Abstract
The present study aimed to explore gene and microRNA (miRNA) expression differences between lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) were identified by analyzing mRNA and miRNA expression data in normal and cancerous lung tissues that were obtained from The Cancer Genome Atlas database. A total of 778 DEGs and 7 DEMs were identified. Altered gene functions and signaling pathways were investigated using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses, which revealed that DEGs were significantly enriched in extracellular matrix organization, cell differentiation, negative regulation of toll signaling pathway, and several other terms and pathways. Transcription factor (TF)-miRNA-gene networks in LUAD and LUSC were predicted using the TargetScan, Miranda, and TRANSFAC databases, which revealed the regulatory links among the TFs, DEMs, and DEGs. The central TFs, i.e., the TFs in the middle of the TF-miRNA-gene network, of LUAD and LUSC were similar. Although LUAD and LUSC shared similar miRNAs in the predicted networks, miR-29b-3p was demonstrated to be upregulated only in LUAD, whereas miR-1, miR-105-5p, and miR-193b-5p were altered in LUSC. These findings may improve our understanding of the different molecular mechanisms in non-small cell lung cancers and may promote new and accurate strategies for prevention, diagnosis, and treatment.
Keywords: lung cancer, differentially expressed genes, differentially expressed microRNAs, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathway analysis, transcription factor-miRNA-gene network
Introduction
Lung cancer is one of the most common malignant tumors and in 2012 accounted for ~1.82 million new cases and ~1.59 million mortalities worldwide (1,2). Despite the advances in treatment methods that have been made available in recent years, including minimally invasive surgical approaches, chemotherapies, and targeted therapies, the 5-year survival of patients with lung cancer is far from satisfying, ranging between 10 and 20% for most geographic areas (3). There are two major pathological subtypes that constitute the majority of lung cancers: lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), which differ in a number of ways (4–7).
LUAD and LUSC originate from different cells and have several major differences not only in biological patterns, but also molecular characteristics and, most importantly, therapeutic strategies (5,8). For example, activating mutations in epidermal growth factor receptor and mutations in ALK fusion proteins usually occur in LUAD, but not LUSC, rendering medications targeted at these genes ineffective for LUSC (9). Therefore, comprehensive investigations into the differences of molecular characteristics and mechanisms of these two major subtypes of lung cancer are required, which will lead to deeper understanding and identification of novel molecular-targeted strategies for lung cancer therapy.
A large amount of high-throughput data on multiple types of cancer have recently been released by The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) database, including mRNA and microRNA (miRNA) sequencing data from hundreds of LUAD and LUSC samples. These data enabled the molecular differences between LUAD and LUSC to be fully investigated. The present study explored differences in gene expression, miRNA expression, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and molecular regulatory networks by bioinformatics analyses, and the results may facilitate a better understanding of the different molecular mechanisms of non-small cell lung cancer (NSCLC) and may promote the discovery and development of new, accurate strategies for lung cancer prevention, diagnosis, and treatment.
Materials and methods
mRNA and miRNA expression data resources and preprocessing
Level 3 RNA sequencing data from 108 normal pulmonary samples and 980 pulmonary carcinoma samples (that is, 490 LUAD with 58 normal control samples, and 490 LUSC with 50 normal control samples), and level 3 miRNA sequencing data from 91 normal pulmonary samples and 966 pulmonary carcinoma samples (that is, 499 LUAD with 46 normal control samples, and 467 LUSC with 45 normal control samples) were released by TCGA prior to April 15, 2015, and were obtained by the present study from the TCGA data portal (https://portal.gdc.cancer.gov). Data preprocessing was carried out as described in previous studies (10).
Identification of differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs)
Genes and miRNAs that are differentially expressed among normal, LUAD, and LUSC sample groups were identified as previously reported (11). DEGs with a fold change (tumor/normal) >2 or <0.5 and DEMs with a fold change >2.5 or <0.4 were qualified for subsequent analyses. A random variance model t-test was used to confirm the DEGs and DEMs, as previously described (10). Following analysis of the significance, fold change, and false discovery rate (FDR), mRNAs and miRNAs that had both P<0.05 and FDR<0.05 were considered to be significantly differentially expressed (10). Only the DEGs and DEMs that were identified in both the LUAD and LUSC groups were included when comparing the differences between gene and miRNA expressions.
