Abstract
The differences in molecular mechanisms between osteochondroma and bizarre parosteal osteochondromatous proliferation (BPOP) remain to be fully elucidated. In the present study, the differentially expressed genes between BPOP and osteochondroma were obtained from the Gene Expression Omnibus online database, and the associations among these genes were analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online bioinformatics software. The results revealed several differentially expressed genes between human BPOP and osteochondroma. These differentially expressed genes were also enriched in different subgroups based on the analysis using DAVID online software, including ‘transforming growth factor β receptor signaling pathway’, ‘BMP signaling pathway’, ‘Wnt receptor signaling pathway’, ‘response to chemical stimulus’, ‘regulation of inflammatory response’, ‘response to stress’, ‘glycosaminoglycan binding’, ‘polysaccharide binding’, ‘extracellular matrix structural constituent’ and ‘growth factors binding’. Taken together, these findings led to the conclusion that different gene regulatory mechanisms exist between BPOP and osteochondroma. Environmental stimulation and inflammation may contribute to BPOP or osteochondroma, and differences in extracellular matrix may contribute to differences in biological characteristics between BPOP and osteochondroma.
Keywords: molecular regulation, osteochondroma, bizarre parosteal osteochondromatous proliferation
Introduction
Osteochondroma is one of the most common types of benign tumor in skeletal bones, including long bones, and jaws at the condylar or coronoid process. Pedunculated osteochondromas contain a stalk, and are long and slender, whereas sessile osteochondromas are flat (1,2). In addition, several osteochondromas exhibit a cauliflower-like shape. Histologically, there is a fibrous perichondrium, which covers the cartilage cap and exhibits continuity with the periosteum of the underlying bone marrow. Bizarre parosteal osteochondromatous proliferation (BPOP) is a rare, benign osteocartilaginous lesion, which can occur in the hands, feet, zygoma, maxilla and mandible. The histological features of BPOP include osteocartilaginous interfaces, a scattering of bizarre enlarged chondrocytes and hypercellular spindle cells (1,3). Previous studies have shown that BPOP arises from periosteal tissues through cartilaginous metaplasia, and can be confused with other benign and malignant lesions of the bone, including osteochondroma (1,3). Magnetic resonance imaging (MRI) and computed tomography (CT) examinations show that the central part of the exophytic bone lesion exhibits continuity with the underlying bone marrow, which is regarded as a typical feature of osteochondroma, compared with BPOP. In addition, MRI shows that the cartilage cap is visible in osteochondroma, and the signal characteristics of the bony regions of the lesion are typical of normal bone (4).
Previous studies have shown that inversion of chromosome 7, and translocation between chromosomes 1 and 2, were observed in BPOP (5,6). The inversion of chromosome 7 has also been observed in osteochondroma (7). In addition, exostosin glycosyltransferase (EXT)1 and EXT2 mutations are contributing factors in many patients with multiple osteochondromatosis (8,9). Although the differential diagnosis between BPOP and osteochondroma can be achieved based on clinical and radiological examinations, the different biological characteristics, including the high rate of recurrence in BPOP (6,10), and the different molecular mechanism between osteochondroma and BPOP remain to be fully elucidated. In the present study, differentially expressed genes between BPOP and osteochondroma were obtained from the Gene Expression Omnibus (GEO) online database, following which the associations among these genes were analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online bioinformatics software to examine the different molecular mechanisms between BPOP and osteochondroma.
