Abstract
Abnormal paired box 9 (PAX9) expression is associated with tumorigenesis, cancer development, invasion and metastasis. The present study investigated the prognostic significance of PAX9 in esophageal squamous cell carcinoma (ESCC) and its role in predicting radiation sensitivity. A total of 52.8% (121/229) ESCC tissues were positive for PAX9. The 1-, 3- and 5-year disease-free survival (DFS) rates were 72.2, 35.2 and 5.6%, respectively, and the overall survival (OS) rates were and 86.1, 44.4, and 23.1%, respectively, in PAX9-positive tumors. In PAX9-negative tumors, the one-, three- and five-year DFS rates were 76.9, 47.9 and 24.0%, and the OS rates were 90.9, 57.9 and 38.8%, respectively. Univariate analysis revealed that PAX9, differentiation, T stage, lymph node metastasis, and tumor-node-metastasis stage were associated with OS. Multivariate analysis of DFS and OS revealed that the hazard ratios for PAX9 were 0.624 (95% CI: 0.472–0.869, P=0.004) and 0.673 (95% CI: 0.491–0.922, P=0.014), respectively. Patients that received adjuvant therapy exhibited significant differences in the 5-year DFS (P<0.001) and OS (P<0.001). PAX9-positive ESCC patients who received post-surgery radiotherapy had a significantly greater 5-year DFS (P=0.011) and OS (P=0.009) than patients who received surgery only. Thus, PAX9 may be an independent prognostic factor for the surgical treatment of ESCC and a possible predictor of radiation sensitivity.
Keywords: esophageal squamous cell carcinoma, paired box 9, immunohistochemistry, prognosis, radiosensitivity
Introduction
Esophageal cancer is one of the most common types of cancer worldwide and has a high incidence of mortality. It is estimated that 455,800 new esophageal cancer cases and 400,200 deaths occurred in 2012 (1). The incidence of esophageal cancer varies depending on location; the highest rates occur in Eastern Asia and Eastern and Southern Africa. Esophageal squamous cell carcinoma (ESCC) accounts for >90% of esophageal cancer cases in China compared with approximately 26% in the United States. Despite progress in clinical diagnosis and treatment modalities, ESCC remains associated with a poor prognosis, and has a 5-year survival rate of <15% (2–6). Traditional methods of characterizing tumors depend on visual information, including the size of the tumor, degree of infiltration and histological features of the tumor, which form the basis of the tumor-node-metastasis (TNM) staging system. Although these parameters provide a way to distinguish between tumor subtypes with distinct biological characteristics, it does not provide sufficient information for the establishment of heterogeneous groupings of tumors and patients for clinical treatment. Therefore, the identification of effective biomarkers that associate with the biological characteristics of ESCC patients is important to predict their prognosis and to improve therapeutic strategies.
Prognostic biomarkers should be chosen based on distinguishing features between benign and malignant tumors or the differentiation/pathological staging status of tumors, which may influence the mode of treatment. Studies have focused on identifying novel ESCC biomarkers and therapeutic targets for adjuvant drug treatments for the enhancement of current surgery, radiotherapy and chemotherapy modalities (7–11). Although research into the pathogenesis and targeted therapy of lung, breast, and colorectal cancer has progressed, there remains a paucity of data on esophageal cancer.
Paired box (PAX) 9 is a member of the paired box gene family, which is composed of nine transcription factors in humans (PAX1-9). This family of genes serves key roles in embryonic development and organogenesis by regulating the expression of genes involved in cell proliferation, apoptotic resistance and cell migration (12,13). PAX genes are usually described as cell lineage-specific regulators of tissues where their expression profiles are finely tuned both spatially and temporally, and are recognized as potentially important factors in cancer progression (14–17). PAX2 is frequently expressed in primary human cancers, including those derived from breast and ovaries, and chromosomal translocations involving PAX3, PAX5, PAX7 and PAX8 genes are present in alveolar rhabdomyosarcoma, B-lymphoid malignancies and thyroid cancer (18). PAX3 is required for the survival of melanoma cell lines and is expressed in breast cancer (19). In addition, PAX6 is highly expressed in cancer cell lines, including those derived from brain and breast tumors (20). Expression of PAX2 and PAX8 has been observed in kidney tissue (21). PAX5 is considered to be a tumor suppressor and is involved in hepatocellular carcinoma carcinogenesis via direct regulation of the tumor protein p53 signaling pathway (22).
Human PAX9 is located within the 14q12-q13 chromosomal region and contains a 128-amino acid DNA binding paired domain, which makes sequence-specific contacts with DNA. Due to the G quadruplex-forming region located near exon 1, which is present in all the known sequenced placental mammals, PAX9 intron 1 serves a key role in splicing efficiency (23,24). As a transcription factor expressed in the tooth mesenchyme during tooth morphogenesis, heterozygous mutations in PAX9 have been associated with non-syndromic tooth agenesis, predominantly in the molars (25–28). In the last decade, the abnormal expression of the PAX9 gene, which is associated with the tumorigenesis, development, invasion and metastasis of many types of cancer, was observed in a variety of malignant human tumors (29,30). However, the expression level and prognostic value of PAX9 in malignant tumor cells remain inconclusive. An experiment comparing genomic abnormalities in a large cohort of patients with esophageal adenocarcinoma (EAC) and ESCC using single nucleotide polymorphism arrays demonstrated that ESCC exhibited greater amplification frequencies in PAX9 (35%) than EAC (4%) (31). Therefore, PAX9 may be an oncogene specifically involved in ESCC.
Prior to the present study, the prognostic value of PAX9 had not been investigated in ESCC. In the present study, the expression of the PAX9 protein in patients with ESCC treated by curative resection was investigated using immunohistochemical staining. In addition, the overall survival (OS) of the ESCC patients was analyzed. The aims of the present study were to investigate the prognostic significance of PAX9 in resectable ESCC and to assess its value in predicting radiation sensitivity by subgroup analysis.
Materials and methods
Patients
A total of 229 ESCC patients who received radical surgery at Qilu Hospital at Shandong University between 1st January 2008 and 31st December 2009 were included in the present study. Patients with distant metastases received neoadjuvant treatment or palliative surgery, and those who were not available for follow-up or lacked sufficient tumor tissue for analysis were excluded. No patients received chemotherapy or radiotherapy prior to surgery. A total of 119 patients received adjuvant treatment with 5,000 cGy/25 fractions radiotherapy and 4–6 cycles of docetaxel and cisplatin. Of these patients, 35 received synchronous radiochemotherapy.
The histological features of the specimens were independently evaluated by two pathologists according to the classification criteria of the World Health Organization. Cohen's Kappa coefficient revealed a strong association between the two pathologists (k=0.86). Patient demographic and clinical data, including age, gender, smoking and alcohol history, tumor location, histological grade, tumor length, T stage, lymph node status, TNM stage and adjuvant treatment, were available. The pathological stages of esophageal cancer were determined according to the pathological (p) TNM staging system (defined by the American Joint Committee on Cancer Staging Manual, 7th edition, 2010) (32). The histological grade was classified according to the degree of differentiation of the tumor by histological examination using hematoxylin and eosin staining. Follow-up visits were performed every 3 months for the first 2 years and every 6 months until death or until the end of the study. Data were censored at the last follow-up visit (December 2014) for patients without recurrence or death. At each visit, a clinical history was collected and a physical examination was performed. Routine diagnostic imaging methods, including upper gastrointestinal barium meal fluoroscopy and computer tomography, were performed. Disease-free survival (DFS) and OS were defined as the interval between the surgery date to the date of recurrence or death, respectively.