GO and KEGG pathway analyses
To investigate the significantly enriched functions and the significant pathways for these DEGs, GO term (http://geneontology.org) and KEGG pathway (http://www.genome.jp/kegg) analyses were conducted as previously reported (12,13). Briefly, the two-tailed Fisher's exact test and the χ2 test were used to classify the GO categories or KEGG pathways, and the FDR was calculated for multiple testing corrections. GO terms or KEGG pathways having both P<0.05 and FDR<0.05 were considered to be significantly different. Enrichment values were calculated to identify those significant terms or pathways that provided the most concrete functional descriptions in this analysis. GO-map and PathNet analyses were conducted to further outline the functional links among the related GO terms and the significant KEGG pathways.
TF-miRNA-gene network
The regulation networks among transcription factors (TFs), DEMs and DEGs were established as previously described (10,14). Briefly, the target DEGs of DEMs were predicted using TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/home.do) (15,16). Subsequently, the TFs that may regulate the expression of DEMs and DEGs were identified using the TRANSFAC database (http://gene-regulation.com/pub/databases.html) (17). Finally, TF-miRNA-gene networks were created in LUAD and LUSC, as previously described (10).
Statistical analysis
Statistical analysis was performed using the software IBM SPSS version 20 (IBM SPSS, Armonk, NY, USA).
Results
Identification of DEGs and DEMs
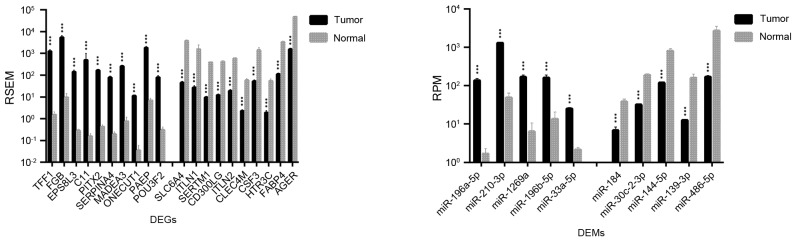
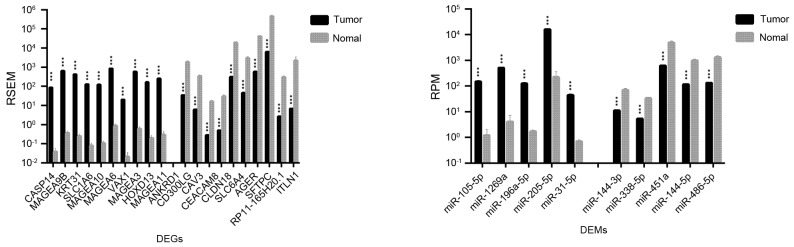
Significant differences in expression levels were detected for certain genes and miRNAs that may be used as biomarkers for the early diagnosis, assessment, and monitoring of lung cancer. As shown in Table I, 1,492 DEGs and 36 DEMs were identified as being significantly different between LUAD and normal lung tissues; for LUSC vs. normal lung tissue, 2,726 DEGs and 45 DEMs were identified. The top 20 DEGs and the top 10 DEMs exhibiting the most significant differential expressions in LUAD and LUSC are shown in Figs. 1 and 2, respectively.
Table I.
DEGs and DEMs identified between LUAD, LUSC and normal samples.
| Comparison | DEGs | DEMs |
|---|---|---|
| LUAD vs. Normal | 1,492 | 36 |
| LUAD>Normal | 611 | 27 |
| LUAD<Normal | 881 | 9 |
| LUSC vs. Normal | 2,726 | 45 |
| LUSC>Normal | 1,052 | 28 |
| LUSC<Normal | 1,674 | 17 |
| LUAD vs. LUSC | 778 | 7 |
| LUAD>LUSC | 340 | 1 |
| LUAD<LUSC | 438 | 6 |
DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs; LUAD, lung adenocarcinoma cells; LUSC, lung squamous cell carcinoma.
Figure 1.
