Abstract
The tricarboxylic acid (TCA) cycle is central to energy production and biosynthetic precursor synthesis in aerobic organisms. There exist few known variations of a complete TCA cycle, with the common notion being that the enzymes involved have already evolved towards optimal performance. Here, we present evidence that an alternative TCA cycle, in which acetate:succinate CoA-transferase (ASCT) replaces the enzymatic step typically performed by succinyl-CoA synthetase (SCS), has arisen in diverse bacterial groups, including microbial symbionts of animals such as humans and insects.
Keywords: citric acid cycle, Krebs cycle, acetate, gut microbiota, Snodgrassella alvi
Main text
The tricarboxylic acid (TCA) cycle arose during early evolution and represents a core process in aerobic respiration and production of carbon-based precursor molecules needed for the biosynthesis of amino acids, nucleotides and cofactors. The enzymes that complete the cycle show homology across Bacteria, Archaea, and Eukaryota, and are highly efficient1,2; consequently, there are relatively few known variations of a complete TCA cycle. Many bacteria have incomplete cycles, while some autotrophs use a reverse (reductive) cycle to fix carbon from CO23. Alternative oxidative cycles have been proposed to operate in Cyanobacteria4, Mycobacterium5, and Helicobacter6. Another variant cycle was identified in Acetobacter aceti, whereby succinyl-CoA is converted to succinate by an acetate:succinate CoA-transferase (ASCT) instead of the typical succinyl-CoA synthetase (SCS)7,8. The variant ASCT pathway is thought to enhance tolerance to the high levels of acetate produced by Acetobacter during fermentation7,8.
In the course of an investigation into the genome of Snodgrassella alvi, a resident gut bacterium of honey bees, we noticed the absence of the two genes encoding SCS subunits (sucC and sucD) from an otherwise complete TCA cycle9. This obligately aerobic heterotroph also lacks pathways for glycolysis and the glyoxylate shunt, implying reliance on the TCA cycle for energy and biosynthesis. We located a gene (hereafter referred to as asct) encoding a protein with sequence similarity to CoA-transferases, which we hypothesized might fulfill the missing role of interconverting succinate and succinyl-CoA (Fig. 1A). This gene has been shown to confer a strong fitness benefit. Based on a recent genome-wide transposon mutagenesis screen10, S. alvi suffered a 38.4-fold fitness reduction in the bee gut when asct was disrupted (padj = 5.5×10−10), which was similar to the reductions caused by disruption of other TCA cycle genes (32.2-fold average, padj < 10−5).
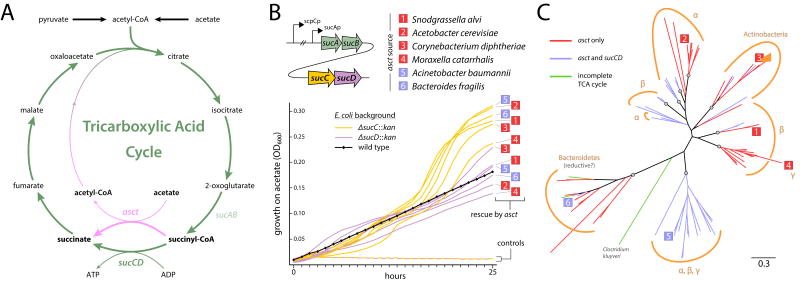
Figure 1.
An acetate-driven TCA cycle in diverse bacteria. (A) The TCA cycle (green) and the asct modification (pink). (B) Restoration of TCA cycle function by asct from diverse bacteria (labeled 1–6) in E. coli succinyl-CoA synthetase (SCS) knockouts. Operon structure of sucCD in E. coli is shown at upper left (not to scale); black arrows indicate promoters. Δ, deletion; ::kan, kanamycin resistance gene insertion. Control strains, carrying a plasmid without asct (pBad-EBFP2), are denoted by dashed lines. Colour of lines indicate E. coli background of asct-complemented or control strains; strains are listed in Supplementary Table 1. Growth curves represent average of three replicates; see Supplementary Table 2 for means and standard deviations at each time point. (C) Phylogeny of asct from representative species of phyla Bacteroidetes, Actinobacteria, and Proteobacteria (α, β, γ; classes Alpha-, Beta-, and Gamma-proteobacteria, respectively). Circles indicate nodes with > 95% bootstrap support from maximum-likelihood analysis. Taxa used to build this tree are shown in Supplementary Fig. 2. Numbered labels indicate positions of taxa in (B), with color scheme reflecting presence (blue) or absence (red) of co-occurring sucCD.
