Abstract
Glioblastoma multiforme is one of the most lethal types of brain cancer. With limited success from conventional therapies, the cancer stem cell theory was developed, and investigation into microRNAs (miRs) has facilitated understanding of this theory. The present study demonstrated that miR-141 is suppressed in sorted cluster of differentiation (CD) 133(+) glioblastoma stem cells (GSCs) compared with CD133(−) non-glioblastoma stem cells (NSCs) from patient samples. In addition, miR-141 overexpression inhibited the sphere formation ability of GSCs in vitro and in vivo. Furthermore, Jagged1 may reverse the effect of miR-141; miR-141 was revealed to target the 3′-untranslated region of Jagged1, thereby inhibiting the stemness of GSCs. Thus, miR-141 may serve as a potent antioncomir targeting cancer stem cells, and may facilitate the development of therapeutic targets to prolong the overall survival of patients with glioblastoma.
Keywords: microRNA-141, Jagged1, glioblastoma, cancer stem cell, self-renewal
Introduction
Glioma is a common type of brain cancer originating from various types of glial cells residing in the brain, including ependymal cells, astrocytes and oligodendrocytes. Glioblastoma multiforme, derived from astrocytes and one of the three types of glioma, is the most common form of malignant brain tumor (1–4). Due to the fast-growing nature of astrocytes and abundant vascular network within glioblastoma, this type of tumor is develops rapidly and aggressively, and accounts for 15.4% of all primary brain tumors and 60–75% of astrocytomas (1,2). Currently, patients with glioblastoma are estimated to have a median survival time of 14.6 months and a 30% two-year survival rate. The primary therapies for glioblastoma are conventional surgery and radiotherapy. However, glioblastoma cells remain active, therefore tumors may recur and develop resistance to radio- and chemotherapy. The median time for disease recurrence following standard therapy is 6.9 months (2,5). Thus, this type of tumor remains one of the most life-threatening; identifying novel therapeutic targets and molecular markers is essential.
Conventional tumor therapies have focused on targeting the mass of homogenously mutated cells using chemo- and radiotherapy. However, multiple combinations of these therapies have yielded limited success; this has led to the development of the cancer stem cell theory, which outlines a hierarchy in tumor cells (6–9). Emerging evidence has indicated the presence of cancer stem cell niches within the tumor, which initiate tumor growth and metastasis, and survive chemo- or radiotherapy, leading to disease recurrence. The first evidence for glioblastoma stem cells (GSCs) was presented in 2003, when Singh et al (6,7) isolated cluster of differentiation (CD) 133(+) brain tumor-initiating cells from patients, and verified the self-renewal and differentiation potential of this subpopulation. Subsequently, further studies demonstrated that GSCs possess markedly greater levels of chemoresistant genes, including O-6-methylguanine-DNA methyltransferase (an enzyme responsible for DNA repair) (10), breakpoint cluster region pseudogene 1, and the anti-apoptosis genes B-cell lymphoma 2, B-cell lymphoma-extra large and the inhibitors of apoptosis proteins (3,11). Therefore, targeting these cell types may provide novel strategies for the treatment of glioblastoma, and the prevention of its recurrence.
Additionally, growing evidence has suggested that micro (mi)RNAs, a large group of non-coding RNAs, regulate numerous pathophysiological processes in humans (12–17). miRNA (miR)-141 has been demonstrated to exhibit antitumor effects in various types of tumors (12,14,16). Chen et al (16) reported that miR-141 may inhibit the proliferation and metastasis of kidney cancer cells via ephrin type-A receptor 2, and Burk et al (18) revealed that the miR-141/200c family may form a feedback loop with zinc finger E-box-binding homeobox 1, thereby regulating epithelial-mesenchymal transition (EMT) and metastasis in cancer. However, whether miR-141 possesses antitumor effects in glioblastoma remains unknown. The present study demonstrated that miR-141 is greatly suppressed in the CD133(+) glioblastoma cell subpopulation compared with CD133(−) cells. Thus, miR-141 may serve as an antioncomir in glioblastoma.