Top 20 genes and top 10 microRNAs identified as the most differentially expressed in LUAD compared with normal lung tissue samples. Data are expressed as the mean ± standard deviation. ***P<0.001 vs. normal. DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs; miR, microRNA; FDR, false discovery rate; LUAD, lung adenocarcinoma; RSEM, RNA-Seq by expectation maximization; RPM, reads per million miRNA mapped.
Figure 2.
Top 20 genes and top 10 microRNAs identified as the most differentially expressed in LUSC compared with normal lung tissue samples. Data are expressed as the mean ± standard deviation. ***P<0.001 vs. normal. DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs; miR, microRNA; FDR, false discovery rate; LUSC, lung squamous cell carcinoma; RSEM, RNA-Seq by expectation maximization; RPM, reads per million miRNA mapped.
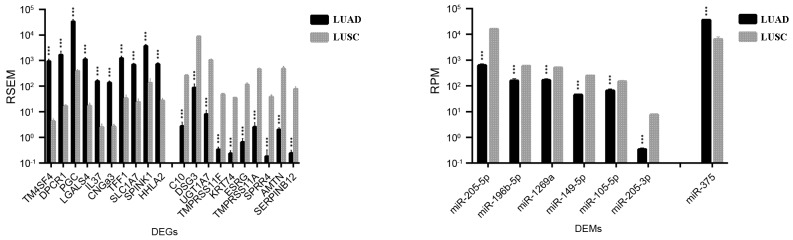
A total of 778 DEGs and 7 DEMs were identified in both LUAD and LUSC (Table I). As demonstrated in Fig. 3, transmembrane 4 L six family member 4 (TM4SF4), diffuse panbronchiolitis critical region 1 (DPCR1), prograstricsin (PGC), galectin 4 (LGALS4), and interleukin 37 (IL37) were the top five DEGs that were more upregulated in LUAD than in LUSC (TM4SF4, 214.5 fold change; IL37, 63.2 fold change, DPCR1, PGC and LGALS4, fold change 63.2–214.5); serpin family B member 12 (SERPINB12), amelotin (AMTN), small proline-rich protein 4 (SPRR4), transmembrane protease, serine 11A (TMPRSS11A), and embryonic stem cell related (ESRG) were the top five DEGs identified as being more upregulated in LUSC compared with LUAD (fold change, 172.0–322.9). As expected, a number of well-established biomarkers for LUAD and LUSC were also identified, including transcription termination factor 1 (TTF1; fold change LUAD/LUSC, 10.5), keratin 7 (KRT7; fold change, 3.63), SRY-box 2 (SOX2; fold change, 0.11), p63 (fold change, 0.03), and KRT5 (fold change, 0.01).
Figure 3.
Top 10 genes and top 5 microRNAs identified as the most differentially expressed between LUAD and LUSC. Data are expressed as the mean ± standard deviation. ***P<0.001 vs. LUSC. DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs; miR, microRNA; FDR, false discovery rate; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; RSEM, RNA-Seq by expectation maximization; RPM, reads per million miRNA mapped.
miRNA miR-375 was the only DEM that was demonstrated to be more upregulated in LUAD compared with LUSC (fold change, 5.62); six other DEMs were revealed to be upregulated in LUSC vs. LUAD, including miR-205-5p, miR-205-3p, miR-149-5p, miR-196b-5p, miR-1269a, and miR-105-5p (fold change, 2.3–25.2; Fig. 3).
Enriched GOs and pathways
GO and KEGG analyses were used in the present study to provide a preliminarily perspective on the altered biological functions and pathways in which the DEGs are enriched. In LUAD vs. Normal lung tissue, DEGS were enriched in 806 GO terms and 84 pathways, whereas in LUSC vs. Normal, DEGs were enriched in 1266 GOs and 146 pathways (Table II).
Table II.