S. alvi’s asct shows sequence homology (55% amino acid identity) to aarC, which encodes the previously characterized acetate:succinate CoA-transferase that completes the TCA cycle in Acetobacter aceti7,8. This homology suggests that the ASCT family of transferases may have a more widespread and substantive role in central carbon metabolism than previously reported. We searched the prokaryotic genome database on NCBI for genomes harbouring ASCT. Of the 5,014 complete genomes queried, 1,067 had positive hits; however, many of these matches may represent CoA-transferases specific for other substrates. Focusing only on genomes that also lacked sucC and/or sucD narrowed these candidates to 180 strains. These represent species from at least 3 phyla, and include several prominent bacteria such as those used in industrial vinegar fermentation (Acetobacter spp.), human commensals (Kingella spp. and Neisseria spp.), and opportunistic pathogens (Moraxella spp. and Corynebacterium diphtheriae – the causative agent of diphtheria).
To determine if the putative asct homologs in these diverse bacteria could functionally replace SCS and complete the oxidative TCA cycle as predicted from their genomes, we cloned and expressed several of these genes in E. coli strains lacking sucC or sucD (Table S1). Upon cultivation in minimal media with acetate as the sole carbon source, only strains complemented with asct were able to grow (Fig. 1B), indicating successful rescue of TCA cycle function. Enzymatic assays confirmed the production of acetyl-CoA in asct-bearing E. coli, which verified that the TCA cycle was being completed via the modified route (Table 1). Interestingly, asct-complemented sucC knockouts (ΔsucC) grew at a faster rate than asct-complemented ΔsucD strains. Given the upstream location of sucC, sucC knockouts are expected to more fully disrupt expression of sucCD and abolish SCS function (Fig. 1B). The faster growth of ΔsucC mutants, and the lower growth of stains co-expressing sucCD and asct (Supplementary Fig. 1) suggests potential competitive inhibition between SCS and ASCT, at least within the E. coli metabolic and gene regulatory framework. Although not tested here, further experiments with double-mutants and tuning of enzyme expression may help uncover these interactions.
Table 1.
Enzyme activities of ASCT and SCS.
| E. coli strain | Activity (nmol product formed/min/mg protein)
|
|
|---|---|---|
| ASCTa | SCSb | |
| CGSC 8788 (ΔsucC) + pWK1 (asct from S. alvi) | 75 ± 8 | — |
| CGSC 8788 (ΔsucC) + pBad-EBFP2 (control) | — | — |
| CGSC 7636 (wild type) + pBad-EBFP2 (control) | — | 234 ± 15 |
Means ± s.d. (n = 3) shown. —, activity not detected.
Succinyl-CoA + Acetate → Succinate + Acetyl-CoA
Succinyl-CoA + Pi + ADP → Succinate + CoA + ATP
The choice of using SCS or ASCT to fulfill TCA cycle function involves a trade-off between the ATP gain with SCS from substrate-level phosphorylation, and the more efficient catabolism of acetate via ASCT7 (Fig. 1A). In acetate-rich environments, such as some animal guts, growth may be enhanced by silencing or deleting sucCD and relying exclusively on ASCT to complete the TCA cycle. However, their presence is not mutually exclusive – in fact, most of the putative asct homologs we detected are found in genomes with complete sucCD genes. A phylogeny of asct shows widespread co-occurrence of ASCT with SCS, as well as multiple evolutionarily independent lineages with dedicated, ASCT-dependent TCA cycles (Fig. 1C, Supplementary Fig. 2). The asct sequences grouped largely according to bacterial clades, suggesting a deep origin and subsequent diversification of this gene. Horizontal gene transfers between distantly related bacteria are also evident, such as in the acquisition of asct by the gammaproteobacterium Moraxella, which has a copy clustering with sequences from Betaproteobacteria. Additionally, some members of the Alpha-, Beta-, and Gammaproteobacteria possess asct from a cluster that tends to co-occur with sucCD.
Taking a closer look at the Neisseriaceae, an obligately aerobic family of Betaproteobacteria, using whole-genome phylogenetics, we find evidence of repeated transitions between SCS- and ASCT-dependent TCA function (Supplementary Fig. 3). At least 14 species in the clade containing Neisseria spp. likely have ASCT-dependent TCA cycles, with the switch from SCS to ASCT occurring at least once and from ASCT to SCS occurring twice. Evolutionary transitions between exclusively ASCT or SCS pathways require both the gain of one enzyme and the loss of the other, a feat that is improbable unless motivated by redundancy and, perhaps, an adaptive advantage in only retaining one pathway.