Materials and methods
Obtaining differentially expressed genes between BPOP and osteochondroma from the GEO database
In the present study, the expression microarray data of differentially expressed genes between BPOP and osteochondroma were obtained from the GEO online database (11). Firstly, on accessing the GEO online database (http://www.ncbi.nlm.nih.gov/gds/), ‘osteochondroma’ was entered into the search box to identify the associated dataset (no. GSE19357). Detailed information regarding this dataset were provided on the website. Briefly, total RNA was extracted from four frozen bone tumor biopsies, comprising two cases of osteochondroma and two cases of BPOP. cDNA was generated, fragmented and end-labelled. The fragmented and biotinylated cDNA was hybridized to the GeneChipR Human Gene 1.0 ST Arrays (Affymetrix, Inc., Santa Clara, CA, USA). In the present study, the GEO data analysis tool, GEO2R, was used to identify the differentially expressed genes between BPOP and osteochondroma. A total of 400 differentially expressed genes (P<0.001) were obtained and used for further analysis. The expression values for certain differentially expressed genes (ACAN, Col2a1, ZFP36, FABP4, PPARG, TGFβ3, FMOD, DKK3, KREMEN1 and ROR2) were obtained and recorded from the GEO data profiles.
Systematic evaluation of differentially expressed genes between BPOP and osteochondroma using DAVID bioinformatics online software
DAVID bioinformatics online software integrates several useful and well-regarded heterogeneous annotation resources, including the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes database, and can perform large-scale gene data analysis to systematically group enriched genes into various functional subgroups based on their biological and functional characteristics (12,13). In the present study, the associations among the differentially expressed genes between BPOP and osteochondroma were systematically analyzed using DAVID bioinformatics online software. Briefly, on accessing the DAVID website (https://david.ncifcrf.gov/), analysis was initiated, followed by copying the names of the 400 differentially expressed genes into ‘box A’ and selecting the official gene symbol as the gene identifier type. Homo sapiens was selected as the annotation, and the list was submitted using the ‘Submit List’ option to perform systematic analysis of the differentially expressed genes between BPOP and osteochondroma based on the analytic modules of DAVID.
Results
Differentially expressed genes between BPOP and osteochondroma
In the present study, several differentially expressed genes were obtained between BPOP and osteochondroma from the GEO online database based on the analysis using GEO2R, and 400 differentially expressed genes (P<0.001) were selected for further analysis. Among these genes, it was found that two chondrocyte markers, collagen type II, α I (Col2a1) and aggrecan (ACAN), showed higher expression levels in BPOP (Fig. 1). By contrast, two inflammatory signaling molecules, fatty acid binding protein 4 and peroxisome proliferator-activated receptor γ, showed lower expression levels in BPOP, whereas ZFP36 ring finger protein, an inflammatory signaling molecule, exhibited a higher expression level in BPOP (Fig. 2). The transforming growth factor (TGF) β3 and fibromodulin genes in the TGFβ signaling pathway, and the dickkopf WNT signaling pathway inhibitor 3, kringle containing transmembrane protein and RTK-like orphan receptor 2 genes in Wnt signaling pathway showed higher expression levels in BPOP (Figs. 3 and 4).
Figure 1.
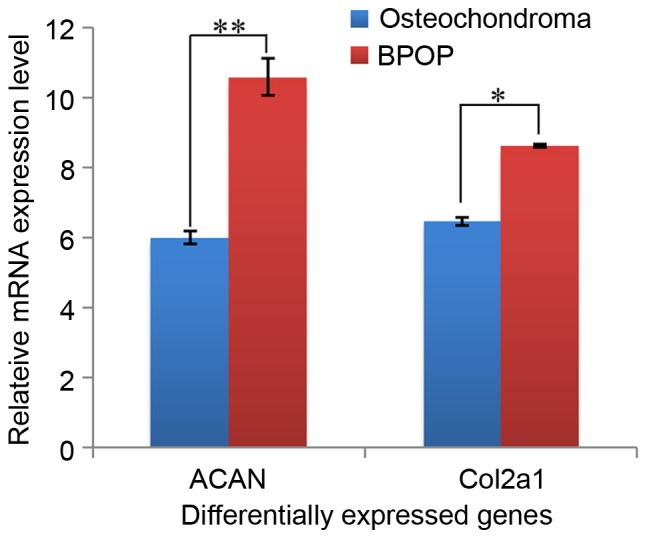
mRNA expression of ACAN and Col2a1 in human BPOP and osteochondroma. *P<0.05, **P<0.01. BPOP, bizarre parosteal osteochondromatous proliferation; Col2a1, collagen type II, α I; ACAN, aggrecan.