Procedures were performed in accordance with The Declaration of Helsinki. Ethical approval was obtained from the Institutional Ethics Committee of Qilu Hospital at Shandong University, which is accredited by the National Council on Ethics in Human Research. The participants provided written informed consent.
Immunohistochemistry
Surgical tissue samples were fixed in 10% neutral buffered formalin at room temperature overnight and then embedded in paraffin. For antigen retrieval, 4 µm thick paraffin sections were deparaffinized in a series of alcohols and microwaved in citrate buffer (pH 6.0). Endogenous peroxidases were blocked with 3% hydrogen peroxide for 20 min at room temperature. Subsequently, tissues were blocked with goat serum (ZLI-9021; ZSGB-Bio, Beijing, China) for 30 min at room temperature. Sections were incubated at −4°C overnight with a rat monoclonal anti-PAX9 antibody (SAB4200083; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) diluted at 1:200 according to the manufacturer's protocol, and probed with HRP-labeled rabbit anti rat secondary antibody at 37°C for one hour (Histostain®-Plus kit; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China). The PAX9 protein was visualized using a substrate solution containing diaminobenzidine and hydrogen peroxide (ZLI-9034; 1:1,000; ZSGB-Bio) at room temperature. The presence of viable tumor was confirmed by hematoxylin and eosin staining. Tissue sections stained in a similar manner with PBS instead of the primary antibody were utilized as negative controls. A total of forty healthy esophageal mucous membranes from esophageal tissue and esophageal cancer tissue microarrays (BN02014; Alenabio, Xi'an, China) were utilized as positive controls.
Immunostaining evaluation
Tissue sections were analyzed with an Olympus IX71S1F-3 Inverted Microscope (Olympus Corporation, Tokyo, Japan) by two independent operators using the blinding method (33). At least five fields of view were observed in each tissue section, and at least 100 cells were counted in each field at a high magnification (×200). Cells that exhibited brown granules in the cytoplasm were considered as positive. Using semi-quantitative analysis, the criteria for scoring the stained sections were: 1, 0–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. According to the intensity of PAX9 staining, tissues were scored as follows: 0, negative; 1, weakly positive; 2, strongly positive. Final scores were calculated by multiplying the percentage of cells stained by the staining intensity of the tissues, and were categorized as PAX9-negative (final score 0–3) or PAX9-positive (final score 4–8) (34). In addition, tumor-adjacent tissues were obtained from all patients, and PAX9 expression in these tissues was determined.
Statistical analysis
Statistical analyses were performed using SPSS software version 19.0 (IBM SPSS, Armonk, NY, USA). Statistical differences between tissue groups were evaluated using the Chi-square test. Kaplan-Meier curves were utilized to analyze the distribution of 5-year DFS and OS, and the log-rank test was performed to compare differences between the survival curves. The same methods were adopted to perform the stratified analysis in which subgroups were divided according to different adjuvant treatment. Variables were subjected to univariate analysis and multivariate survival analysis using the Cox proportional hazard regression model. The statistical tests were two-sided, and P-<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
A total of 229 patients were included in this study. The median age was 60 years (range, 32–84 years), and 80.8% of the patients were male. Tumor locations included the following: cervical, 7 cases; upper thoracic, 16 cases; middle thoracic, 130 cases; and lower thoracic, 76 cases. The tumors ranged from 0.5 cm to 10.0 cm in length, with an average of 4.17 cm. The histopathological stage was well-differentiated in 55 cases, moderately-differentiated in 99 cases, and poorly-differentiated in 75 cases. A total of 88 patients (38.4%) presented with T1/T2 tumors and 141 patients (61.6%) presented with T3/T4 tumors, and 109 patients (47.6%) presented with positive lymph nodes. Twenty-six patients were in pathological stage I, 101 patients were in stage II and 102 patients were in stage III. A total of 110 patients (48.0%) were treated with surgery alone, 53 (23.1%) patients were treated with postoperative chemotherapy, 101 (44.1%) patients were treated with postoperative radiotherapy, and 35 (15.3%) patients were treated with postoperative chemoradiation. Two hundred and thirteen patients (93.0%) experienced tumor recurrence, and 182 (79.5%) patients died during the follow-up period. The estimated 1-, 3-, and 5-year DFS and OS rates were 74.7, 41.9 and 15.3% and 88.6, 51.5 and 31.4%, respectively. The DFSs ranged from 1.0 to 81.4 months (median, 25.9 months), and the OS ranged from 1.7 to 83.2 months (median, 37.0 months).
Immunohistochemical analysis
Examples of PAX9-negative and PAX9-positive tissue staining are shown in Fig. 1. PAX9-positive expression was observed in the nucleus and cytoplasm of tumor and normal esophageal mucosa tissues. In healthy esophageal mucous membranes and tumor-adjacent tissues, the positive expression rates of PAX9 were 99.1% and 97.8%, respectively. In ESCC tissues, 52.8% stained positive for PAX9 (121/229). In ESCC tissues, no significant correlation was identified between the expression of PAX9 and clinicopathological features of the patients, including gender, age, smoking history, drinking history, tumor location, tumor length, differentiation, T stage, N stage, pTNM stage or adjuvant treatment (Table I).
Figure 1.
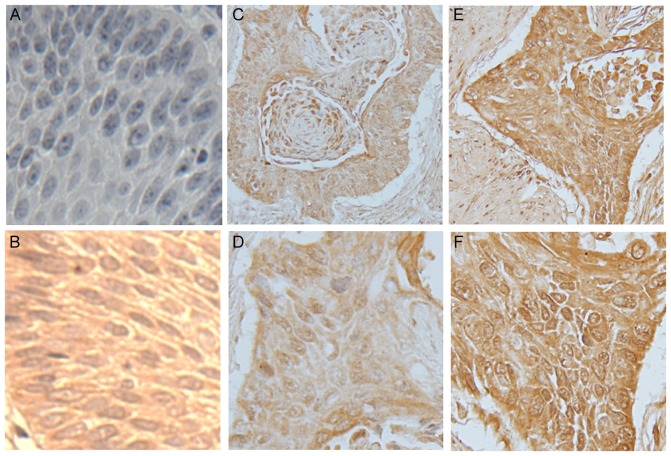
Immunostaining of PAX9 in normal esophageal mucosa and esophageal squamous cell carcinoma tissue. Representative images of (A) negative control for PAX9 in normal mucosa, (B) positive control for PAX9 in normal mucosa, (C and D) low expression of PAX9 in tumor tissue (low rate of positive cells and low staining intensity, score ≤3) and (E and F) high expression of PAX9 in tumor tissue (high rate of positive cells and high staining intensity, score ≥4). The stained sections were observed and images were captured at low magnification (×100) in C and E, and at high magnification (×200) in A, B, D and F. PAX9, paired box 9.
Table I.
