Abstract
Previous studies have suggested that dysregulation of microRNA (miR) −124a is associated with various types of human cancer. However, there are few studies reporting the level of miR-124a expression in non-small cell lung cancer (NSCLC). The present study investigated the association between miR-124a and NSCLC by analyzing the differential expression of miR-124a in NSCLC using the GEO database, as well as subsequently performing reverse transcription-quantitative polymerase chain reaction analysis on 160 NSCLC biopsies, 32 of which were paired with adjacent normal tissues. The results indicated that mir-124a expression levels were decreased in NSCLC tumor biopsies compared with adjacent normal tissues. The overall survival (OS) in patients with a high expression of miR-124a was prolonged relative to patients with low expression of miR-124a. The expression levels of miR-124a were associated with clinical characteristics, including lymph-node metastasis, tumor differentiation, tumor node metastasis (TNM) stage and diameter. Frequently, lymph-node metastasis, TNM stage, diameter and lack of chemotherapy have been associated with a worse prognosis in patients. In addition, the present study identified that high expression of miR-124awith chemotherapy may increase OS. In conclusion, the current study demonstrated that miR-124a was downregulated in NSCLC, and miR-124a was a potential prognostic tumor biomarker response to chemotherapy.
Keywords: microRNA-124, non-small cell lung cancer, chemotherapy, overall survival, disease-free survival, biomarker, prognosis
Introduction
Lung cancer is one of the most common malignancies and is the major cause of cancer-associated mortality, accounting for ~1.38 million deaths each year (1,2). Of all lung carcinomas, non-small cell lung cancer (NSCLC) accounts for ~70–85% (3). Although there have been recent advances in diagnosis and treatment, the prognosis of lung cancer is still unfavorable and the 5-year overall survival (OS) rate remains <15% (4,5). In previous decades, studies reported that microRNAs (miRNAs/miRs) may serve an important role in NSCLC pathogenesis, which provides novel insights into disease biology (6,7). Therefore, improved understanding of detailed mechanisms of NSCLC with miRNAs is necessary for development of effective therapeutic strategies.
miRNAs are a class of 20–24 nucleotide-long non-coding RNAs, which regulate gene expression at the post-transcriptional level through mRNA interference, and are involved in cell development, proliferation, differentiation and apoptosis (8,9). There are currently ~2000 miRNAs that have been identified and this number is rapidly increasing. A large number of miRNAs have been investigated in cancer research as therapeutic targets, with certain miRNAs identified as being associated with the tumor metastasis of NSCLC. For instance, upregulation of miR-24 promotes cell proliferation by targeting nuclear apoptosis inducing factor 1in NSCLC (10). Overexpression of miR-328 has a role in conferring migratory potential to NSCLC cells through targeting protein kinase C alpha (11). miR-34c-3p functions as a tumor suppressor by inhibiting eukaryotic initiation factor-4E expression in NSCLC (12). miR-99a suppresses the metastasis of human NSCLC by targeting AKT serine/threonine kinase 1 (13). However, there are few studies researching the expression of miR-124a in NSCLC and the detailed molecular mechanism of miR-124a in NSCLC requires further investigation.
In the present study, the differential expression of miR-124a in NSCLC was analyzed using the GEO database and the association between miR-124a and NSCLC was subsequently revealed. Furthermore, the expression status of miR-124a in NSCLC was identified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for a group of NSCLC biopsies. Furthermore, the prognostic significance of NSCLC and the response to chemotherapy was assessed.
Materials and methods
Data and date source
All miR-124a expression datasets were downloaded from GEO (www.pubmed.com/geo). Data were retrieved using the keywords ‘miR-124a’ and ‘NSCLC’. The first data set, GSE10021, has samples taken from 16 human cell lines (14), and includes the following cell lines: Human lung carcinoma A549, fibrosarcoma HT1080, cervix carcinoma Henrietta Lacks, cervix carcinoma HeLaS3, hepatocellular carcinoma Huh7, breast adenocarcinoma MCF7, breast adenocarcinoma MDAMB231, embryonal kidney HEK293T, colon adenocarcinoma HT29, hepatocellular carcinoma HepG2, neuroblastoma SKNMC, colon adenocarcinoma Caco2, embryonal kidney HEK293 and colon carcinoma HCT116. The second data set, GSE61741, has samples taken from normal patients and patients with cancer (15), and it has a total of 1,049 samples out of which 94 were normal and 15 were long-lived individuals and 940 patients had been screened for the complete miRNA repertoire. The third data set, GSE63805, has samples taken from normal patients and patients with lung cancer (16), and it has a total of 62 samples out of which 31 were normal and 31 were cancerous. The last data set, GSE17681, has samples taken from normal patients and patients with lung cancer (17), and it has a total of 36 samples out of which 19 were normal and 17 were cancerous.