Basic information of the GOs and KEGG pathways in which the DEGs and DEMs were enriched.
| Comparison | GOs | Pathways |
|---|---|---|
| LUAD vs. Normal | 806 | 84 |
| Upregulated | 237 | 24 |
| Downregulated | 569 | 60 |
| LUSC vs. Normal | 1266 | 146 |
| Upregulated | 468 | 40 |
| Downregulated | 798 | 106 |
| LUAD vs. LUSC | 409 | 47 |
| LUAD>LUSC | 124 | 22 |
| LUAD<LUSC | 285 | 25 |
DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs. GOs, gene oncology terms; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUAD, lung adenocarcinoma cells; LUSC, lung squamous cell carcinoma.
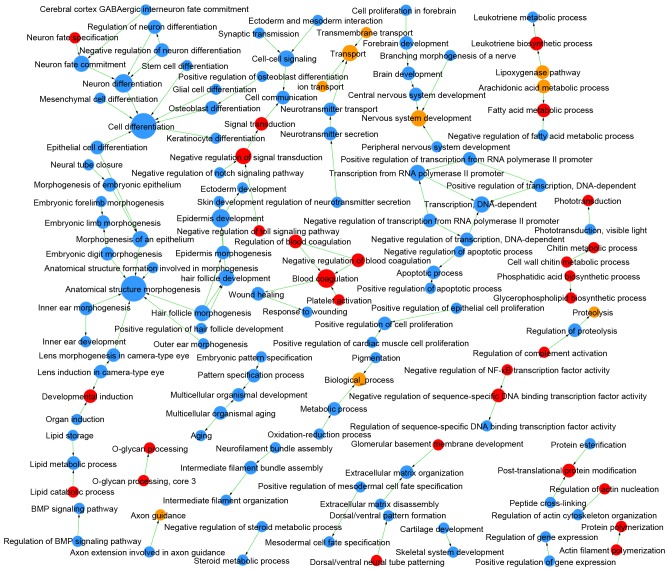
Comparing LUAD and LUSC, DEMs were enriched in 409 GOs and 47 pathways (Table I), and the links among these GOs and pathways are integrated in Figs. 4 and 5, respectively. The DEGs identified as upregulated in LUAD vs. LUSC were enriched in 124 GOs, such as negative regulation of the toll signaling pathway (GO:0045751) and negative regulation of nuclear factor (NF)-κB activity (GO:0032088), and in 22 pathways, such as peroxisome proliferator-activated receptor (PPAR) signaling pathway (id:03320) and glycolysis/gluconeogenesis (id:00010). The upregulated DEGs in LUSC vs. LUAD were enriched in 285 GOs, such as extracellular matrix organization (GO:0030198) and cell differentiation (GO:0030154), and in 25 pathways, such as cell adhesion molecules (id:04514) and p53 signaling pathway (id:04115).
Figure 4.
GO map of LUAD vs. LUSC. Red circles denote GOs in which the upregulated genes in LUAD are significantly enriched; blue circles indicate GOs in which the upregulated genes in LUSC are significantly enriched; yellow circles indicate genes with upregulated expression in both LUAD and LUSC. Arrows denote the relationships between GOs, where the tail end is the source GO and the arrow end is the target GO. The diameter of each circle represents the number of GOs that interact closely with a GO; larger circles indicate more interactions. P<0.05; FDR<0.05. FDR, false discovery rate; GO, gene ontology; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Figure 5.