Materials and methods
Ethical approval
The present study was approved by the Ethics Committee of The First Hospital of Jilin University (Changchun, China) for both the animal experiments and human tissue sample processing. Written consent was obtained from patients donating glioblastoma tissue. This investigation complied with the principles that govern the use of human tissues outlined in the Declaration of Helsinki (19).
Isolation of CD133(+) GSCs and CD133(−) non-glioblastoma stem cells (NSCs). Glioblastoma tissue specimens were obtained from patients from the Department of Neurosurgery of The First Hospital of Jilin University.
Glioblastoma tissue was sectioned into pieces and digested with 0.2% collagenase II (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) in Dulbecco's modified Eagle's medium (DMEM)-high glucose (Corning Life Sciences, Tewksbury, MA, USA) for 2 h at 37°C. The cell suspension was subsequently rinsed with Dulbecco's PBS three times and incubated with Red Blood Cell Lysis buffer (Sigma-Aldrich; Merck Millipore) to eliminate erythrocytes. The remaining cells were incubated with an anti-human-CD133 conjugated with phycoerythrin antibody (cat. no. 130–098-826; dilution, 1:11; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) or a negative control antibody (cat. no. 130–098-845; dilution, 1:11; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) at 4°C for 30 min, rinsed with PBS three times and resuspended in buffer. Following this, CD133(+) and CD133(−) cells were separately isolated via Fluorescence Activated Cell Sorting (FACS). CD133(+) GSCs were maintained in minimal glucose lacking histidine medium, which was composed of: DMEM/nutrient mixture F12 (HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 1X N2 supplement (Thermo Fisher Scientific, Inc., Waltham, MA, USA), 50 µg/ml bovine serum albumin (cat. no. 9048-46-8; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), 25 µg/ml gentamycin (cat. no. G8170; Beijing Solarbio Science & Technology Co., Ltd.), 100 U/ml penicillin/streptomycin (Wuhan Boster Biological Technology, Wuhan, China), 20 ng/ml epidermal growth factor (EGF; PeproTech, Inc., Rocky Hill, NJ, USA) and 20 ng/ml basic fibroblast growth factor (bFGF; PeproTech, Inc.). DMEM/F-12 medium (HyClone; GE Healthcare Life Sciences) supplemented with 20 ng/ml EGF and 20 ng/ml bFGF was prepared for the sphere formation assay.
Sphere formation assay
To evaluate the capability of cancer stem cells to form spheres, cells were trypsinized, pipetted to a single cell suspension and adjusted to a concentration of 500 cells/ml. The single cell suspension (2 µl/well) was seeded into ultra-low attachment 96-well plates (Corning Incorporated, Corning, NY, USA) supplemented with 150 µl/well sphere formation medium, of which an extra 20 µl was added every three days. After two weeks, the total number of spheres formed by each cell type was counted by microscope (1X71 Research Inverted System microscope; Olympus Corporation, Tokyo, Japan) and evaluated as the number of spheres/96 cells.
Subcutaneous tumor xenograft assay
A total of 12 male BALB/c nude mice (5-weeks-old) were obtained from Beijing HFK Bioscience Co., Ltd. (Beijing, China) and kept in a specific pathogen-free vivarium with temperature between 26–34°C, humidity at 30–50%, 12/12 h light/dark cycle and free access to food and water (n=6/group). For the in vivo tumorigenicity assessment of the GSC-vector and GSC-miR-141 cell lines, ~1.0×106 GSCs were cultured, rinsed with precooled PBS three times, and resuspended with precooled Matrigel (BD Biosciences, San Jose, CA, USA) diluted with PBS at a ratio of 1:3. This was subsequently injected into either the right or left axillary fossa of each mouse. From day 7, mouse tumor volumes were measured with calipers by two experienced lab members at different time points throughout the day, every 3 days. The formula to calculate tumor volume was volume=0.5ab2, where ‘a’ is the long axis and ‘b’ is the short axis of the tumor. All animals were sacrificed using general anesthesia by 1.5% isoflurane (Sigma-Aldrich; Merck Millipore) inhalation and tumors (maximum size, 4.7 mm3) were weighed.