Clinical specimens
A total of 160 cases of surgically resected NSCLC and 32 paired normal adjacent lung tissue samples were evaluated for miRNA expression. These specimens were collected from patients from the tissue bank in China-Japan Union Hospital, Jilin University (Changchun, China) between January 2008 and December 2012. All patients gave their informed consent, and the Ethical and Scientific Committees of Shanghai Tenth People's Hospital, Tongji University School of Medicine (Shanghai, China) approved the study. The tumors of NSCLC were staged based on the 7th edition of the AJCC tumor node metastasis (TNM) staging system (18). In addition, several clinical characteristics of these patients were assessed, including age, gender, lymph-node metastasis, tumor differentiation, histological subtypes, TNM stage, invasion of lung membrane, vascular invasion, tumor size, chemotherapy, miR-124a expression status, OS and disease-free survival (DFS). Age was stratified according to ≥60 or <60 years. Tumor size was divided into ≥5 and <5 cm based on the mean tumor diameter. OS represented the date of diagnosis to the date of death, from any cause. DFS was represented the original date of diagnosis to the first date of recurrence, or death. All clinical data were confirmed by the patient or relatives, by medical recording, by the social security record, or by follow-up record.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues using the TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's instructions. RNA concentration and purity was assessed using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.). A total of10 ng total RNA was used for cDNA synthesis using the Taqman MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Reverse transcription with a miR-124a-specific primer was performed using ABI's TaqMan MicroRNA Reverse Transcription kit, miR-124a expression level was detected using a Taqman MicroRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) (19). The RT-qPCR thermocycling conditions were as follows: 94°C for 30 sec (initial denaturation), 94°C for 5 sec (denaturation) and 55°C for 30 sec (annealing), for 40 cycles. U6 expression was used as the internal control. The following primers were used: miR-124a forward, 5′-GGTAAGGCACGCGGT-3′, and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; U6 forward, 5′-CTGGTTAGTACTTGGACGGGAGAC-3′, and reverse, 5′-GTGCAGGGTCCGAGGT-3′. All reactions were performed three times in triplicate, and relative expression of miRNAs was calculated using the standard curve and the ΔΔCq method (20).
Statistical analysis
All statistical data were analyzed using SPSS software (version, 19.0; IBM SPSS; Armonk, NY, USA). Significant differences between the two groups were evaluated using an independent t-test. The expression of miR-124a and other data were presented as the mean ± standard deviation. χ2 test analysis was used to assess differences in patient characteristics. OS rates were calculated actuarially according to the Kaplan-Meier method and results were compared with a log-rank test. To examine which individual characteristics played important roles in survival, univariate and multivariate Cox regression analysis were conducted. P<0.05 was considered to indicate a statistically significant difference and P<0.001 was considered highly significant.
Results
Analysis of miR-124a expression using an online database
The expression of miR-124a in 16 different cell lines was analyzed via the GEO database (Fig. 1A), which revealed that miR-124a exhibited relatively decreased expression in lung cancer A549 cells, when compared with most other cell lines.
Figure 1.
Analysis of miR-124a expression in NSCLC patients using the GEO database. (A) Expression levels of miR-124a in 16 different cell lines derived from the GEO database. (B) Expression levels of miR-124ain normal vs. other cancers derived from the GEO database. (C) Expression levels of miR-124ain 30 paired lung cancer tissues derived from the GEO database. The fold-change for miR-124a expression levels was calculated using thelog2 ratio of paired tumor/normal expression. (D) Expression levels of miR-124ain normal vs. 17 paired lung cancer tissues derived from the GEO database. *P<0.05, **P<0.01 vs. normal tissues. miR, microRNA; NSCLC, non-small cell lung cancer; GEO, Gene Expression Omnibus; FC, fold change.
miR-124a was identified as being relatively expressed in 104 normal samples (114.41±64.55), 120 Wilms' tumor samples (102.75±60.35), 14 gastric cancer samples (98.68±48.73), 62 lung cancer samples (89.27±45.46), 19 glioma samples (81.19±49.43), 33 melanoma samples (79.69±49.62), 20 renal cancer samples (76.57±45.03) and 27 colon cancer samples (72.06±37.34), using the GEO database (Fig. 1B). The results indicated that miR-124a exhibited decreased expression in tumor tissues, when compared with normal tissue.
A total of 30 paired lung cancer tissues were investigated from a dataset on the GEO database (Fig. 1C). The fold-change for relative miR-124a expression levels was calculated using the Log2 ratio of paired tumor/normal expression, and miR-124awas observed to be downregulated in over half of the lung cancer cases [fold change (FC) =0.81; P=0.021].