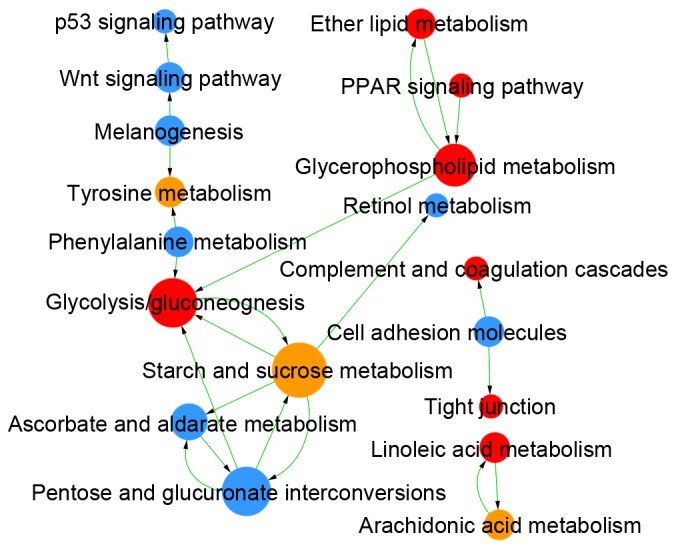
KEGG analysis in LUAD vs. LUSC. Red circles denote GOs in which the upregulated genes in LUAD are significantly enriched; blue circles indicate GOs in which the upregulated genes in LUSC are significantly enriched; yellow circles indicate genes with upregulated expression in both LUAD and LUSC. Arrows denote the relationships between GOs, where the tail end is the source GO and the arrow end is the target GO. The diameter of each circle represents the number of GOs that interact closely with a GO; larger circles indicate more interactions. KEGG analysis results: P<0.05; FDR<0.05. FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
TF-miRNA-gene network
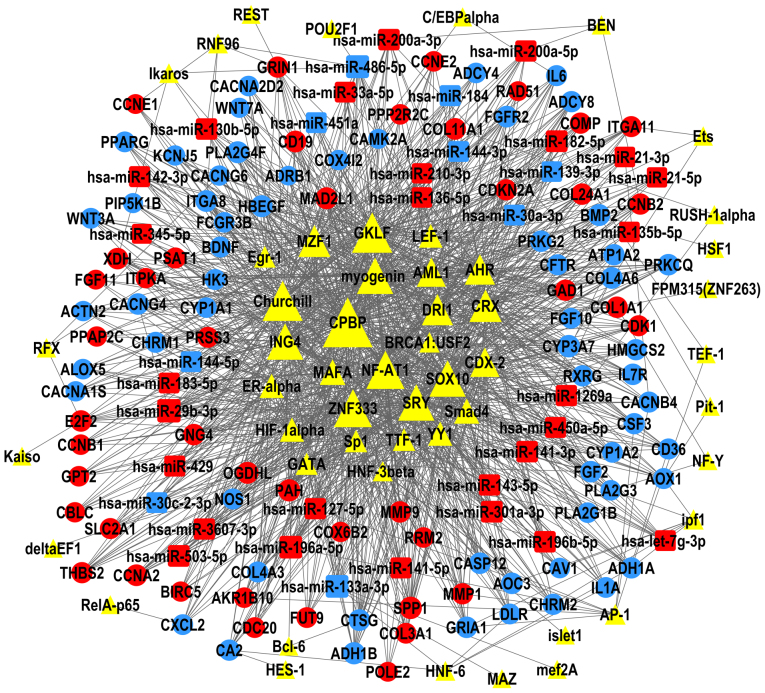
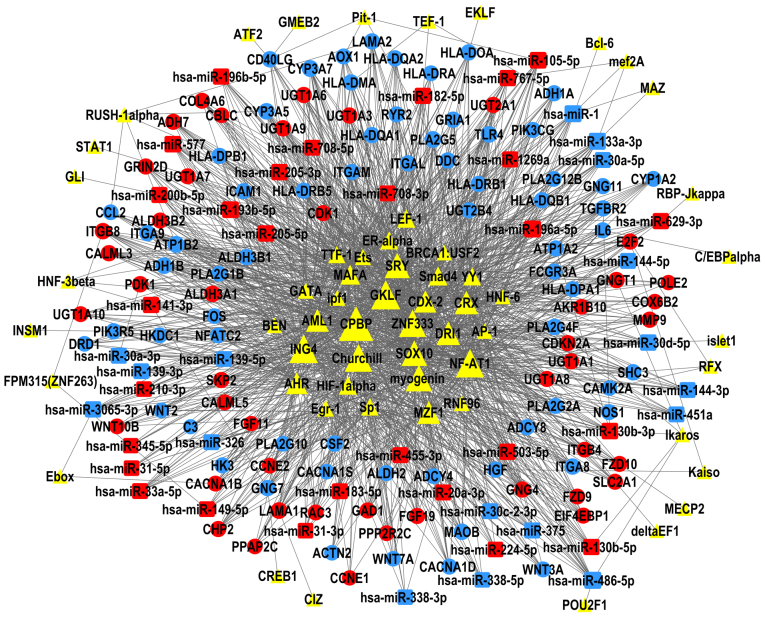
The present study constructed TF-miRNA-gene networks of LUAD and LUSC (Figs. 6 and 7), using the large amount of interrelated expression data of miRNAs and genes in the TCGA database, to predict regulatory networks among the TFs, DEMs, and DEGs. As the proposed networks demonstrated, the central TFs and DEMs, i.e. the TFs and DEMs in the middle of the TF/DEM-miRNA-gene network, in LUAD and LUSC were quite similar. The top six central TFs were core promoter element-binding protein (CPBP), gut-enriched Krüppel-like factor (GKLF), Churchill, nuclear factor of activated T-cells 1 (NF-AT1), zinc finger protein 333 (ZNF333), and inhibitor of growth protein 4 (ING4); these TFs were the same in LUAD and in LUSC, indicating that there are still common regulatory mechanisms shared between these two subtypes of lung cancer. LUAD and LUSC shared 19 DEMs in common (data not shown), of which miR-486-5p, miR-133a-3p, and miR-196a-5p were centrally positioned in the predicted TF-miRNA-gene regulatory networks of both LUAD and LUSC. LUAD and LUSC had different patterns of DEMs. miR-29b-3p was upregulated and was predicted to regulate the most DEGs in the LUAD network, but not in LUSC. miR-1, miR-105-5p, and miR-193b-5p were only in the center of the LUSC network.
Figure 6.
TF-microRNA-gene network in LUAD. The regulatory network of TFs (triangles), DEMs (squares) and DEGs (circles); the size of each shape represents the number of closely interacting factors, with the large shapes indicating a higher number of interactions. Red represents upregulated expression in LUAD; blue represents downregulated expression; and yellow is uncertain. Lines denote the regulatory links among these factors. DEG, differential expressed genes; DEM, differential expressed miRNAs; LUAD, lung adenocarcinoma; TF, transcription factor.