Antibodies and reagents
A Jagged1 rabbit anti-human polyclonal antibody was purchased from Abcam (cat. no. ab7771; Cambridge, MA, USA) and a GAPDH rabbit anti-human polyclonal antibody (cat. no. A00227) was purchased from Wuhan Boster Biological Technology. Horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (cat. no. G1210-2-A) and goat anti-rabbit secondary antibodies (cat. no. G1210-2-B) were obtained from Guge Biotechnology Co., Ltd. (Wuhan, China), and enhanced chemiluminescence (ECL) reagents were purchased from EMD Millipore (Billerica, MA, USA). A quantitative polymerase chain reaction (qPCR) kit was purchased from Takara Bio, Inc. (Otsu, Japan).
Western blot analysis
Whole cell proteins were extracted using radioimmunoprecipitation assay buffer supplemented with an inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA), and subsequently separated by 10% SDS-PAGE with 40 µg loading samples. Following this, proteins were transferred onto polyvinylidene difluoride membranes and blocked with 5% skimmed milk in TBS containing Tween 20 (TBST) for 2 h at room temperature. The membranes were incubated with anti-Jagged1 (1:1,000) at 4°C overnight, and rinsed with TBST three times. The membranes were subsequently incubated with HRP-conjugated secondary antibodies (1:10,000) for 2 h at 37°C. Following the addition of ECL reagents, the protein bands were observed and analyzed using the Bio-Rad imaging system (version, 5.2.1; Bio-Rad Laboratories, Inc., Hercules, CA, USA). GAPDH (1:5,000) served as an internal control.
Plasmids and primers
The hsa-miR-141-3p and Jagged1 overexpression plasmids were modified from a psicoR vector and a vector containing a puromycin resistance gene, respectively (Addgene, Inc., Cambridge, MA, USA). Dual luciferase plasmids of Jagged1 and its corresponding mutant were modified from a psiCHECK2 vector (Promega Corporation, Madison, WI, USA) and dual luciferase assay was undertaken as instructed by the protocol provided by Promega Corporation (20). All primers used are listed in Table I. The primers for qPCR analysis of miR-141 were purchased from GeneCopoeia, Inc. (Rockville, MD, USA). Reverse transcription reagents were purchased and undertaken as the protocol from GeneCopoeia, Inc. (cat. no. QP018), while PCR reagents were also purchased from GeneCopoeia Inc. (cat. no. QP015).
Table I.
Quantitative polymerase chain reaction primers.
| Gene | Sequence (5′-3′) |
|---|---|
| CD133 | F: GGCCCAGTACAACACTACCAA |
| R: ATTCCGCCTCCTAGCACTGAA | |
| Oct-4 | F: CTGGGTTGATCCTCGGACCT |
| R: CCATCGGAGTTGCTCTCCA | |
| Nestin | F: CAACAGCGACGGAGGTCTC |
| R: GCCTCTACGCTCTCTTCTTTGA | |
| C-Myc | F: TCCCTCCACTCGGAAGGAC |
| R: CTGGTGCATTTTCGGTTGTTG | |
| Nanog | F: AAGGTCCCGGTCAAGAAACAG |
| R: CTTCTGCGTCACACCATTGC | |
| Jagged1 | F: GCCGAGGTCCTATACGTTGC |
| R: CCGAGTGAGAAGCCTTTTCAA | |
| TWIST-1 | F: GTCCGCAGTCTTACGAGGAG |
| R: GCTTGAGGGTCTGAATCTTGCT | |
| Vimentin | F: AGTCCACTGAGTACCGGAGAC |
| R: CATTTCACGCATCTGGCGTTC | |
| ZEB-1 | F: TTACACCTTTGCATACAGAACCC |
| R: TTTACGATTACACCCAGACTGC | |
| ZEB-2 | F: GGAGACGAGTCCAGCTAGTGT |
| R: CCACTCCACCCTCCCTTATTTC | |
| N-cadherin | F: AGCCAACCTTAACTGAGGAGT |
| R: GGCAAGTTGATTGGAGGGATG | |
| E-cadherin | F: ATTTTTCCCTCGACACCCGAT |
| R: TCCCAGGCGTAGACCAAGA |
F, forward; R, reverse; Oct-4, octamer-binding transcription factor 4; Nanog, homeobox protein Nanog; TWIST-1, twist family BHLH transcription factor 1; ZEB, zinc finger E-box binding homeobox; N, neural; E, epithelial; CD, cluster of differentiation.
