and Aberrant Angiogenesis In Vivo
To investigate the potential therapeutic relevance of RUNX1, the small molecule Ro5–3335 RUNX1–core binding factor β inhibitor II was tested in vitro and in vivo (20). Treatment with RUNX1 inhibitor reduced total tube length (18% reduction), nodes (35% reduction), and meshes (46% reduction), supporting RUNX1’s role in vascular morphogenesis (Fig. 3C).
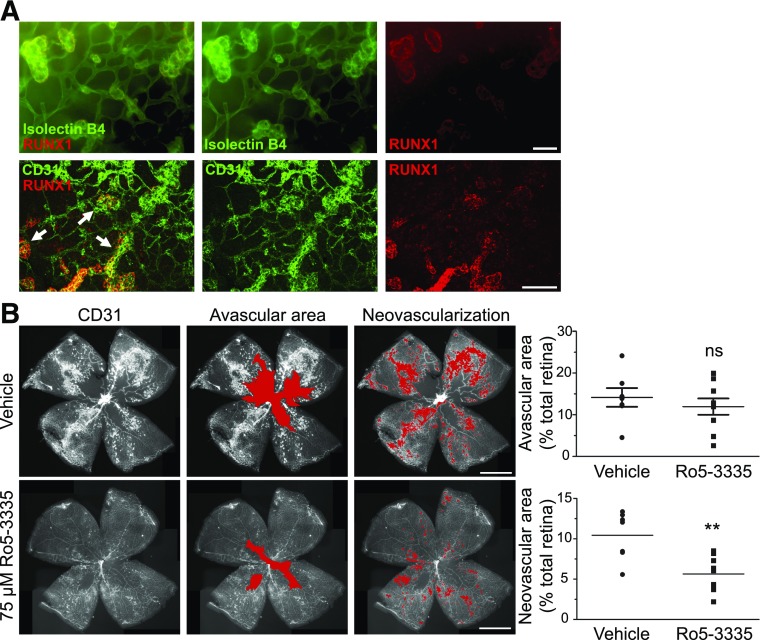
Neovascular tufts of P17 C57BL/6J mice with OIR identified by isolectin B4 and CD31 staining showed increased expression of RUNX1, whereas RUNX1 staining was absent from the underlying normal retinal vasculature, indicating a role for RUNX1 in active pathological angiogenesis (Fig. 4A). In vivo testing of the effects on aberrant angiogenesis of RUNX1 inhibition was conducted by injecting Ro5–3335 intravitreally in mice with OIR. There was no significant change in the extent of vaso-obliteration, and no effect was observed in the contralateral eye (data not shown). Eyes treated with inhibitor exhibited a 50% reduction in neovascularization compared with vehicle-treated eyes (Fig. 4B).
Figure 4.
RUNX1 inhibition reduces neovascularization in the OIR model. A: Retina of P17 C57BL/6J mice with OIR costained for RUNX1 and vessels (IB4 or CD31) showing positive RUNX1 staining conforming to neovascular tufts (arrows) and not to normal underlying vasculature. Scale bars = 50 μm. B: Retinal flat mounts of P17 C57BL/6J mice following OIR induction and intravitreal injection of 75 μmol/L Ro5–3335 or vehicle at P13 and P15 (red overlay identifies the avascular area and neovascularization, respectively). Scale bars = 1 mm. There was a nonsignificant downward trend in avascular area but a significant reduction in neovascularization in the treated group compared with vehicle-treated group (nvehicle = 7; nRo5–3335 = 9). Experiment performed in triplicate. **P < 0.01.
Discussion
Our analyses of CD31+ cells from FVMs of patients with PDR in conjunction with in vitro and in vivo validation assays suggest a role for RUNX1 in the proliferation, migration, and morphogenesis of ECs in vitro as well as in aberrant angiogenesis in vivo. RUNX proteins have pleiotropic functions in vascular development, hematopoiesis, and cancer through direct transcriptional regulation of target genes and complex interactions with fundamental signaling mechanisms including Notch and transforming growth factor-β pathways (21,22). RUNX1 is a major contributor to cancer progression and metastasis, most notably in acute myeloid leukemia, and also plays a role in angiogenesis (18,23–26).