Relative miR-124a expression in 17 lung cancer blood samples vs. 17 adjacent normal tissues was investigated using the GEO database (Fig. 1D). The analysis suggested that miR-124a was downregulated in lung cancer tissues (18.55±6.75), when compared with adjacent normal tissues (40.76±16.48; FC=0.45; P=0.048).
miR-124a expression in NSCLC and normal lung tissue
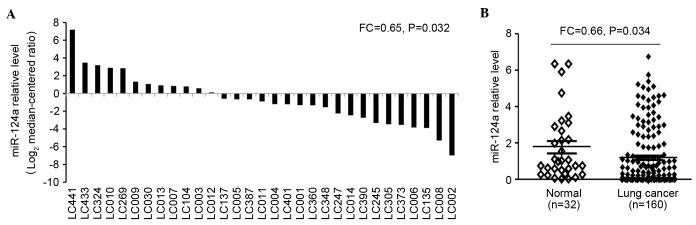
To validate these findings, 32 paired lung cancer tissues were evaluated using RT-qPCR (Fig. 2A). The results demonstrated that miR-124a was expressed at significantly decreased levels in most lung cancer cases (FC=0.64; P=0.032). In addition, the expression levels of miR-124a were examined in tumor (n=160) and adjacent non-neoplastic tissues (n=32) using RT-qPCR (Fig. 2B), which demonstrated that miR-124a expression levels were significantly decreased in NSCLC tumor biopsies (1.14±0.85), when compared with adjacent normal tissues (1.76±0.63; FC=0.65; P=0.034).
Figure 2.
Analysis of miR-124a expression in 32 paired NSCLC tissues using the reverse transcription-quantitative polymerase chain reaction. (A) Expression levels of miR-125a-3p in 32 paired NSCLC tissues. (B) Expression levels of miR-125a-3p in 32 normal vs. 160 lung cancer tissues. NSCLC, non-small cell lung cancer; miR, microRNA; FC, fold change.
Association between miR-124a expression and clinical characteristics
Univariate analysis was used to investigate the relationship between miR-124a expression and clinical characteristics in 160 cases of NSCLC. The results revealed that the expression levels of miR-124a were significantly associated with lymph node metastasis (P=0.021), tumor differentiation (P=0.047), TNM stage (P=0.036) and diameter (P=0.026; Table I). Conversely, the results demonstrated that age, gender, smoking history, histology, invasion of lung membrane, vascular invasion and chemotherapy have no significant association with miR-124a expression (P>0.05; Table I).
Table I.
Univariate analysis of overall patient survival stratified by clinical characteristics.
| Overall survival | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Variable | n | miR-124a expression, mean ± standard deviation | P-value | Months, mean | 95% CI, mean | P-value (log-rank test) |
| Age | |||||||
| ≥60 years | 97 | 1.26±0.63 | 0.281 | 22.34 | 21.06–23.17 | 0.326 | |
| <60 years | 63 | 1.11±0.62 | 25.19 | 22.45–29.63 | |||
| Gender | |||||||
| Male | 97 | 1.19±0.61 | 0.487 | 24.76 | 22.09–26.04 | 0.168 | |
| Female | 63 | 1.21±0.66 | 25.33 | 21.58–28.33 | |||
| Smoking history | |||||||
| Never | 38 | 1.29±0.42 | 0.115 | 26.11 | 24.39–28.63 | 0.091 | |
| Ever | 65 | 1.18±0.79 | 24.86 | 22.77–26.34 | |||
| Unknown | 57 | 1.26±0.67 | 26.34 | 24.91–28.76 | |||
| Lymphnode metastasis | 0.021 | 0.039 | |||||
| Negative | 91 | 1.39±0.81 | 26.38 | 24.73–33.59 | |||
| Positive | 57 | 1.11±0.13 | 20.44 | 17.06–24.31 | |||
| Unknown | 12 | 1.18±0.96 | 22.56 | 20.84–25.69 | |||
| Tumor differentiation | 0.047 | 0.225 | |||||
| Poor | 18 | 0.86±0.25 | 24.03 | 24.60–33.68 | |||
| Moderate | 84 | 1.18±0.13 | 26.68 | 22.28–28.34 | |||
| Well | 58 | 1.39±0.26 | 26.56 | 22.70–29.75 | |||
| Histology | |||||||
| Adenocarcinoma | 55 | 1.19±0.53 | 0.634 | 26.63 | 23.47–29.86 | 0.353 | |
| Squamous cell carcinoma | 105 | 1.27±0.38 | 25.98 | 23.33–28.74 | |||
| TNM stage | |||||||
| I–II | 104 | 1.28±0.38 | 0.036 | 27.58 | 24.19–30.36 | 0.027 | |
| III–IV | 56 | 0.81±0.26 | 23.46 | 18.69–25.43 | |||
| Invasion of lung membrane | 0.068 | 0.088 | |||||
| Negative | 34 | 1.24±0.68 | 30.36 | 22.68–41.33 | |||
| Positive | 114 | 0.99±0.86 | 26.83 | 22.48–28.55 | |||
| Unknown | 12 | 1.18±0.37 | 25.43 | 22.68–26.47 | |||
| Vascular invasion | |||||||
| Negative | 145 | 1.21±0.79 | 0.691 | 26.58 | 21.67–29.23 | 0.318 | |
| Positive | 3 | 0.97±0.83 | 25.24 | 23.36–42.58 | |||
| Unknown | 12 | 1.21±0.36 | 24.37 | 21.48–30.59 | |||
| Chemotherapy | |||||||
| Negative | 80 | 1.17±0.36 | 0.543 | 20.58 | 18.69–24.37 | 0.029 | |
| Positive | 69 | 1.25±0.63 | 26.94 | 23.06–28.94 | |||
| Unknown | 11 | ||||||
| Diameter | |||||||
| ≥5 cm | 39 | 0.88±0.35 | 0.026 | 21.93 | 18.45–26.04 | <0.001 | |
| <5 cm | 121 | 1.36±0.62 | 27.86 | 25.39–29.28 | |||
miR-124a, microRNA-124a; CI, confidence interval; TNM, tumor node metastasis.