Association of clinicopathological variables of ESCC patients and PAX9 expression in tumor tissue samples.
| PAX9 | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | Total patients (n=229) | Negative (n=108) (%) | Positive (n=121) (%) | P-value |
| Gender | 0.801 | |||
| Male | 185 | 88 (47.6 | 97 (52.4 | |
| Female | 44 | 20 (45.5 | 24 (54.5 | |
| Age | 0.383 | |||
| ≤60 | 116 | 58 (50.0 | 58 (50.0 | |
| >60 | 113 | 50 (44.2 | 63 (55.8 | |
| Smoking history | 0.179 | |||
| Never or light | 108 | 56 (51.9 | 52 (48.1 | |
| Heavy | 121 | 52 (43.0 | 69 (57.0 | |
| Drinking history | 0.052 | |||
| Never or light | 118 | 63 (53.3 | 55 (46.7 | |
| Heavy | 111 | 45 (40.5 | 66 (59.5 | |
| Site of tumor | 0.287 | |||
| Cervical | 7 | 2 (28.6 | 5 (71.4 | |
| Upper thoracic | 16 | 9 (56.3 | 7 (43.7 | |
| Middle thoracic | 130 | 56 (43.1 | 74 (56.9 | |
| Lower thoracic | 76 | 41 (53.9 | 35 (46.1 | |
| Tumor length | 0.941 | |||
| <4 cm | 102 | 46 (45.1 | 56 (54.9 | |
| ≥4 cm | 127 | 62 (48.8 | 65 (51.2 | |
| Differentiation | 0.893 | |||
| Well | 55 | 25 (45.5 | 30 (54.5 | |
| Moderate | 99 | 46 (46.5 | 53 (53.5 | |
| Poor | 75 | 37 (49.3 | 38 (50.7 | |
| T stage | 0.150 | |||
| T1 | 23 | 6 (26.1 | 17 (73.9 | |
| T2 | 65 | 31 (47.7 | 34 (52.3 | |
| T3 | 126 | 62 (49.2 | 64 (50.8 | |
| T4 | 15 | 9 (60.0 | 6 (40.0 | |
| N stage | 0.162 | |||
| N0 | 120 | 51 (42.5 | 69 (57.5 | |
| N1 | 87 | 45 (51.7 | 42 (48.3 | |
| N2 | 19 | 9 (47.4 | 10 (52.6 | |
| N3 | 3 | 3 (100.0 | 0 (0.00 | |
| pTNM stage | 0.105 | |||
| I | 26 | 10 (38.5 | 16 (61.5 | |
| II | 101 | 42 (41.6 | 59 (58.4 | |
| III | 102 | 56 (54.9 | 46 (45.1 | |
| Adjuvant treatment | 0.060 | |||
| None | 110 | 55 (50.0 | 55 (50.0 | |
| Radiotherapy | 66 | 24 (36.4 | 42 (63.6 | |
| Chemotherapy | 18 | 7 (38.9 | 11 (61.1 | |
| CRT | 35 | 22 (62.9 | 13 (37.1 | |
T, tumor; N, node; PAX9, paired box 9; TNM, tumor node metastasis; ESCC, esophageal squamous cell carcinoma; CRT, chemoradiotherapy.
The prognostic value of PAX9 and other variables in ESCC tumors was investigated, as demonstrated in Table II. In PAX9-negative tumors, the 1-, 3-, and 5-year DFS and OS rates were 72.2, 35.2 and 5.6%, and 86.1, 44.4, and 23.1%, respectively. By contrast, in PAX9-positive tumors, the 1-, 3-, and 5-year DFS and OS rates were 76.9, 47.9, and 24.0%, and 90.9, 57.9, and 38.8%, respectively. The median OS for the patients in the PAX9-negative group was 29.4 months (range, 1.7–73.1 months) compared with 42.0 months (range, 3.5–83.2 months) for patients in the PAX9-positive group.
Table II.
Univariate analysis of survival of ESCC patients treated with curative surgery.
| DFS | OS | ||||||
|---|---|---|---|---|---|---|---|
| Characteristics | Total patients | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Gender | |||||||
| Male | 185 | 1.000 | Ref. | 1.000 | Ref. | ||
| Female | 44 | 0.695 | 0.933 | 0.695–1.321 | 0.705 | 0.930 | 0.637–1.356 |
| Age | |||||||
| ≤60 | 116 | 1.000 | Ref. | 1.000 | Ref. | ||
| >60 | 113 | 0.608 | 0.932 | 0.712–1.220 | 0.217 | 0.832 | 0.622–1.114 |
| Smoking history | |||||||
| Never or light | 108 | 1.000 | Ref. | 1.000 | Ref. | ||
| Heavy | 121 | 0.960 | 1.007 | 0.769–1.319 | 0.982 | 0.997 | 0.745–1.334 |
| Drinking history | |||||||
| Never or light | 118 | 1.000 | Ref. | 1.000 | Ref. | ||
| Heavy | 111 | 0.545 | 0.920 | 0.702–1.205 | 0.436 | 0.890 | 0.664–1.193 |
| Tumor site | |||||||
| Cervical | 7 | 1.000 | Ref. | 1.000 | Ref. | ||
| Upper thoracic | 16 | 0.879 | 1.077 | 0.413–2.813 | 0.858 | 1.098 | 0.394–3.060 |
| Middle thoracic | 130 | 0.996 | 0.998 | 0.437–2.278 | 0.760 | 0.869 | 0.353–2.139 |
| Lower thoracic | 76 | 0.688 | 1.187 | 0.514–2.744 | 0.956 | 0.974 | 0.392–2.425 |
| Tumor length | |||||||
| <4 cm | 102 | 1.000 | Ref. | 1.000 | Ref. | ||
| ≥4 cm | 127 | 0.428 | 1.117 | 0.850–1.467 | 0.616 | 1.078 | 0.803–1.447 |
| Differentiation | |||||||
| Well | 55 | 1.000 | Ref. | 1.000 | Ref. | ||
| Moderate | 99 | 0.199 | 1.254 | 0.888–1.772 | 0.058 | 1.444 | 0.988–2.111 |
| Poor | 75 | 0.006 | 1.679 | 1.163–2.422 | 0.004 | 1.813 | 1.213–2.711 |
| T stage | |||||||
| T1 | 23 | 1.000 | Ref. | 1.000 | Ref. | ||
| T2 | 65 | 0.025 | 1.874 | 1.084–3.241 | 0.094 | 1.638 | 0.920–2.917 |
| T3 | 126 | 0.003 | 2.198 | 1.307–3.697 | 0.035 | 1.796 | 1.042–3.097 |
| T4 | 15 | <0.001 | 4.343 | 2.123–8.888 | 0.002 | 3.279 | 1.571–6.843 |
| N stage | |||||||
| N0 | 120 | 1.000 | Ref. | 1.000 | Ref. | ||
| N1 | 87 | <0.001 | 2.522 | 1.876–3.389 | <0.001 | 3.102 | 2.247–4.821 |
| N2 | 19 | <0.001 | 5.288 | 3.167–8.831 | <0.001 | 4.839 | 2.873–8.151 |
| N3 | 3 | 0.001 | 7.625 | 2.385–24.376 | <0.001 | 9.859 | 3.042–31.953 |
| pTNM stage | |||||||
| I | 26 | 1.000 | Ref. | 1.000 | Ref. | ||
| II | 101 | 0.056 | 1.633 | 0.988–2.697 | 0.202 | 1.427 | 0.826–2.467 |
| III | 102 | <0.001 | 4.783 | 2.860–7.999 | <0.001 | 4.946 | 2.862–8.548 |
| Adjuvant treatment | |||||||
| None | 110 | 1.000 | Ref. | 1.000 | Ref. | ||
| Radiotherapy | 66 | 0.120 | 0.774 | 0.560–1.069 | 0.152 | 0.774 | 0.545–1.099 |
| Chemotherapy | 18 | 0.181 | 1.410 | 0.853–2.330 | 0.846 | 1.056 | 0.610–0.827 |
| CRT | 35 | 0.631 | 1.101 | 0.743–1.631 | 0.765 | 1.067 | 0.698–1.632 |
| PAX9 | |||||||
| Negative | 108 | 1.000 | Ref. | 1.000 | Ref. | ||
| Positive | 121 | <0.001 | 0.577 | 0.436–0.764 | 0.001 | 0.605 | 0.450–0.813 |
DFS, disease free survival; OS, overall survival; T, tumor; N, node; PAX9, paired box 9; TNM, tumor node metastasis; ESCC, esophageal squamous cell carcinoma; CRT, chemoradiotherapy; CI, confidence interval; Ref., Reference.