Figure 2.
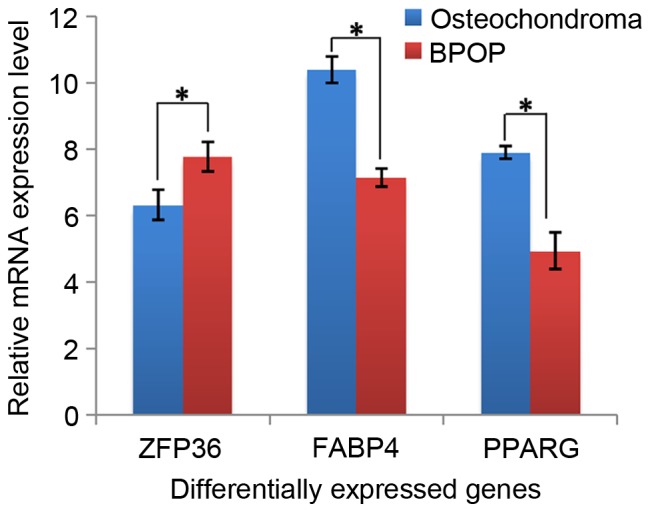
mRNA expression of FABP4, PPARG and ZFP36 in human BPOP and osteochondroma. *P<0.05. BPOP, bizarre parosteal osteochondromatous proliferation; FABP4, fatty acid binding protein 4; PPARG, peroxisome proliferator-activated receptor γ; ZFP36, ZFP36 ring finger protein.
Figure 3.
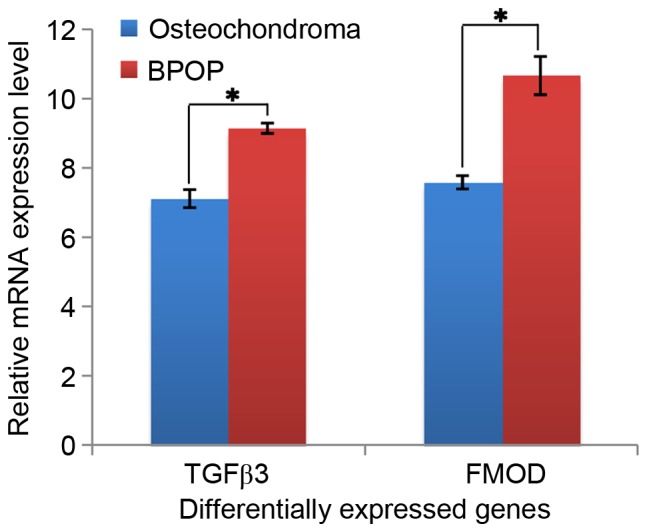
mRNA expression of TGFβ3 and FMOD in human BPOP and osteochondroma. *P<0.05. BPOP, bizarre parosteal osteochondromatous proliferation; TGFβ3, transforming growth factor β3; FMOD, fibromodulin.
Figure 4.
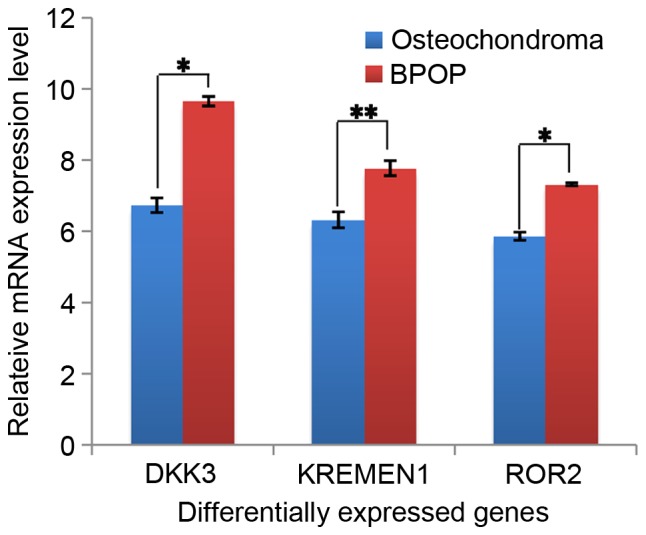
mRNA expression of DKK3, KREMEN1 and ROR2 in human BPOP and osteochondroma. *P<0.05, **P<0.01. BPOP, bizarre parosteal osteochondromatous proliferation; DKK3, dickkopf WNT signaling pathway inhibitor 3; KREMEN1, kringle containing transmembrane protein 1; ROR2, RTK-like orphan receptor 2.