Expression levels of miR-124a area prognostic marker in NSCLC survival
Univariate analysis of OS based on patients stratified by clinical characteristics presented in Table I. These clinical characteristics with univariate analysis included age, gender, smoking history, lymph-node metastasis, tumor differentiation, histology, TNM stage, invasion of lung membrane, vascular invasion, tumor diameter and miR-125a-3p expression. Obviously, there was a meaningful relationship between short OS and several clinical characteristics, including lymph-node metastasis (P=0.039), TNM stage (P=0.027), diameter (P<0.001) and lack of treatment with chemotherapy (P=0.029). Conclusively, it was identified that the patients who exhibited lymph-node metastasis, high TNM stage, tumor size exceeding 0.5 cm or those who had not received chemotherapy exhibited increased mortality (decreased OS).
To determine which clinical characteristics were associated with the prognosis of NSCLC, univariate survival analysis with Kaplan-Meier were performed. Several clinical parameters with Kaplan-Meier survival curves presented a good prognosis for patients with NSCLC. Negative lymph-node metastasis was significantly associated with increased OS (P=0.039; Fig. 3A) and DFS (P=0.018; Fig. 3B) in patients with NSCLC. Similarly, low TNM stage was positively associated with increased OS (P=0.027; Fig. 3A) and DFS (P=0.025; Fig. 3B). In addition, tumor size was significantly associated with increased OS (P<0.001; Fig. 3E) and DFS (P=0.009; Fig. 3F).
Figure 3.
Univariate survival analysis of different clinical parameters in non-small cell lung cancer. Univariate survival analysis of (A) overall survivaland (B) disease-free survival stratified by lymph node metastasis, (C) overall survival and (D) disease-free survival stratified by tumor node metastasis stage, and (E) overall survival and (F) disease-free survival stratified by tumor diameter, as determined by Kaplan-Meier plots estimates.
Univariate analysis with a Cox proportional hazards regression model was used to explore the prognosis of clinical characteristics. The result revealed that there were four parameters significantly associated with good prognosis, including lymph node metastasis [P=0.03; HR=1.72 (1.18–2.96); Table II], TNM stage [P=0.02; HR=1.83 (1.56–2.64); Table II], tumor diameter [P<0.001; HR=2.56 (2.07–5.46); Table II], as well as miR-124a expression [P=0.023; HR=0.68 (0.52–0.73); Table II]. There was no association between prognosis and several parameters, including age, gender, smoking history, tumor differentiation, histology, invasion of lung membrane or vascular invasion. Therefore, these results suggested that miR-124a serves an important role in NSCLC progression. In addition, it was summarized that miR-124a is involved in the prognosis of NSCLC patients.
Table II.
Cox regression model analysis for prognosis based on various clinical characteristics of patients with non-small cell lung cancer.
| miR-124a multivariate analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Factor | HR | 95% CI (univariate) | P-value | HR | 95% CI (multivariate) | P-value | ||||
| Age | 0.96 | 0.69–1.14 | 0.45 | |||||||
| Gender | 0.87 | 0.53–1.16 | 0.18 | |||||||
| Smoking history | 1.21 | 0.71–1.52 | 0.19 | |||||||
| Lymph-node metastasis | 1.72 | 1.18–2.96 | 0.03 | 1.98 | 1.25–3.03 | 0.015 | ||||
| Tumor differentiation | 0.88 | 0.54–1.01 | 0.18 | |||||||
| Histology | 1.18 | 0.86–1.21 | 0.34 | |||||||
| TNM stage | 1.83 | 1.56–2.64 | 0.02 | 2.13 | 1.61–2.89 | 0.009 | ||||
| Invasion of lung membrane | 1.22 | 1.06–1.37 | 0.15 | |||||||
| Vascular invasion | 0.98 | 0.68–1.54 | 0.31 | |||||||
| Diameter | 2.56 | 2.07–5.46 | <0.001 | 3.65 | 2.26–5.43 | <0.001 | ||||
| miR-124a expression | 0.68 | 0.52–0.73 | 0.023 | |||||||
HR, hazard ratio; miR-124a, microRNA-124a; CI, confidence interval; TNM, tumor node metastasis.