qPCR
A total of 106 cells of each group were harvested by TRIzol reagent (cat. no. 15596026, Thermo Fisher Scientific, Inc.) and total RNA was extracted by traditional phenol-chloroform method. The reverse transcription was undertaken according to the protocol form GeneCopoeia, Inc. (cat. no. QP018). A total of 1 µg RNA for each group was mixed with the following 25 µl reaction mixture: 1 µg total RNA, 1 µl 2.5 U/µl Poly A Polymerase, 1 µl RTase Mix, 5 µl 5X PAP/RT buffer and RNase/DNase-free water to make the final 25 µl in total; cat. no. QP018, GeneCopoeia, Inc.) and was incubated as 37°C of 60 min followed with enzyme inactivation by inbucation at 85°C for 5 min. Then, the reverse transcribed cDNA was mixed with other qPCR reagents in a kit (cat. no. QP015; GeneCopoeia, Inc.) and was analyzed using the Mx3000P qPCR system (cat. no. 401512, Agilent Technologies, Santa Clara, USA) with the following thermocycling conditions: 95°C for 10 min (1 cycle), 95°C for 10 sec followed with 60°C for 20 sec and 72°C for 10 sec (40 cycles). Data processing and normalization was undertaken as reported by Livak and Schmittgen (21).
Transfection
The lentivirus of has-miR-141-3p and Jagged1 from respective plasmids or empty plasmids were packaged by Hanbio Co., Ltd. (Shanghai, China). GSCs were transfected with the virus and selected with puromycin at 1 µg/ml for 1 week for subsequent in vitro and in vivo studies.
Statistical analysis
Statistical analyses were performed using SPSS software version 20.0 (IBM SPSS, Armonk, NY, USA). A one-way analysis of variance and Fisher's exact test were used to determine significant differences between groups. Data are presented as the mean ± standard error unless otherwise indicated. P<0.05 was considered to indicate a statistically significant difference.
Results
CD133(+) GSCs are self-renewing
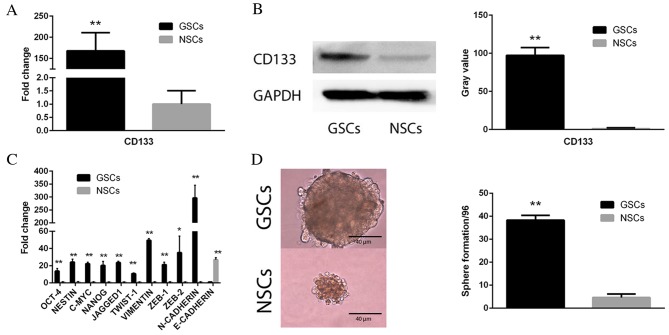
In order to verify the FACS sorting result of CD133(+) GSCs and CD133(−) NSCs, qPCR (Fig. 1A; P<0.01) and western blot (Fig. 1B; P<0.01) analyses were performed to analyze the comparative expression levels of CD133 in these two cell subpopulations. The sorted CD133(+) cells demonstrated markedly increased expression levels of CD133, compared with CD133(−) cells. In addition, qPCR was performed on five stemness-associated [octamer-binding transcription factor 4 (Oct-4), nestin, c-Myc, homeobox protein Nanog (Nanog) and Jagged1] and six EMT-associated [twist family BHLH transcription factor 1 (TWIST-1), vimentin, zinc finger E-box binding homeobox (ZEB)-1, ZEB-2, neural (N)-cadherin and epithelial (E)-cadherin] genes to determine differences in mRNA expression levels between the two cell subopulations. As presented in Fig. 1C, the CD133(+) GSC subpopulation exhibited upregulated expression levels of all five stemness genes (Oct-4, nestin, c-Myc, Nanog and Jagged; all P<0.01) and five mesenchymal-associated genes (TWIST-1, P<0.01; vimentin, P<0.01; ZEB-1, P<0.01; ZEB-2, P<0.05; and N-cadherin, P<0.01) compared with NSCs, which had upregulated E-cadherin expression levels compared with GSCs. A sphere formation assay was performed to assess the self-renewal of cells. It revealed that CD133(+) GSCs exhibited an increased potential to duplicate themselves compared with CD133(−) NSCs (Fig. 1D; P<0.01).