According to univariate analysis, which deals with one predictor variable, PAX9, differentiation, T stage, N stage and pTNM stage were statistically associated with DFS and OS (Table II). To further exclude confounding factors and obtain more reliable results, multivariate analysis was then performed, which deals with multiple predictor variables. In a multivariate analysis of DFS and OS, neither differentiation or T stage were independent prognostic factors (Table III). According to the Cox proportional hazard regression model, PAX9 was an independent predictor for DFS (HR=0.641, 95% CI: 0.472–0.869, P=0.004) and OS (HR=0.673, 95% CI: 0.491–0.922, P=0.014; Table III). Lymph node metastasis (N stage) was associated with decreased DFS (N1: HR=1.988, 95% CI: 1.118–3.534, P=0.019; N2: HR=3.357, 95% CI: 1.612–6.993, P=0.001; N3: HR=4.029, 95% CI: 1.001–16.224, P=0.049), and decreased OS (N1: HR=1.988, 95% CI: 1.118–3.534, P=0.019; N2: HR=3.357, 95% CI: 1.612–6.993, P=0.001; N3: HR=4.029, 95% CI: 1.001–16.224, P=0.049). In addition, pTNM stage III was an independent prognostic factor for DFS (HR=2.552, 95% CI: 1.290–7.319, P=0.031) and OS (HR=2.537, 95% CI: 1.274–7.365, P=0.037), respectively (Table III). Therefore, the results showed that PAX9, N stage and pTNM stage III were prognostic factors of survival in ESCC.
Table III.
Multivariate analyses of prognostic variables of ESCC treated with curative surgery.
| DFS | OS | |||||
|---|---|---|---|---|---|---|
| Characteristics | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Gender | ||||||
| Male | 1.000 | Ref. | 1.000 | Ref. | ||
| Female | 0.715 | 0.933 | 0.641–1.356 | 0.458 | 0.852 | 0.559–1.299 |
| Age | ||||||
| ≤60 | 1.000 | Ref. | 1.000 | Ref. | ||
| >60 | 0.747 | 1.048 | 0.789–1.392 | 0.786 | 0.958 | 0.704–1.304 |
| Smoking history | ||||||
| Never or light | 1.000 | Ref. | 1.000 | Ref. | ||
| Heavy | 0.155 | 1.251 | 0.919–1.703 | 0.409 | 1.150 | 0.825–1.604 |
| Drinking history | ||||||
| Never or light | 1.000 | Ref. | 1.000 | Ref. | ||
| Heavy | 0.917 | 1.016 | 0.751–1.375 | 0.946 | 0.989 | 0.713–1.371 |
| Tumor site | ||||||
| Cervical | 1.000 | Ref. | 1.000 | Ref. | ||
| Upper thoracic | 0.389 | 0.638 | 0.229–1.774 | 0.234 | 0.509 | 0.167–1.549 |
| Middle thoracic | 0.118 | 0.489 | 0.200–1.200 | 0.101 | 0.442 | 0.167–1.172 |
| Lower thoracic | 0.366 | 0.658 | 0.266–1.629 | 0.244 | 0.557 | 0.208–1.492 |
| Tumor length | ||||||
| <4 cm | 1.000 | Ref. | 1.000 | Ref. | ||
| ≥4 cm | 0.646 | 1.071 | 0.799–1.437 | 0.413 | 1.145 | 0.828–1.582 |
| Differentiation | ||||||
| Well | 1.000 | Ref. | 1.000 | Ref. | ||
| Moderate | 0.462 | 0.861 | 0.577–1.284 | 0.780 | 1.063 | 0.691–1.636 |
| Poor | 0.741 | 1.077 | 0.694–1.670 | 0.335 | 1.261 | 0.786–2.024 |
| T stage | ||||||
| T1 | 1.000 | Ref. | 1.000 | Ref. | ||
| T2 | 0.701 | 1.157 | 0.550–2.435 | 0.948 | 0.976 | 0.469–2.030 |
| T3 | 0.633 | 1.214 | 0.548–2.688 | 0.999 | 1.001 | 0.458–2.185 |
| T4 | 0.301 | 1.766 | 0.601–5.194 | 0.607 | 1.332 | 0.446–3.978 |
| N stage | ||||||
| N0 | 1.000 | Ref. | 1.000 | Ref. | ||
| N1 | 0.019 | 1.988 | 1.118–3.534 | 0.004 | 2.407 | 1.315–4.405 |
| N2 | 0.001 | 3.357 | 1.612–6.993 | 0.009 | 2.824 | 1.298–6.147 |
| N3 | 0.049 | 4.029 | 1.001–16.224 | 0.029 | 4.642 | 1.169–18.432 |
| pTNM stage | ||||||
| I | 1.000 | Ref. | 1.000 | Ref. | ||
| II | 0.321 | 1.491 | 0.677–3.286 | 0.399 | 1.415 | 0.632–3.171 |
| III | 0.031 | 2.552 | 1.290–7.319 | 0.037 | 2.537 | 1.274–7.365 |
| Adjuvant treatment | ||||||
| None | 1.000 | Ref. | 1.000 | Ref. | ||
| Radiotherapy | 0.064 | 0.646 | 0.455–1.016 | 0.072 | 0.641 | 0.438–1.038 |
| Chemotherapy | 0.563 | 1.183 | 0.669–2.092 | 0.437 | 0.776 | 0.409–1.472 |
| CRT | 0.081 | 0.658 | 0.411–1.052 | 0.075 | 0.584 | 0.354–1.062 |
| PAX9 | ||||||
| Negative | 1.000 | Ref. | 1.000 | Ref. | ||
| Positive | 0.004 | 0.641 | 0.472–0.869 | 0.014 | 0.673 | 0.491–0.922 |
DFS, disease free survival; OS, overall survival; T, tumor; N, node; PAX9, paired box 9; TNM, tumor node metastasis; ESCC, esophageal squamous cell carcinoma; CRT, chemoradiotherapy; CI, confidence interval; Ref., Reference.
Considering adjuvant therapy associations, different stratified analyses were further performed to compare the survival curves between the PAX9-negative and PAX9-positive groups. For patients that did not receive adjuvant therapy, no significant differences in the 5-year DFS (P=0.494) and OS (P=0.663) were observed. However, there was a significant difference in the 5-year DFS and OS (P<0.001) in patients that received adjuvant therapy (Fig. 3). In addition, subgroup analysis was performed according to PAX9 positivity. In the PAX9-positive group, the patients who received adjuvant radiotherapy had an improved prognosis compared with patients who did not receive adjuvant radiotherapy, as demonstrated by significant differences in the 5-year DFS (P=0.011) and OS (P=0.009) (Fig. 4).
Figure 3.