Figure 7.
TF-microRNA-gene network in LUSC. The regulatory network of TFs (triangles), DEMs (squares) and DEGs (circles); the size of each shape represents the number of closely interacting factors, with the large shapes indicating a higher number interactions. Red represents upregulated expression in LUAD; blue represents downregulated expression; and yellow are uncertain. Lines denote the regulatory links among these factors. DEGs, differentially expressed genes; DEMs, differentially expressed microRNAs; LUSC, lung squamous cell carcinoma; TF, transcription factor.
Discussion
The present study analyzed the differences in DEG and DEM expressions in LUAD and LUSC compared with normal lung tissue and, most importantly, the differences between LUAD and LUSC. To further elucidate the functions of these DEGs and DEMs, GO and KEGG pathway analyses were performed. In addition, TF-miRNA-gene networks were constructed for LUAD and LUSC; however, further independent validation with experimental data is still required.
Cytology and pathology have traditionally been used in the differential diagnosis of LUAD and LUSC; however, in some cases, such as small biopsy samples or aspiration cytology samples, additional tests of molecular characteristics were required (18). Although several genes have been used as biomarkers in lung cancer diagnosis and differential diagnosis, more accurate and convenient biomarkers are still needed. The present study identified 778 DEGs and 7 DEMs that were differentially expressed between LUAD and LUSC. These DEGs and DEMs may be possible candidates for differential diagnosis between LUAD and LUSC, and several have already been used in clinical practice, such as TTF-1, SOX2, p63, KRT5, and KRT7 (6,19). Although a high fold change is not the only criteria for biomarkers, those exhibiting significant differences in expression, such as TM4SF4, DPCR1, SERPINB12, and AMTN, may be worth further investigation. In our previous study, several of the DEGs, such as melanophilin (MLPH), transmembrane channel-like 5 (TMC5), surfactant associated 3 (SFTA3), desmoglein 3 (DSG3), desmocollin 3 (DSC3), and calmodulin-like 3 (CALML3), were confirmed to be differentially expressed in LUAD and LUSC by immunohistochemical staining (7). miR-205-5p expression levels were previously reported to be significantly higher in LUSC compared with LUAD, both in serum and tissue (20); miR-375 was also demonstrated to be highly expressed in LUAD (21), which was consistent with the current data. The present study offers a list of DEGs and DEMs in which better biomarkers may exist.