Figure 1.
CD133 expression and self-renewal following Fluorescence Activated Cell Sorting of GSCs. (A) qPCR and (B) western blot analysis demonstrating markedly increased expression levels of CD133 in CD133(+) GSCs. Gray value indicates protein optical density as analyzed by ImageJ software. GAPDH served as an internal control. (C) qPCR analysis revealed an array of stemness- and epithelial-mesenchymal transition-associated genes were upregulated in CD133(+) GSCs compared with CD133(−) NSCs. (D) A sphere formation assay demonstrated increased sphere formation in CD133(+) GSCs compared with CD133(−) NSCs. Data are presented as the mean ± standard error. *P<0.05 and **P<0.01 vs. NSCs. GSCs, glioblastoma stem cells; NSCs, non-glioblastoma stem cells; Oct-4, octamer-binding transcription factor 4; Nanog, homeobox protein Nanog; TWIST-1, twist family BHLH transcription factor 1; ZEB, zinc finger E-box binding homeobox; N, neural; E, epithelial; qPCR, quantitative polymerase chain reaction; CD133, cluster of differentiation 133.
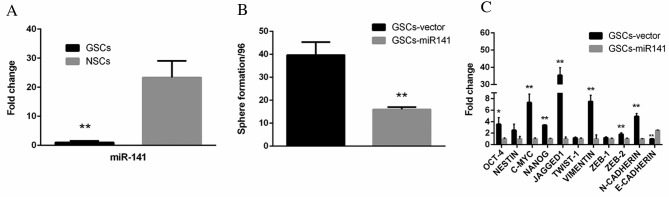
miR-141 expression levels are suppressed in GSCs and inhibit the self-renewal of GSCs in vitro
To investigate the role of miR-141 in GSCs, qPCR analysis was performed. Compared with NSCs, GSCs exhibited markedly reduced expression levels of miR-141 (Fig. 2A; P<0.01). GSCs transfected with miR-141 exhibited significantly reduced sphere formation ability, compared with vector-transfected cells (Fig. 2B; P<0.01). In addition, a set of stemness (Oct-4, P<0.05; nestin, P>0.05; c-Myc, P<0.01; Nanog, P<0.01; Jagged, P<0.01; Twist-1, P>0.05) and EMT-associated genes (vimentin, P<0.01; Zeb-1, P>0.05; Zeb-2, P<0.01; N-cadherin, P<0.01; E-cadherin, P<0.01) were downregulated in GSCs-miR-141 compared with GSCs-vector, whereas E-cadherin was upregulated (Fig. 2C).
Figure 2.
miR-141 expression pattern and function in GSCs. (A) Quantitative polymerase chain reaction analysis of miR-141 expression levels demonstrated that GSCs exhibited reduced expression levels of miR-141. (B) GSCs transfected with miR-141 downregulated of an array of stemness- and epithelial-mesenchymal transition-associated genes, and (C) had inhibited sphere formation ability. Data are presented as the mean ± standard error. *P<0.05; **P<0.01. miR-141, microRNA-141; GSCs, glioblastoma stem cells; Oct-4, octamer-binding transcription factor 4; Nanog, homeobox protein Nanog; TWIST-1, twist family BHLH transcription factor 1; ZEB, zinc finger E-box binding homeobox; N, neural; E, epithelial.