Previous studies in model organisms have shown that glucose levels trigger RUNX1 expression via reactive oxygen species–mediated upregulation of hypoxia-inducible factor 1 and regulate hematopoietic stem cell production by endothelial to hematopoietic transition in a RUNX1-dependent manner (27,28). Consistent with these observations, our results demonstrate that high glucose regulates RUNX1 and implicates RUNX1 in aberrant retinal angiogenesis.
Previous studies of PDR have mainly focused on gene expression in FVMs or biomarkers in the vitreous (29,30). Although FVMs from PDR may be highly informative pathological tissues, they are largely unstudied. The window for processing samples postsurgery is narrow, and intraoperative confounders such as FVM size, location, and involvement of sensory retina further complicate successful sample acquisition. CD31+ cells from FVMs do not grow readily in culture, making additional experimentation difficult. Limited sample size may also skew our gene ontology analysis.
We report elevated RUNX1 expression in ECs of patient-derived FVMs from patients with PDR. Also, we demonstrate a role for RUNX1 in EC migration, proliferation, and vascular morphogenesis. Furthermore, we report selectively enhanced expression of RUNX1 in neovascular tufts in an experimental model of OIR and that inhibition of RUNX1 function reduced retinal neovascularization. These findings, including the high glucose–dependent expression of RUNX1, identify a novel pathway of potential therapeutic interest and implicate RUNX1 in aberrant angiogenesis in multiple conditions.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Magali Saint-Geniez for insightful discussions and providing mouse retinal samples, Dr. Eric Ng, Mark Graham, and Dr. Arturo Machuca-Parra for helpful discussions, and Dr. Christina Marko for technical contributions (all from Schepens Eye Research Institute, Boston, MA). The authors also thank Kristin Waraska and Andrew Gagne of the Harvard Biopolymers Facility, Harvard Medical School, and the Ocular Genetics Institute core facility, Massachusetts Eye and Ear (Boston, MA) for their insights and work.
Funding. The work performed in the manuscript was supported by National Institutes of Health grants R01-EY-005318 (to P.A.D.), R00-EY-021624 (to J.F.A.-V.), UH2NS100121-01 (to J.F.A.-V.), R21-EY-027061 (to L.A.K.), K12-EY-16335 (to L.A.K.), and Center Core grant P30-EY-003790; American Diabetes Association Innovation Award 7–12-IN-11 (to P.A.D.); Massachusetts Lions Eye Research Fund (to J.F.A.-V.); E. Matilda Ziegler Foundation for the Blind (to L.A.K.); Karl Kirchgessner Foundation (to L.A.K.); Department of Ophthalmology, Harvard Medical School (to J.F.A.-V. and L.A.K.); the Howard Hughes Medical Institute Medical Research Fellows Program (to D.J.O.); and the Vascular Biology Program at Boston Children’s Hospital (to D.R.B.).
Duality of Interest. P.A.D., L.A.K., and J.F.A.-V. have filed a provisional patent application on RUNX1 regulation for the treatment of aberrant angiogenesis and PDR. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.D.L. and D.J.O. performed the experiments, analyzed the data, and wrote the manuscript. L.L.W., D.A., C.P.-W., A.V.S., and J.C.-V. performed experiments. D.M., A.O.S.-R., D.E., and D.R.B. provided tissue samples and reviewed and edited the manuscript. T.v.Z., L.S., and X.G. obtained data and reviewed the manuscript. P.A.D., L.A.K., and J.F.A.-V. conceived the project, analyzed the data, and wrote the manuscript. P.A.D., L.A.K., and J.F.A.-V. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1035/-/DC1.
J.D.L. and D.J.O contributed equally to this work.
J.F.A.-V., L.A.K., and P.A.D. contributed equally to this work.