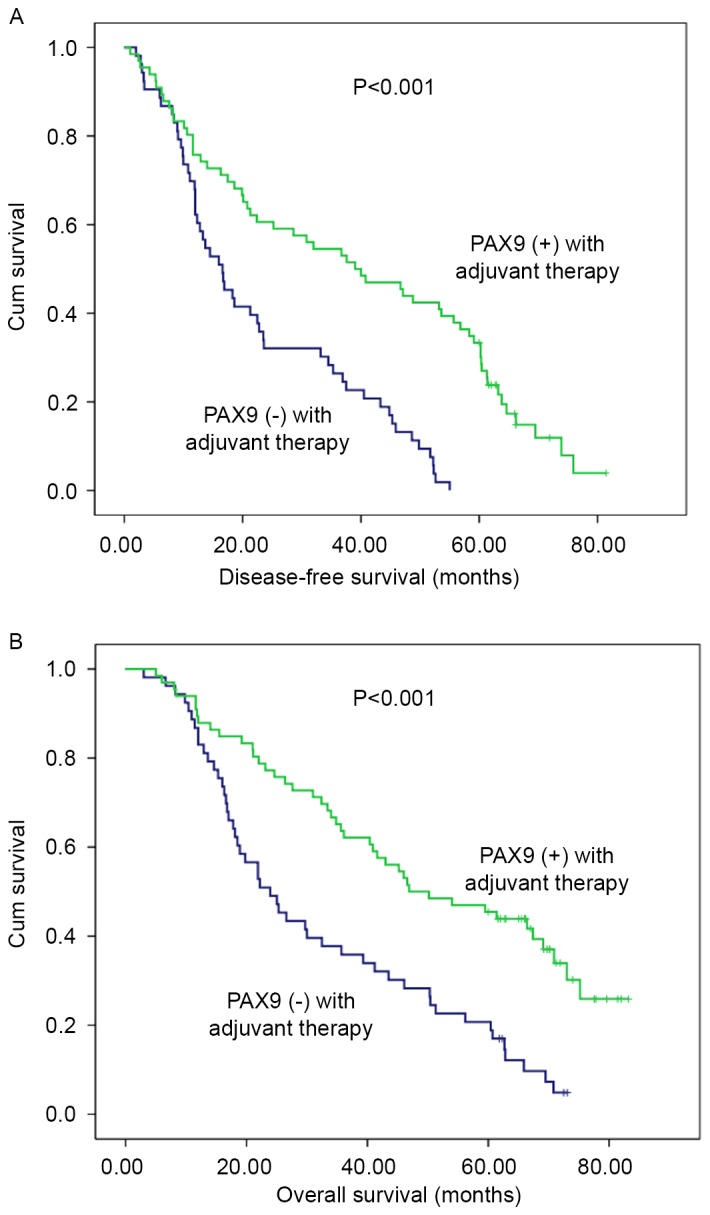
Kaplan-Meier analysis of PAX9 expression in 119 patients with esophageal squamous cell carcinoma who received adjuvant therapy following surgery. Negative PAX9 expression was significantly associated with (A) poor disease-free survival and (B) poor overall survival. P<0.001 vs. PAX9 positive. PAX9, paired box 9.
Figure 4.
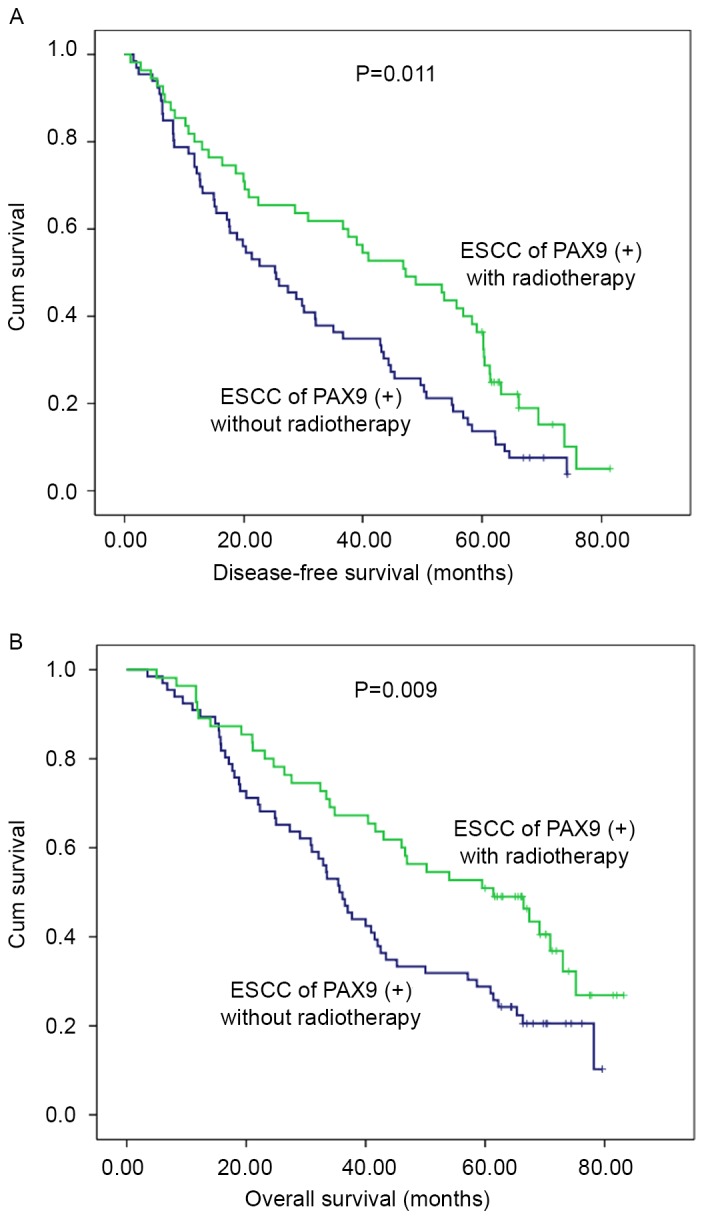
Kaplan-Meier analysis of 121 patients with esophageal squamous cell carcinoma with PAX9 positivity who did or did not receive postoperative radiotherapy. Patients who did not receive radiotherapy were significantly associated with (A) poor disease-free survival (P=0.011 vs. with radiotherapy) and (B) poor overall survival (P=0.009 vs. with radiotherapy). PAX9, paired box 9.
Discussion
The morbidity and mortality rates for ESCC increase annually; however, advances in therapeutic strategies that significantly alter patient outcome remains to be achieved. This may be due to the complexity of the oncogenic process, which involves somatic acquisition of large numbers of mutations that gives rise to complex, heterogenic genetic profiles among patients, making it difficult to characterize and treat effectively. Although previous reports focus on PAX9 expression in different tumors (35–37), there is a paucity of data describing PAX9 expression in esophageal cancer. PAX9 may be a key gene involved in ESCC. In the present study, the correlation between PAX9 expression and survival in 229 ESCC patients who received curative surgery was investigated. In addition, associations between PAX9 and ESCC radiosensitivity was determined.
A greater tumor length, advanced T stage and poor differentiation were associated with low DFS and OS rates, as determined by univariate analysis; however, these factors were not found to be independent predictors of prognosis by multivariate analysis, which is more important in the analysis of prognostic factors. In addition, lymph node metastasis and advanced pTNM stage were independent prognostic factors, which concurred with previous studies (7,38). The results of the present study demonstrated that negative PAX9 expression is significantly associated with poor prognosis in ESCC patients. In addition, in a stratified analysis, the patients were divided into two subgroups according to whether postoperative adjuvant therapy was given in the PAX9 positive or negative groups, respectively. PAX9-positive patients in the adjuvant group exhibited a significantly prolonged prognosis compared with PAX9-negative patients; however, this was not the case in the surgery group. It was hypothesized that PAX9 expression may best predict survival in patients that received postoperative treatment. Adjuvant radiotherapy benefited patients in the PAX9-positive group; however, this was not the case in the PAX9-negative group, compared with patients who did not receive adjuvant radiotherapy. To the best of our knowledge, similar studies have not been conducted in ESCC. Abnormal expression levels of PAX9 suggested that it may be involved in the development of ESCC. In the present study, the PAX9 protein was expressed in the majority of healthy epithelia and the mucosa adjacent to esophageal tumors. In malignant squamous cells, loss or decreased expression levels of PAX9 were observed. These results are consistent with previous findings (39). However, the results of the present study did not demonstrate statistical differences between PAX9 expression and the degree of cell differentiation, which contradicts findings reported in the literature (39). This may be due to differences in the case numbers, enrollment criteria and ESCC TNM staging standards. Knockdown of PAX9 resulted in the loss or disorganization of squamous epithelium and downregulation of the differentiation markers, Krt4 and Krt5, in zebrafish esophagus (40). PAX9 expression levels may vary in different tumors. Decreased PAX9 expression serves a role in congenital tooth agenesis and as a high risk factor for epithelial ovarian cancer (35). PAX9 is significantly highly expressed in malignant melanomas and nodular melanomas (41).