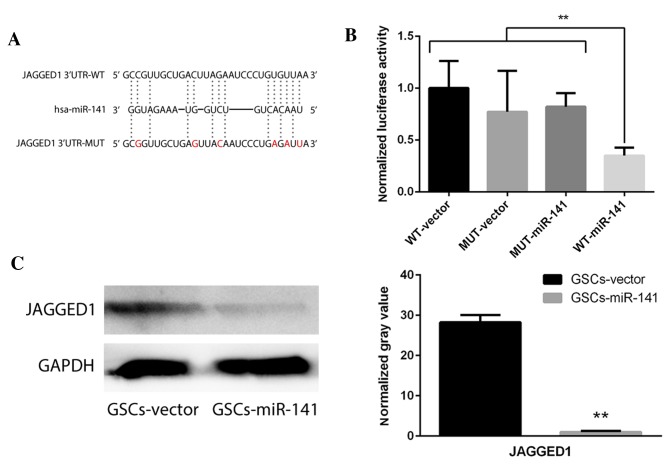
Jagged1 may be inhibited via direct binding with miR-141
To investigate the potential targets of miR-141, two dual luciferase constructs were made containing a Jagged1 3′-untranslated region and a mutated version of this region (Fig. 3A). Following transfection with miR-141, luminescence significantly decreased in the wild-type compared with the mutant group (Fig. 3B; P<0.01). Western blot analysis revealed that Jagged1 expression levels were significantly decreased following overexpression of miR-141 in GSCs (Fig. 3C; P<0.01). To verify the effect of Jagged1 on self-renewal, Jagged1 overexpression constructs were cloned to upregulate its expression levels in GSCs-miR-141. The results demonstrated that the reduced sphere formation ability in GSCs-miR-141 was recovered (Fig. 4; P<0.01). These results were compared to a GSC-vector-vector group, and this demonstrated a similar sphere formation ability, whereby it was only partially recovered following Jagged1 overexpression (data not shown).
Figure 3.
miR-141 target gene investigation via dual-luciferase assay. (A) Dual luciferase constructs for WT Jagged1 3′-UTR and a mutated version of this sequence. (B) Quantification of the dual luciferase assay demonstrated a statistically significant decrease of luminescence in the WT-miR-141 group. **P<0.01. (C) Western blot analysis demonstrating downregulated Jagged1 protein expression levels following miR-141 overexpression, compared with vector-transfected GSCs. Normalized gray value indicates protein optical density as analyzed by ImageJ software. GAPDH served as an internal control. Data are presented as the mean ± standard error. **P<0.01 vs. GSCs-vector. WT, wild-type; MUT, mutant; miR-141, microRNA-141; GSCs, glioblastoma stem cells; UTR, untranslated region.
Figure 4.
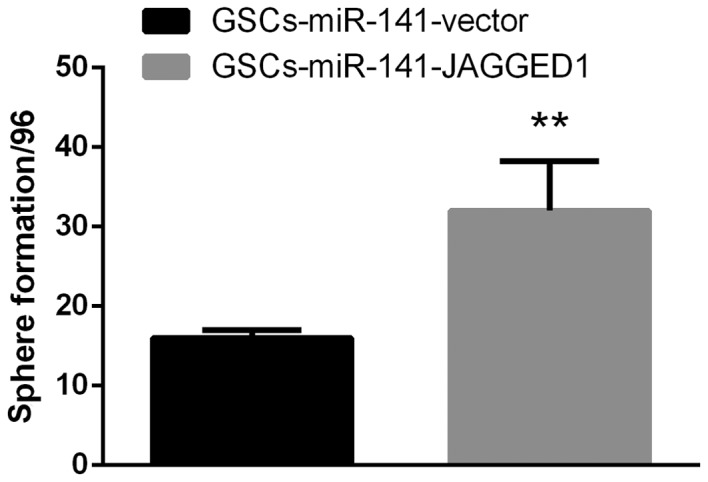
Jagged1 overexpression may restore sphere formation ability in GSCs. Artificial overexpression of Jagged1 in GSCs overexpressing miR-141 may abrogate the miR-141-mediated inhibitory effect on sphere formation ability. Data are presented as the mean ± standard error. **P<0.01 vs. GSCs-miR-141-vector. miR-141, microRNA-141; GSCs, glioblastoma stem cells.