References
- 1.Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol 2011;27:563–584 [DOI] [PubMed] [Google Scholar]
- 2.Kim LA, D’Amore PA. A brief history of anti-VEGF for the treatment of ocular angiogenesis. Am J Pathol 2012;181:376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu YJ, Hsieh YT, Yeh PT, Huang JY, Yang CM. Combined tractional and rhegmatogenous retinal detachment in proliferative diabetic retinopathy in the anti-VEGF era. J Ophthalmol 2014;2014:917375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuiper EJ, Van Nieuwenhoven FA, de Smet MD, et al. The angio-fibrotic switch of VEGF and CTGF in proliferative diabetic retinopathy. PLoS One 2008;3:e2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saint-Geniez M, Maharaj AS, Walshe TE, et al. Endogenous VEGF is required for visual function: evidence for a survival role on müller cells and photoreceptors. PLoS One 2008;3:e3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sang DN, D’Amore PA. Is blockade of vascular endothelial growth factor beneficial for all types of diabetic retinopathy? Diabetologia 2008;51:1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo DH, Cho CS, Kim JH, Jun HO, Kim JH. Animal models of diabetic retinopathy: doors to investigate pathogenesis and potential therapeutics. J Biomed Sci 2013;20:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim LA, Wong LL, Amarnani DS, et al. Characterization of cells from patient-derived fibrovascular membranes in proliferative diabetic retinopathy. Mol Vis 2015;21:673–687 [PMC free article] [PubMed] [Google Scholar]
- 9.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith LE, Wesolowski E, McLellan A, et al. Oxygen-induced retinopathy in the mouse. Invest Ophthalmol Vis Sci 1994;35:101–111 [PubMed] [Google Scholar]
- 12.Connor KM, Krah NM, Dennison RJ, et al. Quantification of oxygen-induced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat Protoc 2009;4:1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007;2:329–333 [DOI] [PubMed] [Google Scholar]
- 14.Gebäck T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 2009;46:265–274 [DOI] [PubMed] [Google Scholar]
- 15.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Diabetes Control and Complications Trial Research Group The relationship of glycemic exposure (HbA1c) to the risk of development and progression of retinopathy in the diabetes control and complications trial. Diabetes 1995;44:968–983 [PubMed] [Google Scholar]
- 17.McLeod DS, Hasegawa T, Baba T, et al. From blood islands to blood vessels: morphologic observations and expression of key molecules during hyaloid vascular system development. Invest Ophthalmol Vis Sci 2012;53:7912–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwatsuki K, Tanaka K, Kaneko T, et al. Runx1 promotes angiogenesis by downregulation of insulin-like growth factor-binding protein-3. Oncogene 2005;24:1129–1137 [DOI] [PubMed] [Google Scholar]
- 19.Namba K, Abe M, Saito S, et al. Indispensable role of the transcription factor PEBP2/CBF in angiogenic activity of a murine endothelial cell MSS31. Oncogene 2000;19:106–114 [DOI] [PubMed] [Google Scholar]
- 20.Cunningham L, Finckbeiner S, Hyde RK, et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFβ interaction. Proc Natl Acad Sci U S A 2012;109:14592–14597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev 2005;19:2331–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr Opin Genet Dev 2003;13:43–47 [DOI] [PubMed] [Google Scholar]
- 23.Doll A, Gonzalez M, Abal M, et al. An orthotopic endometrial cancer mouse model demonstrates a role for RUNX1 in distant metastasis. Int J Cancer 2009;125:257–263 [DOI] [PubMed] [Google Scholar]
- 24.Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J 2012;31:4124–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planagumà J, Díaz-Fuertes M, Gil-Moreno A, et al. A differential gene expression profile reveals overexpression of RUNX1/AML1 in invasive endometrioid carcinoma. Cancer Res 2004;64:8846–8853 [DOI] [PubMed] [Google Scholar]
- 26.Nagamachi A, Htun PW, Ma F, et al. A 5′ untranslated region containing the IRES element in the Runx1 gene is required for angiogenesis, hematopoiesis and leukemogenesis in a knock-in mouse model. Dev Biol 2010;345:226–236 [DOI] [PubMed] [Google Scholar]
- 27.Harris JM, Esain V, Frechette GM, et al. Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood 2013;121:2483–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imanirad P, Solaimani Kartalaei P, Crisan M, et al. HIF1α is a regulator of hematopoietic progenitor and stem cell development in hypoxic sites of the mouse embryo. Stem Cell Res (Amst) 2014;12:24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa K, Yoshida S, Kobayashi Y, et al. Microarray analysis of gene expression in fibrovascular membranes excised from patients with proliferative diabetic retinopathy. Invest Ophthalmol Vis Sci 2015;56:932–946 [DOI] [PubMed] [Google Scholar]
- 30.McAuley AK, Sanfilippo PG, Hewitt AW, et al. Vitreous biomarkers in diabetic retinopathy: a systematic review and meta-analysis. J Diabetes Complications 2014;28:419–425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.