To further investigate the association between miR-124a expression and the prognosis of patients with NSCLC, multivariate Cox proportional hazards regression analysis was conducted. The model included all of the characteristics relative to predicted OS in the univariate analysis of the entire patients as presented in Table I. There were three characteristics that presented a significant association with prognosis, including lymph-node metastasis [P=0.015; HR=1.98 (1.25–3.03); Table II], TNM stage [P=0.009; HR=2.13 (1.61–2.89); Table II] and tumor diameter [P<0.001; HR=3.65 (2.26–5.43); Table II]. Multivariable Cox regression model analysis revealed that increased expression of miR-124a was determined to be a predictor of increased OS in patients with NSCLC.
Association amongst survival, chemotherapy and miR-124a expression
Chemotherapy is a major treatment of NSCLC, thus it was analyzed as an influential factor inOS and DFS. According to Table III, chemotherapy was demonstrated to significantly prolong OS (29.94±1.57 vs. 22.58±1.96; P=0.029) and DFS (26.43±3.56 vs. 20.06±3.78; P=0.032) in the entire group. By comprehensive analysis of chemotherapy and miR-124a expression level, it was concluded that patients who received chemotherapy with high expression of miR-124a exhibited an increased OS (32.78±6.96 vs. 20.43±3.58; P=0.001) and DFS (29.33±2.65 vs. 20.06±3.98, P<0.001), compared with those who did not receive chemotherapy with low expression of miR-124a.
Table III.
OS and DFS of patients with non-small cell lung cancer stratified by chemotherapy alone, or chemotherapy and miR-124a expression.
| OS | DFS | |||||
|---|---|---|---|---|---|---|
| Mean ± SD | 95% CI | P-value | Mean ± SD | 95% CI | P-value | |
| Chemotherapy | ||||||
| Positive | 29.94±1.57 | 23.06–33.94 | 0.029 | 26.43±3.56 | 25.43–28.04 | 0.032 |
| Negative | 22.58±1.96 | 18.69–24.37 | 20.06±3.78 | 18.78–25.33 | ||
| Chemotherapy+expression | ||||||
| P+H | 32.78±6.96 | 26.79–36.43 | 0.001 | 29.33±2.65 | 26.04–33.41 | <0.001 |
| N+L | 20.43±3.58 | 16.98–22.54 | 20.06±3.98 | 18.07–24.49 | ||
OS, overall survival; DFS, disease-free survival; SD, standard deviation; CI, confidence interval; P+H, chemotherapy and high miR-124a expression; N+L, no chemotherapy and low miR-124a expression.
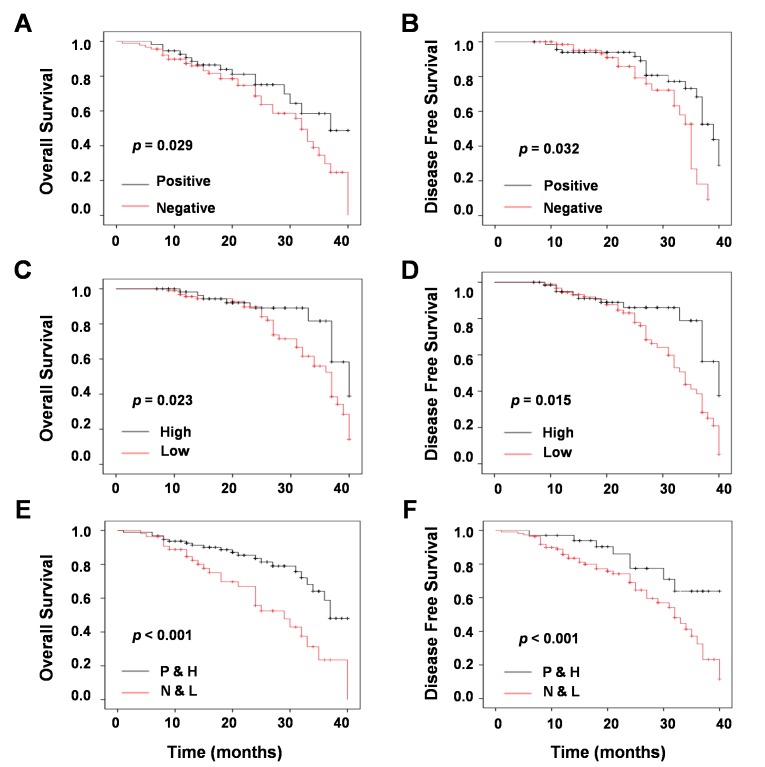
To further identify whether chemotherapy or with miR-124a expression was associated with OS and DFS, univariate and multivariate survival analysis with Kaplan-Meier estimates were conducted. The result of univariate survival analysis indicated that increased OS (P=0.029; Fig. 4A) and DFS (P=0.032; Fig. 4B) was significantly associated with chemotherapy compared with the results from untreated patients. In addition, high expression of miR-124a increased OS (P=0.023; Fig. 4C) and DFS (P=0.015; Fig. 4D) when compared with low expression. The result of multivariate survival analysis demonstrated that chemotherapy with high expression of miR-124a prolonged OS (P<0.001; Fig. 4E) and DFS (P<0.001; Fig. 4F).