Various gene functions and pathways that are greatly altered in LUAD and LUSC have been identified in the present study, suggesting that these GOs and pathways serve primary roles in lung cancer pathogenesis, as do the DEGs that participate in these GOs and pathways. Although LUAD and LUSC share a lot of common activated GOs and pathways, they also display their own features. According to the results, genes related to extracellular matrix organization, such as matrix metalloproteinase (MMP) 3, MMP10 and MMP12, were upregulated in LUAD and even significantly higher in LUSC compared with in normal tissue. MMPs are key factors in the development of the tumor microenvironment and drive cancer progression and metastasis, and have been identified as prognostic factors for poor survival in many types of cancer (22–24). In the present study, the PPAR pathway was demonstrated to be activated in LUAD, but not in LUSC. PPARs have been reported to be associated with breast, ovary, prostate, bladder, gastric and colon adenocarcinoma carcinogenesis, as well as in leukemia (25). Tsubouchi et al (26) proposed that a PPARγ agonist may be a useful therapeutic agent in the treatment of human lung cancer. The p53 signaling pathway was also identified as upregulated in LUSC; a critical role of the p53 mutation in malignant transformation, histologic progression, invasion, and metastasis has been previously demonstrated in both in vitro and in vivo models of lung cancer (27–29). Smoking was revealed to be closely related to p53 mutation (4,30), which may explain the prevalence of p53 alterations in LUSC.
miR-29b acts as a tumor suppressor in breast cancer and is a potential marker for recurrence and metastasis (31). miR-29b-3p in peripheral blood mononuclear cells was reported to be a novel target for the diagnosis of NSCLC (32). miR-1 was revealed to be downregulated in various types of cancers, including LUSC, and could act as a tumor suppressor (33). Previous studies have suggested that miR-1 functions through the regulation of oncogenic coronin 1C (34), and the silencing miR-1 resulted in sensitization of LUSC to traditional chemotherapeutics (35). miR-375 appears to serve many different roles in carcinogenesis, and functions as an oncogene or a tumor suppressor depending on the type of cancer (36). miR-375 was previously revealed to inhibit cell proliferation, invasion and motility in several types of cancer, including NSCLC (37), whereas upregulated miR-375 expression may stimulate cell proliferation in thyroid carcinoma, small-cell lung, breast and prostate cancers (38). Conversely, miR-375 was reported to be downregulated in NSCLC, but the prognostic significance remains unclear (39). Further research into these TFs and miRNAs may lead to novel treatment of NSCLC.
In conclusion, the present study investigated the differences between the gene and miRNA expression patterns in LUAD and LUSC, and explored their different biological characteristics. Further understanding of these differences may promote the discovery and development of new, accurate strategies for the prevention, diagnosis and treatment of lung cancer. Further experiments are required to validate the results of the present bioinformatics analysis.
Acknowledgements
The present study was supported by The National Natural Science Foundation of China (grant nos. 81401875 and 81472225) and The Natural Science Foundation of Shanghai, China (grant no. 14ZR1406000). The results are based upon data generated by the TCGA Research Network. The authors would like to thank the language-editing service, International Science Editing Co., for editing this manuscript.