Differentially expressed genes between BPOP and osteochondroma are enriched in various biological process and signaling pathway subgroups
The 400 differentially expressed genes were enriched in various subgroups based on GO terms or pathways, determined through analysis using the DAVID bioinformatics online software. For the GO term ‘biological process’, the 400 differentially expressed genes were enriched into 256 subgroups. The three signaling pathway subgroups, ‘transforming growth factor β receptor signaling pathway’, ‘BMP signaling pathway’ and ‘Wnt receptor signaling pathway’ were identified (Table I). This suggested that these signaling pathways may exhibit different roles between BPOP and osteochondroma. In addition, several subgroups were associated with environmental stimulation, including ‘response to chemical stimulus’, ‘response to wounding’, ‘response to hormone stimulus’, ‘regulation of inflammatory response’ and ‘response to stress’ (Table I). This suggested that environmental stimulation and inflammation may contribute to BPOP and osteochondroma.
Table I.
Differentially expressed genes between human bizarre parosteal osteochondromatous proliferation and osteochondroma, enriched into different biological process subgroups.
| Term | P-value | Genes |
|---|---|---|
| Transforming growth factor β receptor signaling pathway | 1.31E-04 | FMOD, FOS, JUN, COL3A1, TGFB3, TGFBR3, TGFB2, ACVR1 |
| Collagen fibril organization | 1.91E-04 | COL3A1, ACAN, COL12A1, COL2A1, ADAMTS3, TGFB2 |
| Lipid homeostasis | 3.82E-04 | LPL, INS-IGF2, LCAT, PPARG, LIPG, FABP4, SCARB1 |
| Positive regulation of ossification | 9.87E-04 | ACVR2A, ADRB2, TGFB3, TGFB2, ACVR1 |
| Response to chemical stimulus | 0.00355 | ADCY4, TF, INS-IGF2, TACR1, S100A9, SNCA, COL3A1, PPARG, TGFB3, AQP7, TGFB2, FOS, PRKAR2B, DEFA1B, PLOD2, LCAT, IPCEF1, SEMA3C, SLC30A5, SCARB1, TIE1, SEMA3A, NDUFS2, EGR1, APOLD1, LIFR, MMP14, ADIPOQ, ITPR1, S100A12, PCK1, RPS6KA5, BTG2, SQLE, JUN, LIPG, FABP4, TGFBR3, DEFA1, KLF4 |
| Response to wounding | 0.005836 | ITGAL, TF, S100A8, INS-IGF2, TACR1, S100A9, COL3A1, TGFB3, AFAP1L2, MECOM, LMAN1, S100A12, TGFB2, CD9, VWF, FOS, SCARB1, CFD, ACVR1, AOC3 |
| BMP signaling pathway | 0.009385 | ACVR2A, CHRDL1, TGFBR3, UBE2D1, ACVR1 |
| Response to hormone stimulus | 0.01025 | ADCY4, INS-IGF2, TACR1, PPARG, TGFB3, MMP14, ADIPOQ, TGFB2, PCK1, PRKAR2B, FOS, BTG2, LCAT, TGFBR3, FABP4 |
| Response to stress | 0.010368 | ITGAL, TF, S100A8, INS-IGF2, TACR1, SNCA, COL3A1, S100A9, PPARG, TGFB3, AFAP1L2, INTS3, LMAN1, TGFB2, CD9, FOS, LGALS3BP, DEFA1B, PLOD2, IPCEF1, LTF, SCARB1, CFD, NDUFS2, SHPRH, APOLD1, CIDEA, MMP14, MECOM, ADIPOQ, EEPD1, ITPR1, S100A12, RPS6KA5, VWF, RECQL, EYA1, BPI, ADRB2, BTG2, JUN, DEFA3, ROR2, DEFA1, CTSG, ACVR1, AOC3 |
| Wnt receptor signaling pathway | 0.01325 | DKK3, CCDC88C, KREMEN1, ROR2, FZD3, FRZB, FZD4, TCF7L1 |