At present, no effective predictive biomarkers have been identified that accurately estimate sensitivity to radiation therapy in ESCC tumors. The present study demonstrated that adjuvant radiotherapy may benefit patients with PAX9-positive tumors and suggests that PAX9 may be a predictor of radiation sensitivity in ESCC patients. The ability to identify ESCC patients with a poor prognosis, which was observed in patients with PAX9-positive tumors who did not receive radiotherapy after surgery, suggested that radiotherapy, the standard of care for unresectable patients, may be beneficial in PAX9-positive patients. Therefore, the present study revealed the clinical significance and predictive value of PAX9 in ESCC. In the clinic, ESCC patients frequently receive radiation therapy or concurrent chemoradiation, generally at high doses of 50.4–64.8 Gy, which is administered in fractions of 1.8–2.0 Gy per day (42,43). However, the curative effects vary largely in patients with the same pathological diagnosis and radiotherapy dose, and this remains to be elucidated. Tumor-associated factors that contribute to the efficacy of radiotherapy include oxygen status, proliferative potential and capacity to repair radiation damage. However, there remains a lack of understanding of the differences in ESCC patient responses to radiotherapy. The findings of the present study may additionally aid the generation of a rational strategy for targeted therapy in PAX9-positive ESCC patients.
Biomarkers have been utilized for predicting tumorigenesis, progression and prognosis (44,45). In ESCC, PAX9 downregulation may influence the cell cycle via by maintaining tumor cells in the G0 stage and that may cause cells to be more resistant to cellular radiosensitivity. Cells in the G2 and M phases of the cell cycle are sensitive to ionizing radiation, whereas those in the G0 phase are resistant (46). Studies on PAX9 expression in other tumor types have been published over the last 10 years. For example, Lee et al revealed that PAX9 inhibition causes the induction of apoptosis with enhanced cleavage of caspase-3 and poly ADP-ribose polymerase, increased expression levels of Bax and reduced expression levels of Bcl-2 in oral squamous cell carcinoma. In addition, cells transfected with PAX9 siRNA and cells infected with adenovirus-mediated expression of dominant-negative c-myb have been reported to display cell cycle arrest at the G0 phase (30). The findings of the present study may support this mechanism.
PAX9 has been demonstrated to be involved in numerous signaling pathways, including the Janus kinase/signal transducers and activators of transcription (JAK/STAT), wingless-type MMTV integration site family (Wnt), bone morphogenetic protein (BMP) and phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) pathways. However, the involvement of PAX9 in carcinogenesis is not likely to be limited to these signaling pathways. PAX9 may serve different roles via different mechanisms in various tumor tissues. In the mothers against DPP homolog (Smad) 3 and Smad4 signaling pathways, PAX9 serves an important role in the suppression of microRNA-450b-5p by transforming growth factor-β1 (36). In a study conducted by Kendall et al (29), PAX9 was identified as a lung cell lineage addiction oncogene, representing a fundamental tumor survival mechanism with important therapeutic implications; in addition to thyroid transcription factor 1 (TTF1) and NK2 homeobox 8 (NKX2-8), PAX9 serves a role in the maintenance of squamous cell carcinoma tumor cells that exhibit the 14q13.3 amplification, which is a recurrent amplicon in lung cancer. The amplification of these genes frequently results in the promotion of lung cancer and cell proliferation. Hsu et al demonstrated that this three-gene signature is associated with genes and pathways involved in embryonal tissue development (JAK/STAT, Wnt, BMP and Hedgehog) and lung development (mitogen activated protein kinase, PI3 K and JAK/STAT) (47). The majority of the adenocarcinoma samples exhibited a higher probability of PAX9 and TTF1 activation relative to SCC cells. In addition, the specific patterns of coactivation of developmental transcription factors (PAX9, TTF-1 and NKX2-8) were independent of KRAS or epidermal growth factor mutational events. PAX9 activation was associated with cisplatin sensitivity. However, the specific mechanism of action of PAX9 requires further investigation.
ESCC has been widely considered a genetic disease resulting from synergistic action of tumor suppressor genes/oncogenes, and there is interconnectivity between multiple signaling pathways. High-throughput technologies, including cDNA microarrays, proteomics, transcriptomics, genome-wide association studies and microRNA arrays, have allowed access to a host of cancer genomes, thereby increasing understanding of cancer-associated pathobiology and signaling networks. Combined analysis of the activity spectra and proteomics data revealed that the primary signaling pathways involved in ESCC were mitogen-activated protein kinase, Wnt and Akt pathways, which led to the identification of regulatory networks, cross-transcription factors, microRNAs and target genes (44,48,49). Driven by heritable and somatic alterations in DNA, ESCC displays genetic and epigenetic alterations, including DNA methylation, histone deacetylation, chromatin remodeling, gene imprinting and noncoding RNA regulation, which underpin carcinogenesis, progression and metastasis. Protein-based biomarkers are possibly more appropriate biomarkers than DNA- or RNA-based markers. Therefore, proteomic markers are closer and more relevant to disease state initiation and progression (50).
The present study has some limitations. The number of cases assessed was limited. Despite its preliminary nature, the present study suggests that PAX9 is an independent prognostic factor for ESCC. More studies involving a larger number of samples are required to further confirm these results. In addition, the patients selected for the present study were post-operative. The selection of non-surgery patients with observable and measurable tumors may be beneficial to investigate the association between PAX9 expression and the short-term curative effects of radiotherapy. In vitro and in vivo experiments are required to ascertain the predictive value of PAX9 in radiation sensitivity in ESCC.
In the present study, low expression levels of PAX9 were significantly associated with poor survival in ESCC patients following surgery. PAX9 may be an independent prognostic factor for ESCC patient survival. Additionally, PAX9 may serve as a possible predictor of radiation sensitivity, which may assist clinicians in developing a rational approach to personalized treatment for ESCC patients. However, the role of PAX9 in ESCC requires further investigation.
Figure 2.