Surprisingly, we also detected putative asct homologs in a number of obligate anaerobes, such as Bacteroides species, which are abundant in the gut communities of mammals, including humans. Here, these genes may operate in a reductive manner, converting succinate to succinyl-CoA for synthesis of 2-oxoglutarate or further degradation11. It is notable that, as with TCA-oxidizing bacteria, some of these reverse-TCA utilizers have lost SCS and instead appear to rely on ASCT (Fig. 1C). We tested the function of the asct homologs present in Bacteroides fragilis, an obligate anaerobe, and Acinetobacter baumannii, an obligate aerobe, both of which also carry SCS, and discovered that both enzymes can successfully rescue TCA cycle function (Fig. 1B). These findings support our conclusion that ASCT can replace the function of SCS, but also that the retention of both ASCT and SCS in some organisms may be beneficial.
Further analyses will be required to better characterize ASCT and its interactions with other components of cellular metabolism. The fulfillment of critical metabolic duties by ASCT is not unprecedented: in the hydrogenosomes of some protozoans, ATP production is mediated by an ASCT in a coupled cycle with SCS12. In bacteria, the repeated evolution of ASCT-dependent TCA cycles may be a consequence of niche-specialization, reflecting adaptation to life in acetate-rich environments7,9,13. Our results demonstrate the inherent diversity and flexibility of the TCA cycle14, and should prompt a re-evaluation of the role of unorthodox enzymes in central metabolic processes.
Methods
Heterologous expression of ASCT
The genes coding for ASCT in S. alvi wkB2, Acetobacter cerevisiae ATCC 23765, C. diphtheriae NCTC 13129, Moraxella catarrhalis ATCC 25240, Acinetobacter baumannii ATCC 19606, and Bacteroides fragilis NCTC 9343 were amplified by PCR from genomic DNA and cloned into the plasmid pBad-EBFP215 (Table S1). The existing blue fluorescent protein gene, EBFP2, was excised by digesting with NdeI and EcoRI (New England BioLabs) prior to cloning in the ASCT genes via blunt-end ligation with T4 DNA ligase (Promega). The plasmids were transformed into the Keio collection E. coli strains CGSC 8788 (ΔsucC) and CGSC 11810 (ΔsucD)16; the parent strain, CGSC 7636, was used as a control. To test for rescue of TCA cycle function, strains were grown in M9 minimal media (1% w/v M9 salts, 1mM MgSO4, 0.1 mM CaCl2) supplemented with 0.3% w/v sodium acetate. Growth curves of cultures, grown at 37°C aerobically, were quantified by measuring absorbance at 600 nm on a Tecan Spark 10M. Growth assays were conducted in triplicate, with average values plotted in Fig. 1B. pBad-EBFP2 and pWK31 (Supplementary Table 1) were used as vector controls.
Phylogenetic analysis
Prokaryotic genomes were obtained from the NCBI Genomes database. To identify bacteria with putative ASCT, genomes were searched by BLASTP using the ASCT of Acetobacter aceti 1023 (AarC) as the query. To determine the presence/absence of SCS, genomes were searched by BLASTP using the SucC and SucD sequences of E. coli MG1655 as queries. For select strains of interest, searches for other genes of the TCA cycle were also conducted to verify cycle completeness. BLASTP hits with over 75% query coverage and 40% similarity were retained.
The phylogeny of asct genes (Fig. 1C) was built with codon-aligned nucleotide sequences in RAxML version 8 using the maximum-likelihood algorithm and a GTRGAMMAI substitution model with 1,000 bootstraps17. For the whole-genome Neisseriaceae phylogeny, single copy orthologs were identified from representative genomes for all available Neisseriaceae species using OrthoFinder18. Their amino-acid sequences were then extracted and aligned using MUSCLE19, concatenated, and degapped. The tree was built using the maximum-likelihood algorithm in RAxML version 8 with the PROTCATLG model and 1,000 bootstraps.
Enzyme activity assays
E. coli strains were were grown at 37°C in 2-liter flasks containing 1 liter of LB medium with ampicillin for 2 hours, and induced with 0.02% arabinose at OD600nm of 0.2–0.4. Cells were grown continuously until OD600nm = 1.0, and harvested by centrifugation. The harvested cells were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5; 100 mM NaCl) and sonicated 20 times with bursts of 10 s followed by 10 s for cooling. Cell debris was removed by centrifugation at 4°C for 20 min at 20,000 g (fast spin lysate) or 10 min at 2,700 g (slow spin lysate). The protein in the supernatant was quantified with the Quick StartTM Bradford Protein Assay kit using the Bovine Serum Albumin Standard Set (Bio-Rad Inc.).