| Regulation of inflammatory response | 0.014485 | ZFP36, ADRB2, INS-IGF2, PPARG, FABP4, ADIPOQ |
For the GO term ‘molecular function’, the 400 differentially expressed genes were enriched into 57 subgroups. It was found that several subgroups were associated with extracellular matrix, including ‘glycosaminoglycan binding’, ‘polysaccharide binding’ and ‘extracellular matrix structural constituent’ (Table II). This suggested that the extracellular matrix may be different between BPOP and osteochondroma. In addition, growth factors exhibited different roles between BPOP and osteochondroma.
Table II.
Differentially expressed genes between human bizarre parosteal osteochondromatous proliferation and osteochondroma, enriched into different molecular function subgroups.
| Term | P-value | Genes |
|---|---|---|
| Growth factor binding | 3.50E-05 | ACVR2A, LTBP1, KLB, INS-IGF2, COL3A1, LIFR, TGFB3, TGFBR3, COL2A1, FGFBP2, ACVR1 |
| Glycosaminoglycan binding | 8.50E-05 | LPL, HAPLN1, COMP, LIPG, ACAN, TGFBR3, DCN, ADAMTS3, EPYC, THBS2, PRELP, THBS4 |
| Polysaccharide binding | 1.99E-04 | LPL, HAPLN1, COMP, LIPG, ACAN, TGFBR3, DCN, ADAMTS3, EPYC, THBS2, PRELP, THBS4 |
| Calcium ion binding | 8.86E-04 | ITGAL, REPS2, LTBP1, S100A8, S100A9, SNCA, ARSJ, MMP7, SPOCK1, EDIL3, CALU, TMEM37, PLCB4, SLC25A25, NPTX2, COMP, AIF1L, ANO2, THBS2, THBS3, THBS4, MATN2, SVEP1, PADI4, MMP14, ITPR1, S100A12, ATP2A2, S100B, SULF1, NOTCH4, RCN3, AOC3 |
| Heparin binding | 0.00378 | LPL, COMP, LIPG, TGFBR3, ADAMTS3, THBS2, PRELP, THBS4 |
| Transforming growth factor β receptor activity | 0.004672 | ACVR2A, LTBP1, TGFBR3, ACVR1 |
| Extracellular matrix structural constituent | 0.006326 | LAMA1, COMP, COL3A1, ACAN, COL12A1, COL2A1, PRELP |
| SMAD binding | 0.011707 | FOS, JUN, COL3A1, TGFBR3, ACVR1 |
| Transforming growth factor β receptor binding | 0.041652 | TGFB3, TGFBR3, TGFB2 |
| Lipid binding | 0.054158 | LPL, RBP7, EPB41, NCF4, SNCA, PPARG, APOLD1, ITPR1, ZCCHC14, BPI, SDPR, FABP4, SCARB1, ARAP2, APOL4 |
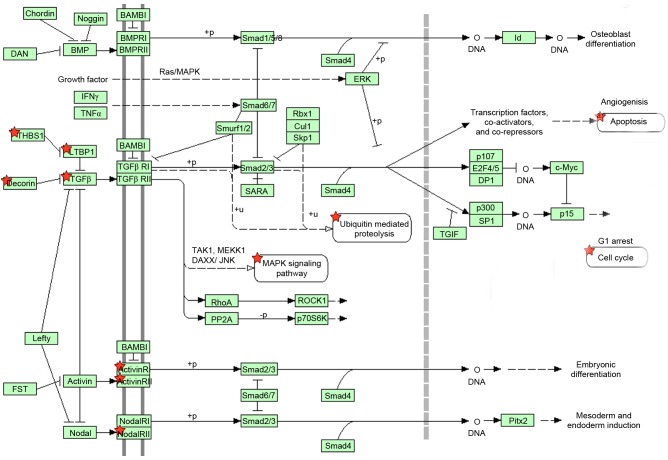
In addition to GO term enrichment, DAVID online software was also used to enrich large gene lists into signaling pathway subgroups. In the present study, it was found that the 400 differentially expressed genes were enriched into different signaling pathway subgroups. There were 10 genes, including activin A receptor, type I (ACVR1) activin A receptor, type 2A (ACVR2A), cartilage oligomeric matrix protein (COMP), decorin (DCN), latent TGFβ binding protein 1 (LTBP1), thrombospondin (THBS)2, THBS3, THBS4, TGFβ2 and TGFβ3, enriched in the TGFβ signaling pathway. The position of these 10 genes in the TGFβ signaling pathway are shown in Fig. 5.
Figure 5.
Ten differentially expressed genes between human bizarre parosteal osteochondromatous proliferation and osteochondroma enriched in the TGFβ signaling pathway. Red stars indicate these associated genes. TGFβ, transforming growth factor β.