Glossary
Abbreviations
- DEGs
differentially expressed genes
- DEMs
differentially expressed microRNAs
- FDR
false discovery rate
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- NSCLC
non-small cell lung cancer
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.1186/s40880-015-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis WD, Rekhtman N, Riley GJ, Geisinger KR, Asamura H, Brambilla E, Garg K, Hirsch FR, Noguchi M, Powell CA, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: A paradigm shift. J Thorac Oncol. 2010;5:411–414. doi: 10.1097/JTO.0b013e3181d57f6e. [DOI] [PubMed] [Google Scholar]
- 7.Zhan C, Yan L, Wang L, Sun Y, Wang X, Lin Z, Zhang Y, Shi Y, Jiang W, Wang Q. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J Thorac Dis. 2015;7:1398–1405. doi: 10.3978/j.issn.2072-1439.2015.07.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network: Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG, Ladanyi M. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: Lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan C, Yan L, Wang L, Jiang W, Zhang Y, Xi J, Jin Y, Chen L, Shi Y, Lin Z, Wang Q. Landscape of expression profiles in esophageal carcinoma by the cancer genome atlas data. Dis Esophagus. 2016;29:920–928. doi: 10.1111/dote.12416. [DOI] [PubMed] [Google Scholar]
- 11.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan L, Zhan C, Wu J, Wang S. Expression profile analysis of head and neck squamous cell carcinomas using data from the Cancer Genome Atlas. Mol Med Rep. 2016;13:4259–4265. doi: 10.3892/mmr.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: Strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. doi: 10.1055/s-0031-1272866. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz AM, Rezaei MK. Diagnostic surgical pathology in lung cancer: Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143:e251S–e262S. doi: 10.1378/chest.12-2356. (Suppl 5) [DOI] [PubMed] [Google Scholar]
- 20.Jiang M, Zhang P, Hu G, Xiao Z, Xu F, Zhong T, Huang F, Kuang H, Zhang W. Relative expressions of miR-205-5p, miR-205-3p, and miR-21 in tissues and serum of non-small cell lung cancer patients. Mol Cell Biochem. 2013;383:67–75. doi: 10.1007/s11010-013-1755-y. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehner C, Miller E, Nassar A, Bamlet WR, Radisky ES, Radisky DC. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer. 2015;6:480–489. doi: 10.18632/genesandcancer.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol. 2010;15:201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S, Roman J. Peroxisome proliferator-activated receptor gamma: A novel target for cancer therapeutics? Anticancer Drug. 2007;18:237–244. doi: 10.1097/CAD.0b013e328011e67d. [DOI] [PubMed] [Google Scholar]
- 26.Tsubouchi Y, Sano H, Kawahito Y, Mukai S, Yamada R, Kohno M, Inoue K, Hla T, Kondo M. Inhibition of human lung cancer cell growth by the peroxisome proliferator-activated receptor-gamma agonists through induction of apoptosis. Biochem Bioph Res Commun. 2000;270:400–405. doi: 10.1006/bbrc.2000.2436. [DOI] [PubMed] [Google Scholar]
- 27.Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower LA. A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53-deficient mice. Nat Genet. 1995;9:305–311. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- 28.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-X. [DOI] [PubMed] [Google Scholar]
- 29.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 30.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinden Y, Iguchi T, Akiyoshi S, Ueo H, Ueda M, Hirata H, Sakimura S, Uchi R, Takano Y, Eguchi H, et al. miR-29b is an indicator of prognosis in breast cancer patients. Mol Clin Oncol. 2015;3:919–923. doi: 10.3892/mco.2015.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J, Lin Y, Zhan M, Mann DL, Stass SA, Jiang F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest. 2015;95:1197–1206. doi: 10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: Dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mataki H, Enokida H, Chiyomaru T, Mizuno K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T, Nakagawa M, et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J Hum Genet. 2015;60:53–61. doi: 10.1038/jhg.2014.111. [DOI] [PubMed] [Google Scholar]
- 35.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Jonsdottir K, Janssen SR, Da Rosa FC, Gudlaugsson E, Skaland I, Baak JP, Janssen EA. Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS One. 2012;7:e48692. doi: 10.1371/journal.pone.0048692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao X, Mei W, Weng W, Qin J, Zhou J, Liu J, Cheng J. Mir-375 enhances ruthenium-derived compound Rawq01 induced cell death in human ovarian cancer. Int J Clin Exp Patho. 2013;6:1095–1102. [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikawa E, Osada H, Okazaki Y, Arima C, Tomida S, Tatematsu Y, Taguchi A, Shimada Y, Yanagisawa K, Yatabe Y, et al. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71:6165–6173. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 39.Yu H, Jiang L, Sun C, Li Guo L, Lin M, Huang J, Zhu L. Decreased circulating miR-375: A potential biomarker for patients with non-small-cell lung cancer. Gene. 2014;534:60–65. doi: 10.1016/j.gene.2013.10.024. [DOI] [PubMed] [Google Scholar]