miR-141 may inhibit the tumorigenecity of GSCs in vivo
To investigate the role of miR-141 in GSCs in vivo, two groups of mice were transfected with GSCs-vector or GSCs-miR-141. As presented in Fig. 5A, the tumors produced by GSCs-miR-141 were markedly reduced in size compared with GSCs-vector. Furthermore, significantly reduced tumor volumes (Fig. 5B; P<0.01) and weight (Fig. 5C) were observed in mice implanted with miR-141 overexpressing GSCs, compared with those implanted with vector-transfected cells, suggesting that miR-141 expression may inhibit tumor growth in vivo (Fig. 5B; P<0.01).
Figure 5.
miR-141 inhibits glioblastoma growth in vivo. (A) Tumor size in mice implanted with GSCs-vector was markedly increased compared with those implanted with GSCs-miR-141. Tumor (B) volume and (C) weight; miR-141 overexpression may inhibit tumor cell growth. Data are presented as the mean ± standard error. **P<0.01 vs. GSCs-vector. miR-141, microRNA-141; GSCs, glioblastoma stem cells.
Discussion
The present study demonstrated a direct interaction between Jagged1 and miR-141 in GSCs. Clement et al (20) reported in 2011 that miR-141 may inhibit Jagged1, thereby inhibiting the metastasis of prostate cancer cells; however, the effect of miR-141 on the self-renewal of cancer stem cells, particularly in glioblastoma, remains unclear.
Jagged1 is a protein mutated in 60–75% of patients with Alagille Syndrome (22). It is encoded by the JAG1 gene, and the translation product is the ligand for the receptor Notch-1/3 (22). A previous study by Chen et al (23) revealed that Jagged1 promoted the pathogenesis of ovarian cancer by activating Notch-3. Furthermore, the canonical stemness-associated Wnt/β-catenin signaling pathway may upregulate the expression levels of Jagged1, thus facilitating the development of ovarian cancer. Notably, Jagged1 expression has been identified to be negatively correlated with outcome in glioblastoma patients (24). Qiu et al (24) reported that in 82 glioblastoma patients, reduced expression levels of Jagged1 were consistent with slower tumor progression and increased overall survival. Multivariate survival analysis additionally revealed that increased Jagged1 expression levels in tumor or endothelial cells are an independent prediction factor for poor prognosis. Consistent with these findings, the present study demonstrated that miR-141 overexpression results in decreased Jagged1 expression levels and reduced self-renewal in GSCs. These results may facilitate the development of therapeutic targets to prolong the overall survival rate of patients with glioblastoma.
Furthermore, Jagged1 is closely associated with the Notch signaling pathway. The Notch signaling pathway is involved in numerous physiological and pathophysiological processes, including differentiation and stemness maintenance. There are primarily four types of Notch family receptors: Notch-1, -2, -3 and -4 (12,23,25–27), which may promote proliferative signaling during neuron development. The Notch receptor ligands in mammals primarily include delta-like proteins and jagged proteins (23). There has been increasing evidence to suggest Notch's involvement in cancer cell growth; Farnie et al (26) demonstrated that a Notch-4-neutralizing antibody may markedly reduce the self-renewal of breast cancer stem cells. Furthermore, Fan et al (27) identified a feedback loop between human epidermal growth factor receptor 2/proto-oncogene Neu and Notch in breast cancer stem cells. In glioblastoma, Fan et al (27) demonstrated that the Notch signaling pathway blockade by gamma-secretase inhibitors may deplete CD133(+) glioblastoma cells. The results of the present study suggested that the signal attenuation of the Notch signaling pathway via inhibition of Jagged1 by miR-141 may serve as a potential strategy to eradicate glioblastoma cells within tumors.