The ASCT activity assay was performed as described by Mullins et al.7. In brief, the reaction was performed in 50 mM potassium phosphate buffer (pH 8.0) with 100 mM KCl, 350 mM potassium acetate, and 0.5 mM succinyl-CoA. The reaction was initiated by adding 150–200 μg of the fast spin cell lysate. After 5 min, 20 μl of the reaction mixture was used to measure the amount of produced acetyl-CoA using the Acetyl-Coenzyme A Assay Kit (Sigma-Aldrich Inc.). Acetyl-CoA Quencher supplied in the kit was used to correct for background created by free Coenzyme A or succinyl-CoA in the samples. Succinyl-CoA synthetase activity was assayed using the Succinyl-CoA Synthetase Activity Colorimetric Assay Kit (BioVision Inc.) with the slow spin lysate following the kit protocol. Detection limits for ASCT and SCS assays are 0.2 and 20 nmol product/min/mg protein, respectively.
Data availability
Sequence data used are available from GenBank and accession numbers are referenced in the supplementary materials.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Natural Sciences and Engineering Research Council Postgraduate Scholarship award PGSD-3-420434-2012 (to W.K.K.), and the U.S. National Science Foundation Dimensions of Biodiversity awards 1046153 and 1415604 and the U.S. National Institutes of Health award 1R01GM108477-01 (to N.A.M.).
Footnotes
Author Contributions
Conceptualization, Formal analysis, Visualization, and Writing – original draft, W.K.K.; Methodology, Investigation, W.K.K., H.Z.; Writing – review & editing, W.K.K., H.Z., N.A.M.; Resources and Supervision, N.A.M.
Additional Information
Supplementary information, comprising three supplementary figures and two supplementary tables, are available online.
Competing interests
The authors declare no competing interests.
References
- 1.Meyer FM, Gerwig J, Hammer E, Herzberg C, Commichau FM, Völker U, Stülke J. Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab Eng. 2011;13(1):18–27. doi: 10.1016/j.ymben.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Wu F, Minteer S. Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross-linking and mass spectrometry. Angew Chem Int Ed Engl. 2015;54(6):1851–1854. doi: 10.1002/anie.201409336. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan BB, Arnon DI. A reverse KREBS cycle in photosynthesis: consensus at last. Photosynth Res. 1990;24:47–53. [PubMed] [Google Scholar]
- 4.Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334(6062):1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 5.Baughn AD, Garforth SJ, Vilchèze C, Jacobs WR., Jr An anaerobic-type alpha-ketoglutarate ferredoxin oxidoreductase completes the oxidative tricarboxylic acid cycle of Mycobacterium tuberculosis. PLoS Pathog. 2009;5(11):e1000662. doi: 10.1371/journal.ppat.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kather B, Stingl K, van der Rest ME, Altendorf K, Molenaar D. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J Bacteriol. 2000;182(11):3204–3209. doi: 10.1128/jb.182.11.3204-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullins EA, Francois JA, Kappock TJ. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J Bacteriol. 2008;190(14):4933–4940. doi: 10.1128/JB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullins EA, Kappock TJ. Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity determinants and illustrate the mechanism used by class I CoA-transferases. Biochemistry. 2012;51(42):8422–8434. doi: 10.1021/bi300957f. [DOI] [PubMed] [Google Scholar]
- 9.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A. 2014;111(31):11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci U S A. 2016;113(48):13887–13892. doi: 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Söhling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178(3):871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Grinsven KW, Rosnowsky S, van Weelden SW, Pütz S, van der Giezen M, Martin W, van Hellemond JJ, Tielens AG, Henze K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J Biol Chem. 2008;283(3):1411–1418. doi: 10.1074/jbc.M702528200. [DOI] [PubMed] [Google Scholar]
- 13.Wertz JT, Breznak JA. Physiological ecology of Stenoxybacter acetivorans, an obligate microaerophile in termite guts. Appl Environ Microbiol. 2007;73(21):6829–6841. doi: 10.1128/AEM.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meléndez-Hevia E, Waddell TG, Cascante M. The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. J Mol Evol. 1996;43(3):293–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]
- 15.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46(20):5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 16.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data used are available from GenBank and accession numbers are referenced in the supplementary materials.