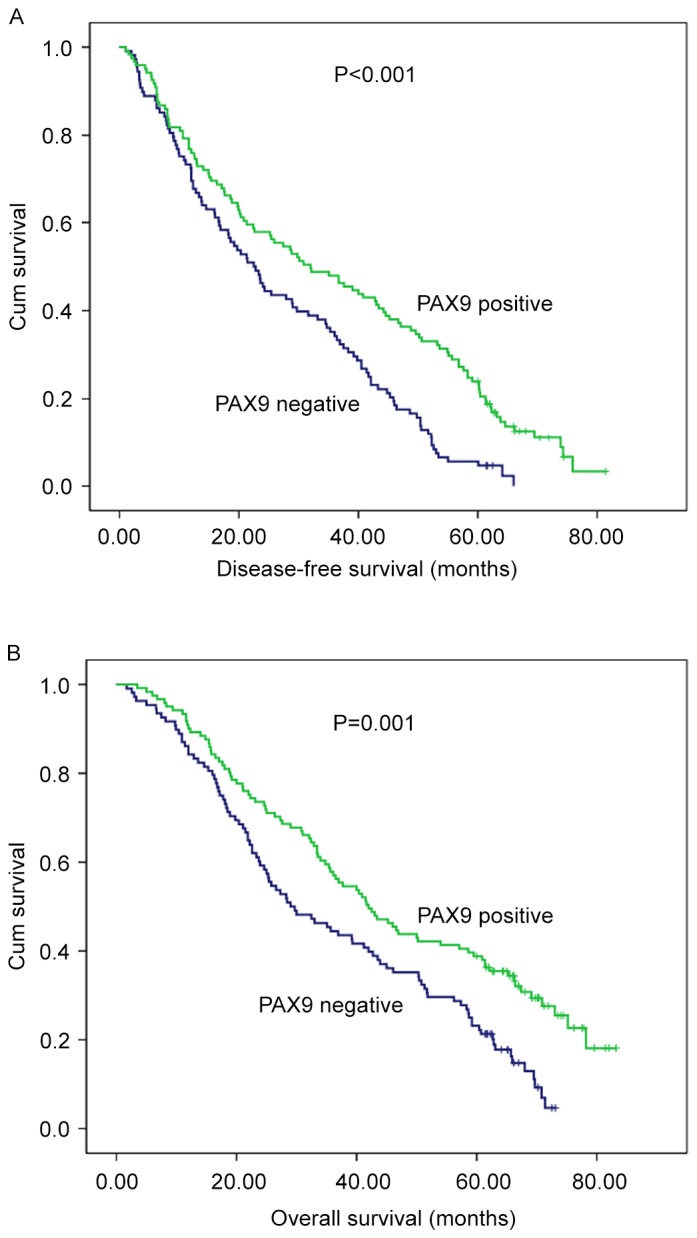
Kaplan-Meier analysis of PAX9 expression in 229 patients with esophageal squamous cell carcinoma who underwent surgery. Negative PAX9 expression was significantly associated with (A) poor disease-free survival (P<0.001 vs. PAX9 positive) and (B) poor overall survival (P=0.001 vs. PAX9 positive). PAX9, paired box 9.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81572958) and Ji'nan Municipal Science and Technology Bureau Fund (grant no. 26010105081212).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Dutton SJ, Ferry DR, Blazeby JM, Abbas H, Dahle-Smith A, Mansoor W, Thompson J, Harrison M, Chatterjee A, Falk S, et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): A phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol. 2014;15:894–904. doi: 10.1016/S1470-2045(14)70024-5. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Galais MP, Raoul JL, Bouché O, Gourgou-Bourgade S, Douillard JY, Etienne PL, Boige V, Martel-Lafay I, Michel P, et al. Definitive chemoradiotherapy with FOLFOX versus fluorouracil and cisplatin in patients with oesophageal cancer (PRODIGE5/ACCORD17): Final results of a randomised, phase 2/3 trial. Lancet Oncol. 2014;15:305–314. doi: 10.1016/S1470-2045(14)70028-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhu HD, Guo JH, Mao AW, Lv WF, Ji JS, Wang WH, Lv B, Yang RM, Wu W, Ni CF, et al. Conventional stents versus stents loaded with (125)iodine seeds for the treatment of unresectable oesophageal cancer: A multicentre, randomised phase 3 trial. Lancet Oncol. 2014;15:612–619. doi: 10.1016/S1470-2045(14)70131-7. [DOI] [PubMed] [Google Scholar]
- 5.Graf D, Vallböhmer D, Knoefel WT, Budach W, Häussinger D. Multimodal treatment of esophageal carcinoma. Dtsch Med Wochenschr. 2014;139:2141–2147. doi: 10.1055/s-0034-1370297. [DOI] [PubMed] [Google Scholar]
- 6.Paul S, Altorki N. Outcomes in the management of esophageal cancer. J Surg Oncol. 2014;110:599–610. doi: 10.1002/jso.23759. [DOI] [PubMed] [Google Scholar]
- 7.Wang N, Liu F, Cao F, Jia Y, Wang J, Ma W, Tan B, Wang K, Song Q, Cheng Y. RACK1 predicts poor prognosis and regulates progression of esophageal squamous cell carcinoma through its epithelial-mesenchymal transition. Cancer Biol Ther. 2015;16:528–540. doi: 10.1080/15384047.2015.1016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urakawa N, Utsunomiya S, Nishio M, Shigeoka M, Takase N, Arai N, Kakeji Y, Koma Y, Yokozaki H. GDF15 derived from both tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Lab Inves. 2015;95:491–503. doi: 10.1038/labinvest.2015.36. [DOI] [PubMed] [Google Scholar]
- 9.Nishi T, Takeuchi H, Matsuda S, Ogura M, Kawakubo H, Fukuda K, Nakamura R, Takahashi T, Wada N, Saikawa Y, et al. CXCR2 expression and postoperative complications affect long-term survival in patients with esophageal cancer. World J Surg Oncol. 2015;13:232. doi: 10.1186/s12957-015-0658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usui A, Hoshino I, Akutsu Y, Sakata H, Nishimori T, Murakami K, Kano M, Shuto K, Matsubara H. The molecular role of Fra-1 and its prognostic significance in human esophageal squamous cell carcinoma. Cancer. 2012;118:3387–3396. doi: 10.1002/cncr.26652. [DOI] [PubMed] [Google Scholar]
- 11.Forghanifard MM, Khales Ardalan S, Javdani-Mallak A, Rad A, Farshchian M, Abbaszadegan MR. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med Oncol. 2014;31:922. doi: 10.1007/s12032-014-0922-7. [DOI] [PubMed] [Google Scholar]
- 12.Paixão-Côrtes VR, Salzano FM, Bortolini MC. Evolutionary history of chordate PAX genes: Dynamics of change in a complex gene family. PLoS One. 2013;8:e73560. doi: 10.1371/journal.pone.0073560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dressler GR. Patterning and early cell lineage decisions in the developing kidney: The role of Pax genes. Pediatr Nephrol. 2011;26:1387–1394. doi: 10.1007/s00467-010-1749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li CG, Eccles MR. PAX Genes in Cancer; Friends or Foes? Front Genet. 2012;3:6. doi: 10.3389/fgene.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Fang WH, Krupinski J, Kumar S, Slevin M, Kumar P. Pax genes in embryogenesis and oncogenesis. J Cell Mol Med. 2008;12:2281–2294. doi: 10.1111/j.1582-4934.2008.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: Roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Muratovska A, Zhou C, He S, Goodyer P, Eccles MR. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22:7989–7997. doi: 10.1038/sj.onc.1206766. [DOI] [PubMed] [Google Scholar]
- 19.Kubic JD, Little EC, Lui JW, Iizuka T, Lang D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene. 2015;34:4964–4974. doi: 10.1038/onc.2014.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curto GG, Gard C, Ribes V. Structures and properties of PAX linked regulatory networks architecting and pacing the emergence of neuronal diversity. Semin cell Dev Biol. 2015;44:75–86. doi: 10.1016/j.semcdb.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Mentrikoski MJ, Wendroth SM, Wick MR. Immunohistochemical distinction of renal cell carcinoma from other carcinomas with clear-cell histomorphology: Utility of CD10 and CA-125 in addition to PAX-2, PAX-8, RCCma, and adipophilin. Appl Immunohistochem Mol Morphol. 2014;22:635–641. doi: 10.1097/PAI.0000000000000004. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G, Li L, Dai N, Si J, Tao Q, et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology. 2011;53:843–853. doi: 10.1002/hep.24124. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro MM, Teixeira GS, Martins L, Marques MR, de Souza AP, Line SR. G-quadruplex formation enhances splicing efficiency of PAX9 intron 1. Hum Genet. 2015;134:37–44. doi: 10.1007/s00439-014-1485-6. [DOI] [PubMed] [Google Scholar]