EMT has been associated with cancer stem cell maintenance and development (28). Previous studies have demonstrated that the activation of EMT confers stemness traits to healthy and neoplastic cells, thus EMT- and stemness-associated genes may be upregulated in parallel or sequentially (12,26,28). The present study revealed that the EMT-associated genes TWIST-1, vimentin, ZEB-1, ZEB-2 and N-cadherin, were co-upregulated with the stemness genes Oct-4, nestin, C-Myc, Nanog and Jagged1, in GSCs compared with NSCs. miR-141-overexpressing GSCs exhibited downregulated EMT- and stemness-associated gene expression, and upregulation of E-cadherin. Therefore, miR-141 may additionally serve a role in EMT in glioblastoma and reduce tumor aggression, leading to tumor demarcation that may facilitate surgical resection.
In conclusion, the present study demonstrated that miR-141 may serve as an antioncomir in GSCs and markedly inhibit their self-renewal via downregulating Jagged1 expression levels in vitro and in vivo. Similar to commercially available small interfering RNA treatment agents, it is hypothesized that miR-141 may be utilized for glioblastoma treatment. Targeting GSCs or the source of glioblastoma remains a promising strategy to impede metastasis and resistance to chemotherapy or radiotherapy. The potential of miR-141 to inhibit cell metastasis requires further investigation, to facilitate the development of novel treatment strategies for glioblastoma.
Acknowledgements
The authors thank all the patients who participated in this program and all coworkers who contributed to this study.
References
- 1.Luo JW, Wang X, Yang Y, Mao Q. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med Pharmacol Sci. 2015;19:1630–1639. [PubMed] [Google Scholar]
- 2.Rachet B, Mitry E, Quinn MJ, Cooper N, Coleman MP. Survival from brain tumours in England and Wales up to 2001. Br J Cancer. 2008;99:S98–S101. doi: 10.1038/sj.bjc.6604603. (Suppl 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho DY, Lin SZ, Yang WK, Lee HC, Hsu DM, Lin HL, Chen CC, Liu CL, Lee WY, Ho LH. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transplant. 2013;22:731–739. doi: 10.3727/096368912X655136. [DOI] [PubMed] [Google Scholar]
- 4.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji J, Black KL, Yu JS. Glioma stem cell research for the development of immunotherapy. Neurosurg Clin N Am. 2010;21:159–166. doi: 10.1016/j.nec.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Li Y, Kong D, Ahmad A, Banerjee S, Sarkar FH. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292:141–148. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, deCarvalho AC, Slavin S, et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4:665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong JW. MicroRNA-induced silencing of glioma progression. J Neurosci. 2010;30:3868–3869. doi: 10.1523/JNEUROSCI.0442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu N, Xiao L, Zhao X, Zhao J, Wang J, Wang F, Cao S, Lin X. miR-125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS Lett. 2012;586:3831–3839. doi: 10.1016/j.febslet.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu X, Xiao P, Shi H, Wang R, Chen L, et al. miR-141 is A key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res. 2014;20:2617–2630. doi: 10.1158/1078-0432.CCR-13-3224. [DOI] [PubMed] [Google Scholar]
- 17.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B, Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 20.Clement T, Salone V, Rederstorff M. Dual luciferase gene reporter assays to study miRNA function. Methods Mol Biol. 2015;1296:187–198. doi: 10.1007/978-1-4939-2547-6_17. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Spinner NB, Colliton RP, Crosnier C, Krantz ID, Hadchouel M, Meunier-Rotival M. Jagged1 mutations in alagille syndrome. Hum Mut. 2001;17:18–33. doi: 10.1002/1098-1004(2001)17:1<18::AID-HUMU3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Stoeck A, Lee SJ, Shih Ie M, Wang MM, Wang TL. Jagged1 expression regulated by Notch3 and Wnt/b-catenin signaling pathways in ovarian cancer. Oncotarget. 2010;1:210–218. doi: 10.18632/oncotarget.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu XX, Wang CH, You N, Chen BJ, Wang XF, Chen YP, Lin ZX. High Jagged1 expression is associated with poor outcome in primary glioblastoma. Med Oncol. 2015;32:341. doi: 10.1007/s12032-014-0341-9. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ. Novel cell culture technique for primary ductal carcinoma in situ: Role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 27.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]