Figure 4.
Survival analysis of chemotherapy and miR-124a expression in non-small cell lung cancer. Univariate survival analysis of (A) overall survival and (B) disease-free survival stratified by chemotherapy, (C) overall survival and (D) disease-free survival stratified by miR124-a expression, and (E) overall survival and (F) disease-free survival stratified by chemotherapy and miR-124a expression, as determined by Kaplan-Meier plots estimates. miR, microRNA; P&H, chemotherapy and high miR-124a expression; N&L, no chemotherapy and low miR-124a expression.
Discussion
Evidence has demonstrated that miRNAs regulated cell proliferation, cell metastasis and apoptosis at a post transcriptional level. There are previous studies that have reported that miRNAs serve a crucial role in the development of chemosensitivity or chemoresistance in NSCLC (21,22). In addition, miRNAs expression level has been identified to be associatedwith tumor development (23). In recent years, miRNAs have been used to detect early diagnosis, prognosis and therapeutic evaluation (24).
miR-124a has been identified to be a novel suppressor for cancer, and has been reported to be associated with the suppressive effects of a variety of human cancers, including breast, glioma, gastric cancer and colitis (25). Certain previous studies reported that methylation of miR-124a is associated with aggressive and advanced breast cancer disease (26,27). Furthermore, miR-124a has been investigated to potentially inhibit glioma cell proliferation and invasion by blocking the expression of a particular gene (28,29). In addition, the methylation of miR-124a was identified early in colorectal carcinogenesis (30,31) and was epigenetically silenced in the development of uveal melanoma (32).
In the present study, the expression of miR-124a in 160 NSCLC tissues and 32 paired normal tissues was evaluated. Several previous papers reported that miR-124a expression level in normal tissues is higher than those in lung cancer tissues (33–35). Consistently, miR-124a expression levels were lower in NSCLC tumor biopsies, when compared to adjacent normal tissues. The OS in patients with high expression of miR-124a was prolonged relative to patients with low expression of miR-124a. Therefore, miR-124a may bea deregulated gene in NSCLC. The expression levels of miR-124a were associated with clinical characteristics, including lymph-node metastasis, tumor differentiation, TNM stage and diameter. Frequently, lymph-node metastasis, TNM stage, diameter and chemotherapy are associated with a worse prognosis in patients. Therefore, there was a significant association between miR-124a and prognosis. In addition, there was a high expression of miR-124a identified with chemotherapy that may increase OS. Therefore, it was concluded that miR-124a may act as a biomarker for response to chemotherapy in NSCLC.
Furthermore, the authors explored the biomarker role of miR-124a in NSCLC. To the best of the authors' knowledge, it is the first attempt to identify the status of miR-124a for chemotherapy in NSCLC. Generally, tumor metastasis is a major cause of high mortality of NSCLC. In recent decades, chemotherapy has remained as the central therapeutic mainstay in metastatic lung cancer, despite response rates being maintained at 30–40% and the median survival being 7–12 months (36). For the past few years, characteristic molecules began to play an important role in cancer treatment (37). It is reported that molecular biomarkers successfully respond to NSCLC chemotherapy as diagnostic biomarkers and prognostic factors. For instance, Perez-Carbonell et al (38) demonstrated that miR-320e was a novel prognostic biomarker associated with adverse clinical outcome in patients with stage III colorectal cancer treated with 5-FU-based adjuvant chemotherapy. In addition, miR-22, miR-24, miR-34a and miR-638 were investigated as novel predictive biomarkers (39,40). Furthermore, miR-200c, miR-744 and miR-34a, have been reported to be prognostic biomarkers in esophageal cancer, pancreatic cancer and breast cancer (41–43).
In conclusion, the present study reported the clinical and prognostic relevance of miR-124a in patients with NSCLC. These data revealed that the OS and DFS of patients undergoing chemotherapy were prolonged in comparison with those not receiving chemotherapy. Furthermore, patients who exhibited increased expression of miR-124a with chemotherapeutic treatment presented the longest OS and DFS, compared with those who exhibited decreased expression of miR-124a without chemotherapy. Conclusively, miR-124a is a predictive biomarker for the prognosis of NSCLC with chemotherapy. In future studies, bioinformatic and transcriptomic approaches, as well as functional analysis, will be necessary to investigate the mechanistic role of miR-124a in NSCLC and to confirm the present results.