Discussion
BPOP is a rare disease, which usually presents as a parosteal mass in the short bones of the hands, feet and jaws. BPOP usually presents as a mushroom-shaped mass, and is often confused with osteochondroma (1). Histopathologically, cartilage is present at the margins of the lesion in BPOP, and the marginal cartilage is fibrocartilage, compared with the hyaline cartilage usually present in the cartilage cap of osteochondromas (1,14). Therefore, it is possible to distinguish a BPOP from an osteochondroma based on histopathological findings. In addition, MRI and CT examinations show that the central region of the exophytic bone lesion in osteochondroma exhibits continuity with the underlying bone marrow, which is not the case in BPOP (15). This can also assist in distinguishing a BPOP from an osteochondroma.
BPOP has been reported to have a relatively high rate of recurrence following surgical resection (10), which is different to that of osteochondroma, and suggests a different molecular mechanism between osteochondroma and BPOP. In the present study, it was found that BPOP exhibited more cartilage characteristics, compared with BPOP, and the TGFβ receptor signaling pathway, BMP signaling pathway and Wnt signaling pathway may have different roles between osteochondroma and BPOP. In addition, the results showed that several genes were associated with environmental stimulation and inflammation, for example ‘response to chemical stimulus’, ‘response to wounding’, ‘regulation of inflammatory response’ and ‘response to stress’, which may contribute to BPOP or osteochondroma. The findings also showed that the extracellular matrix potentially differs between BPOP and osteochondroma, which may contribute to the different biological characteristics between BPOP and osteochondroma.
In conclusion, several differentially expressed genes between human BPOP and osteochondroma were obtained from the GEO online database. These differentially expressed genes was enriched into different subgroups based on analysis using DAVID online software, which included the ‘transforming growth factor β receptor signaling pathway’, ‘BMP signaling pathway’, ‘Wnt receptor signaling pathway’, ‘response to chemical stimulus’, ‘regulation of inflammatory response’, ‘response to stress’, ‘glycosaminoglycan binding’, ‘polysaccharide binding’, ‘extracellular matrix structural constituent’ and ‘growth factors binding’. Taken together, the results of the present study suggested that there are different gene regulatory mechanisms between BPOP and osteochondroma. Environmental stimulation and inflammation may contribute to BPOP or osteochondroma, and differences in extracellular matrix may contribute to the different biological characteristics between BPOP and osteochondroma.