- 24.Narasimhan K, Hilbig A, Udayasuryan B, Jayabal S, Kolatkar PR, Jauch R. Crystallization and preliminary X-ray diffraction analysis of the Pax9 paired domain bound to a DC5 enhancer DNA element. Acta crystallogr F Struct Biol Commun. 2014;70:1357–1361. doi: 10.1107/S2053230X14017415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsui SN, Yasue A, Masuda K, Watanabe K, Horiuchi S, Imoto I, Tanaka E. Novel PAX9 mutations cause non-syndromic tooth agenesis. J Dent Res. 2014;93:245–249. doi: 10.1177/0022034513519801. [DOI] [PubMed] [Google Scholar]
- 26.Boeira BR, Jr, Echeverrigaray S. Novel missense mutation in PAX9 gene associated with familial tooth agenesis. J Oral Pathol Med. 2013;42:99–105. doi: 10.1111/j.1600-0714.2012.01193.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Yang X, Zhang C, Ge L, Zheng S. A novel nonsense mutation in PAX9 is associated with sporadic hypodontia. Mutagenesis. 2012;27:313–317. doi: 10.1093/mutage/ger080. [DOI] [PubMed] [Google Scholar]
- 28.Liang J, Song G, Li Q, Bian Z. Novel missense mutations in PAX9 causing oligodontia. Arch Oral Biol. 2012;57:784–789. doi: 10.1016/j.archoralbio.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JC, Sharma M, Lee YH, Lee NH, Kim SY, Yun JS, Nam SY, Hwang PH, Jhee EC, Yi HK. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochem Funct. 2008;26:892–899. doi: 10.1002/cbf.1522. [DOI] [PubMed] [Google Scholar]
- 31.Bandla S, Pennathur A, Luketich JD, Beer DG, Lin L, Bass AJ, Godfrey TE, Litle VR. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann Thorac Surg. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manualand the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 33.Zeidan AM, Salem MR, Mazoit JX, Abdullah MA, Ghattas T, Crystal GJ. The effectiveness of cricoid pressure for occluding the esophageal entrance in anesthetized and paralyzed patients: An experimental and observational glidescope study. Anesth Analq. 2014;118:580–586. doi: 10.1213/ANE.0000000000000068. [DOI] [PubMed] [Google Scholar]
- 34.Prasad B, Kashyap B, Babu GS, Kumar GR, Manyam R. Expression of podoplanin in different grades of oral squamous cell carcinoma. Ann Med Health Sci Res. 2015;5:299–304. doi: 10.4103/2141-9248.160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonds J, Pollan-White S, Xiang L, Mues G, D'Souza R. Is there a link between ovarian cancer and tooth agenesis? Eur J Med Genet. 2014;57:235–239. doi: 10.1016/j.ejmg.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun MM, Li JF, Guo LL, Xiao HT, Dong L, Wang F, Huang FB, Cao D, Qin T, Yin XH, et al. TGF-beta1 suppression of microRNA-450b-5p expression: A novel mechanism for blocking myogenic differentiation of rhabdomyosarcoma. Oncogene. 2014;33:2075–2086. doi: 10.1038/onc.2013.165. [DOI] [PubMed] [Google Scholar]
- 37.Harris T, Pan Q, Sironi J, Lutz D, Tian J, Sapkar J, Perez-Soler R, Keller S, Locker J. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin Cancer Res. 2011;17:690–699. doi: 10.1158/1078-0432.CCR-10-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang WP, Wang Z, Jia Y. CEP55 overexpression predicts poor prognosis in patients with locally advanced esophageal squamous cell carcinoma. Oncol Lett. 2017;13:236–242. doi: 10.3892/ol.2016.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerber JK, Richter T, Kremmer E, Adamski J, Höfler H, Balling R, Peters H. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J Pathol. 2002;197:293–297. doi: 10.1002/path.1115. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, Beasley A, Hu Y, Chen X. A Zebrafish model for studies on esophageal epithelial biology. PLoS One. 2015;10:e0143878. doi: 10.1371/journal.pone.0143878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- 42.Cooper JS, Guo MD, Herskovic A, Macdonald JS, Martenson JA, Jr, Al-Sarraf M, Byhardt R, Russell AH, Beitler JJ, Spencer S, et al. Chemoradiotherapy of locally advanced esophageal cancer: Long-term follow-up of a prospective randomized trial (RTOG 85–01). Radiation therapy oncology group. JAMA. 1999;281:1623–1627. doi: 10.1001/jama.281.17.1623. [DOI] [PubMed] [Google Scholar]
- 43.Minsky BD, Pajak TF, Ginsberg RJ, Pisansky TM, Martenson J, Komaki R, Okawara G, Rosenthal SA, Kelsen DP. INT 0123 (Radiation Therapy Oncology Group 94–05) phase III trial of combined-modality therapy for esophageal cancer: High-dose versus standard-dose radiation therapy. J Clin Oncol. 2002;20:1167–1174. doi: 10.1200/JCO.20.5.1167. [DOI] [PubMed] [Google Scholar]
- 44.Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, Huang E, Xiong S, Xu F, Liu G, et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21:854–863. doi: 10.1158/1078-0432.CCR-14-1740. [DOI] [PubMed] [Google Scholar]
- 45.Lee KW, Sung CO, Kim JH, Kang M, Yoo HY, Kim HH, Um SH, Kim SH. CD10 expression is enhanced by Twist1 and associated with poor prognosis in esophageal squamous cell carcinoma with facilitating tumorigenicity in vitro and in vivo. Int J Cancer. 2015;136:310–321. doi: 10.1002/ijc.29006. [DOI] [PubMed] [Google Scholar]
- 46.Hein AL, Ouellette MM, Yan Y. Radiation-induced signaling pathways that promote cancer cell survival (review) Int J Oncol. 2014;45:1813–1819. doi: 10.3892/ijo.2014.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu DS, Acharya CR, Balakumaran BS, Riedel RF, Kim MK, Stevenson M, Tuchman S, Mukherjee S, Barry W, Dressman HK, et al. Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc Natl Acad Sci USA. 2009;106:5312–5317. doi: 10.1073/pnas.0900827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Kwong DL, Cao T, Hu Q, Zhang L, Ming X, Chen J, Fu L, Guan X. Esophageal squamous cell carcinoma (ESCC): Advance in genomics and molecular genetics. Dis Esophagus. 2015;28:84–89. doi: 10.1111/dote.12088. [DOI] [PubMed] [Google Scholar]
- 49.Bu F, Liu X, Li J, Chen S, Tong X, Ma C, Mao H, Pan F, Li X, Chen B, et al. TGF-β1 induces epigenetic silence of TIP30 to promote tumor metastasis in esophageal carcinoma. Oncotarget. 2015;6:2120–2133. doi: 10.18632/oncotarget.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra A, Verma M. Cancer biomarkers: Are we ready for the prime time? Cancers (Basel) 2010;2:190–208. doi: 10.3390/cancers2010190. [DOI] [PMC free article] [PubMed] [Google Scholar]