Acknowledgements
The present study was partially supported by grants from the National Natural Science Foundation of China (grant nos. 81201535, 81472202 and 81302065), Shanghai Natural Science Foundation (grant nos. 12ZR1436000 and 16ZR1428900) and Shanghai Municipal Commission of Health and Family Planning (grant nos. 201440398 and 201540228), Jiangxi Province Department of Science Plan Funded Projects (grant no. 2011BBG70046), Hunan Natural Science Fund for Distinguished Young Scholars Project (grant no. 2015JJ1009) and the Jilin Provincial Science and Technology Department (grant nos. 20130727029YY, 20140f14061GH and 20150204057SF).
References
- 1.Hamamoto J, Soejima K, Yoda S, Naoki K, Nakayama S, Satomi R, Terai H, Ikemura S, Sato T, Yasuda H, et al. Identification of microRNAs differentially expressed between lung squamous cell carcinoma and lung adenocarcinoma. Mol Med Rep. 2013;8:456–462. doi: 10.3892/mmr.2013.1517. [DOI] [PubMed] [Google Scholar]
- 2.Meng W, Ye Z, Cui R, Perry J, Dedousi-Huebner V, Huebner A, Wang Y, Li B, Volinia S, Nakanishi H, et al. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin Cancer Res. 2013;19:5423–5433. doi: 10.1158/1078-0432.CCR-13-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E, Grossi F, Recchia F, Zanelli F, Fontanini G, et al. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108:1695–1703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Mitra R, Edmonds MD, Sun J, Zhao M, Yu H, Eischen CM, Zhao Z. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014;20:1356–1368. doi: 10.1261/rna.042754.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J, Yu F, Li D, Zhu X, Zhang X, Lv Z. MicroRNA-218 regulates cisplatin (DPP) chemosensitivity in non-small cell lung cancer by targeting RUNX2. Tumour Biol. 2016;37:1197–1204. doi: 10.1007/s13277-015-3831-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhou YL, Xu YJ, Qiao CW. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int J Clin Exp Pathol. 2015;8:6312–6322. [PMC free article] [PubMed] [Google Scholar]
- 8.Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, Martin-Subero JI, Cordeu L, Gárate L, San José-Eneriz E, Abizanda G, Rodríguez-Otero P, Fortes P, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 9.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90:1–7. doi: 10.1007/s11060-008-9624-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhao G, Liu L, Zhao T, Jin S, Jiang S, Cao S, Han J, Xin Y, Dong Q, Liu X, Cui J. Upregulation of miR-24 promotes cell proliferation by targeting NAIF1 in non-small cell lung cancer. Tumour Biol. 2015;36:3693–3701. doi: 10.1007/s13277-014-3008-4. [DOI] [PubMed] [Google Scholar]
- 11.Arora S, Ranade AR, Tran NL, Nasser S, Sridhar S, Korn RL, Ross JT, Dhruv H, Foss KM, Sibenaller Z, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer. 2011;129:2621–2631. doi: 10.1002/ijc.25939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Wang X, Li J, Gu K, Lv L, Zhang S, Che D, Cao J, Jin S, Yu Y. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015;48:582–592. doi: 10.1111/cpr.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu SH, Zhang CL, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116:268–276. doi: 10.1002/jcb.24965. [DOI] [PubMed] [Google Scholar]
- 14.Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J Hum Genet. 2008;53:515–523. doi: 10.1007/s10038-008-0279-x. [DOI] [PubMed] [Google Scholar]
- 15.Keller A, Leidinger P, Vogel B, Backes C, ElSharawy A, Galata V, Mueller SC, Marquart S, Schrauder MG, Strick R, et al. miRNAs can be generally associated with human pathologies as exemplified for miR-144. BMC Med. 2014;12:224. doi: 10.1186/s12916-014-0224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robles AI, Arai E, Mathé EA, Okayama H, Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10:1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, Scheffler M, Huwer H, Lenhof HP, Meese E. miRNAs in lung cancer-studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Hou LK, Ma YS, Han Y, Lu GX, Luo P, Chang ZY, Xie RT, Yang HQ, Chai L, Cai MX, et al. Association of microRNA-33a molecular signature with non-small cell lung cancer diagnosis and prognosis after chemotherapy. PLoS One. 2017;12:e0170431. doi: 10.1371/journal.pone.0170431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faversani A, Amatori S, Augello C, Colombo F, Porretti L, Fanelli M, Ferrero S, Palleschi A, Pelicci PG, Belloni E, et al. miR-494-3p is a novel tumor driver of lung carcinogenesis. Oncotarget. 2017;8:7231–7247. doi: 10.18632/oncotarget.13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, Long L, Zhou T, Tian J, Zhou B. Demethylation of MicroRNA-124a genes attenuated proliferation of rheumatoid arthritis derived fibroblast-like synoviocytes and synthesis of tumor necrosis factor-α. PLoS One. 2016;11:e0164207. doi: 10.1371/journal.pone.0164207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben Gacem R, Ben Abdelkrim O, Ziadi S, Ben Dhiab M, Trimeche M. Methylation of miR-124a-1, miR-124a-2, and miR-124a-3 genes correlates with aggressive and advanced breast cancer disease. Tumour Biol. 2014;35:4047–4056. doi: 10.1007/s13277-013-1530-4. [DOI] [PubMed] [Google Scholar]