Acknowledgements
The present study work was supported by Nanchong applied technology research and development program (grant no. 15A0019).
References
- 1.Kim SM, Myoung H, Lee SS, Kim YS, Lee SK. Bizarre parosteal osteochondromatous proliferation in the lingual area of the mandibular body versus osteochondroma at the mandibular condyle. World J Surg Oncol. 2016;14:35. doi: 10.1186/s12957-016-0777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roychoudhury A, Bhatt K, Yadav R, Bhutia O, Roychoudhury S. Review of osteochondroma of mandibular condyle and report of a case series. J Oral Maxillofac Surg. 2011;69:2815–2823. doi: 10.1016/j.joms.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Khan SA, Kumar Sampath V, Sharma MC. Bizarre parosteal osteochondromatous proliferation (Nora's lesion) of phalanx in a child. BMJ Case Rep. 2014;2014 doi: 10.1136/bcr-2013-201714. pii: bcr2013201714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitsoulis P, Galani V, Stefanaki K, Paraskevas G, Karatzias G, Agnantis NJ, Bai M. Osteochondromas: Review of the clinical, radiological and pathological features. In Vivo. 2008;22:633–646. [PubMed] [Google Scholar]
- 5.Sakamoto A, Imamura S, Matsumoto Y, Harimaya K, Matsuda S, Takahashi Y, Oda Y, Iwamoto Y. Bizarre parosteal osteochondromatous proliferation with an inversion of chromosome 7. Skeletal Radiol. 2011;40:1487–1490. doi: 10.1007/s00256-011-1173-8. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson M, Domanski HA, Mertens F, Mandahl N. Molecular cytogenetic characterization of recurrent translocation breakpoints in bizarre parosteal osteochondromatous proliferation (Nora's lesion) Hum Pathol. 2004;35:1063–1069. doi: 10.1016/j.humpath.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Panagopoulos I, Bjerkehagen B, Gorunova L, Taksdal I, Heim S. Rearrangement of chromosome bands 12q14~15 causing HMGA2-SOX5 gene fusion and HMGA2 expression in extraskeletal osteochondroma. Oncol Rep. 2015;34:577–584. doi: 10.3892/or.2015.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanteles GA, Nicolaou M, Neocleous V, Shammas C, Loizidou MA, Alexandrou A, Ellina E, Patsia N, Sismani C, Phylactou LA, Christophidou-Anastasiadou V. Genetic screening of EXT1 and EXT2 in Cypriot families with hereditary multiple osteochondromas. J Genet. 2015;94:749–754. doi: 10.1007/s12041-015-0564-3. [DOI] [PubMed] [Google Scholar]
- 9.Sarrión P, Sangorrin A, Urreizti R, Delgado A, Artuch R, Martorell L, Armstrong J, Anton J, Torner F, Vilaseca MA, et al. Mutations in the EXT1 and EXT2 genes in Spanish patients with multiple osteochondromas. Sci Rep. 2013;3:1346. doi: 10.1038/srep01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting BL, Jupiter JB. Recurrent bizarre parosteal osteochondromatous proliferation of the ulna with erosion of the adjacent radius: Case report. J Hand Surg Am. 2013;38:2381–2386. doi: 10.1016/j.jhsa.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abramovici L, Steiner GC. Bizarre parosteal osteochondromatous proliferation (Nora's lesion): A retrospective study of 12 cases, 2 arising in long bones. Hum Pathol. 2002;33:1205–1210. doi: 10.1053/hupa.2002.130103. [DOI] [PubMed] [Google Scholar]
- 15.Torreggiani WC, Munk PL, Al-Ismail K, O'Connell JX, Nicolaou S, Lee MJ, Masri BA. MR imaging features of bizarre parosteal osteochondromatous proliferation of bone (Nora's lesion) Eur J Radiol. 2001;40:224–231. doi: 10.1016/S0720-048X(01)00362-X. [DOI] [PubMed] [Google Scholar]