- 27.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tivnan A, Zhao J, Johns TG, Day BW, Stringer BW, Boyd AW, Tiwari S, Giles KM, Teo C, McDonald KL. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumour Biol. 2014;35:1459–1465. doi: 10.1007/s13277-013-1200-6. [DOI] [PubMed] [Google Scholar]
- 29.Lu SH, Jiang XJ, Xiao GL, Liu DY, Yuan XR. miR-124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and β-catenin. Oncol Rep. 2014;32:2104–2110. doi: 10.3892/or.2014.3455. [DOI] [PubMed] [Google Scholar]
- 30.Deng G, Kakar S, Kim YS. MicroRNA-124a and microRNA-34b/c are frequently methylated in all histological types of colorectal cancer and polyps, and in the adjacent normal mucosa. Oncol Lett. 2011;2:175–180. doi: 10.3892/ol.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda Y, Ando T, Nanjo S, Ushijima T, Sugiyama T. DNA methylation of microRNA-124a is a potential risk marker of colitis-associated cancer in patients with ulcerative colitis. Dig Dis Sci. 2014;59:2444–2451. doi: 10.1007/s10620-014-3193-4. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, He D, Dong XD, Dong F, Wang J, Wang L, Tang J, Hu DN, Yan D, Tu L. MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci. 2013;54:2248–2256. doi: 10.1167/iovs.12-10977. [DOI] [PubMed] [Google Scholar]
- 33.He T, Feng G, Chen H, Wang L, Wang Y. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation. 2009;4:112–118. doi: 10.6026/97320630004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YH, Lee WK, Lee EB, Son JW, Kim DS, Park JY. Combined effect of metastasis-related MicroRNA, miR-34 and miR-124 family, methylation on prognosis of non-small-cell lung cancer. Clin Lung Cancer. 2017;18:e13–e20. doi: 10.1016/j.cllc.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao R, Cui L. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46:798–808. doi: 10.3892/ijo.2014.2786. [DOI] [PubMed] [Google Scholar]
- 36.Paz-Ares L, de Marinis F, Dediu M, Thomas M, Pujol JL, Bidoli P, Molinier O, Sahoo TP, Laack E, Reck M, et al. Maintenance therapy with pemetrexed plus best supportive care versus placebo plus best supportive care after induction therapy with pemetrexed plus cisplatin for advanced non-squamous non-small-cell lung cancer (PARAMOUNT): A double-blind, phase 3, randomised controlled trial. Lancet Oncol. 2012;13:247–255. doi: 10.1016/S1470-2045(12)70063-3. [DOI] [PubMed] [Google Scholar]
- 37.Lee SH, Jung SH, Kim TM, Rhee JK, Park HC, Kim MS, Kim SS, An CH, Lee SH, Chung YJ. Whole-exome sequencing identified mutational profiles of high-grade colon adenomas. Oncotarget. 2017;8:6579–6588. doi: 10.18632/oncotarget.14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Carbonell L, Sinicrope FA, Alberts SR, Oberg AL, Balaguer F, Castells A, Boland CR, Goel A. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer. 2015;113:83–90. doi: 10.1038/bjc.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Lou JF, Cao Y, Shi XH, Wang P, Xu J, Xie EF, Xu T, Sun RH, Rao JY, et al. miR-638 is a new biomarker for outcome prediction of non-small cell lung cancer patients receiving chemotherapy. Exp Mol Med. 2015;47:e162. doi: 10.1038/emm.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M, Doki Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20:S607–S615. doi: 10.1245/s10434-013-3093-4. (Suppl 3) [DOI] [PubMed] [Google Scholar]
- 42.Miyamae M, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H, Shiozaki A, et al. Plasma microRNA profiles: Identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113:1467–1476. doi: 10.1038/bjc.2015.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frères P, Josse C, Bovy N, Boukerroucha M, Struman I, Bours V, Jerusalem G. Neoadjuvant chemotherapy in breast cancer patients induces miR-34a and miR-122 expression. J Cell Physiol. 2015;230:473–481. doi: 10.1002/jcp.24730. [DOI] [PubMed] [Google Scholar]